Abstract
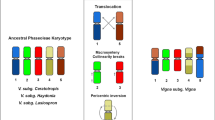
Polyploidy and dysploidy have been reported as the main events in karyotype evolution of plants. In the genus Phaseolus L. (2n = 22), a small monophyletic group of three species, the Leptostachyus group, presents a dysploid karyotype with 2n = 20. It was shown in Phaseolus leptostachyus that the dysploidy was caused by a nested chromosome fusion (NCF) accompanied by several translocations, suggesting a high rate of karyotype evolution in the group. To verify if this karyotype restructuring was a single event or occurred progressively during the evolution of this group, we analysed P. macvaughii, sister to Phaseolus micranthus + P. leptostachyus. Twenty-four genomic clones of P. vulgaris previously mapped on P. leptostachyus, in addition to the 5S and 35S rDNA probes, were used for fluorescence in situ hybridization. Only a single rearrangement was common to the two species: the nested chromosome fusion (NCF) involving chromosomes 10 and 11. The translocation of chromosome 2 is not the same found in P. leptostachyus, and pericentric inversions in chromosomed 3 and 4 were exclusive of P. macvaughii. The other rearrangements observed in P. leptostachyus were not shared with this species, suggesting that they occurred after the separation of these lineages. The presence of private rearrangements indicates a progressive accumulation of karyotype changes in the Leptostachyus group instead of an instant genome-wide repatterning.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genome stability is important for species survival and reproduction, but a degree of variability is essential for adaptation to changing environments. Therefore, the evolution of the genomes aims to establish a balance between stability and plasticity through strategies such as increasing or reducing genome size or chromosomes number, as long as no essential genes are lost on the way (Schubert and Vu 2016).
The haploid chromosome number varies widely in plants, from species with n = 2, as in Rhynchospora tenuis Link (Vanzela et al. 1996), to species with approximately n = 700 as in representatives of the genus Ophioglossum L. (Khandelwal 1990). Different events may lead to variation in this number; however, only polyploidy and dysploidy seem to be involved in karyotype evolution (Guerra 2000). Polyploidy consists of the multiplication of the entire chromosome complement (Guerra 2008), whereas dysploidy is the increase or reduction of the original haploid number without significant chromatin gain or loss. Dysploidy is usually related to events of fusion (Robertsonian fusion) or centric fission, resulting respectively in descending and ascending dysploidy (Schubert and Lysak 2011). The reduction of chromosome number can also be caused by a nested chromosome fusion (NCF) event, observed in species of Triticeae L. (Luo et al. 2009), Brachypodium P. Beauv. (International Brachypodium Initiative 2010; Idziak et al. 2014), and recently detected in Coffea canephora Pierre ex A. Froehner. Of the ten fusion events that gave rise to x = 11 in coffee, three of them were NCF (Li et al. 2019).
Dysploidy can be detected by analysing chromosome number variation in a given group of species and better interpreted when this information is examined in a phylogenetic context. In Marantaceae R. Brown., dysploidy and also polyploidy seem to be the main factors in chromosome number evolution, which may be associated with species diversification and geographical patterns (Winterfeld et al. 2020). In species of Araceae Juss., chromosome number reductions were predominant, while polyploidization occurred less frequently (Cusimano et al. 2012). The same was observed for a group of high mountain Artemisia L. when compared to the rest of the genus (Mas de Xaxars et al. 2015). However, the detection of structural rearrangements involved in the dysploid event requires comparative genomics analyses or comparative genetic or cytogenetic mapping, such as by BAC-FISH technique.
Synteny conservation analyses in crucifers revealed that the main mechanisms behind dysploid events are structural rearrangements such as inversions and translocations (Yogeeswaran et al. 2005; Lysak et al. 2003, 2006). In the model plant Arabidopsis thaliana (L.) Heynh., considered as a paleopolyploid, the n = 5 was derived from an ancestral karyotype with n = 8, found in several other Brassicaceae genera (Yogeeswaran et al. 2005). This strong reduction in chromosome number was promoted by an accelerated rate of rearrangements, mainly inversions and translocations (Lysak et al. 2006). However, since the species of this group have undergone several cycles of polyploidization and diploidization, it is possible that the high rates of chromosome rearrangements may be associated with their polyploid origin.
An event of dysploidy was also observed in a group of Phaseolus L., a legume genus that includes common bean (P. vulgaris L.) and lima bean (P. lunatus L.), as well as three other species of economic importance (Broughton et al. 2003). The genus is exclusively diploid and shows mostly 2n = 22 (Mercado-Ruaro and Delgado-Salinas 1998), as well as a relative structural karyotype stability (Fonsêca and Pedrosa-Harand 2013; Bonifácio et al. 2012). However, Leptostachyus, a small monophyletic group composed of three species (P. macvaughii Delgado, P. micranthus Hook. and Arn. and P. leptostachyus Benth.) from Mexico and Central America, originated around 2.5 mya, presents a dysploid karyotype with 2n = 20 (Mercado-Ruaro and Delgado-Salinas 1998; Delgado-Salinas et al. 2006). The results of the comparative cytogenetic mapping between P. leptostachyus and P. vulgaris revealed that numerous structural rearrangements, including a NCF that gave rise to the dysploid number and several translocations, occurred during the divergence of this lineage, suggesting a high rate of karyotype evolution in the Leptostahyus group (Fonsêca et al. 2016).
To determine if the karyotype repatterning seen in P. leptostachyus was a single event or the result of multiple and successive events during evolution of this group, we comparatively mapped P. macvaughii, sister to the other two species of the group. If the rearrangements present in P. leptostachyus were shared with P. macvaughii, they probably occurred before the differentiation of these species. If this were the case, it would suggest a single moment of great genomic restructuring. Alternatively, there was an acceleration of chromosome mutation rate in Leptostachyus group or in P. leptostachyus after dysploidy, with a progressive accumulation of rearrangements, which would be, at least in part, exclusive of one or the other species.
Materials and methods
Plant material
Seeds of P. macvaughii (G40656) and P. leptostachyus (179671), obtained from the germplasm banks of CIAT (Colombia) and Embrapa Genetic Resources and Biotechnology - CENARGEN (Brasília, DF), were germinated in Petri dishes with moistened filter paper. The roots were collected, pretreated in 2 mM 8-hydroxyquinoline for 20 h at 10 °C and fixed in methanol:acetic acid (3:1, v/v). Specimens were maintained on soil in the experimental garden or on vermiculite with modified nutrient solution of Hoagland and Arnon (1950) in the growing room of the Laboratory of Plant Cytogenetics and Evolution at Federal University of Penambuco for seed multiplication.
Mitotic preparations
Root meristems were digested with 2% cellulase (Onozuka) and 20% pectinase (Sigma) solution for 1 h and 30 min at 37 °C in humid chamber. Slides were prepared following a standard squashing technique (Guerra and Souza 2002), or by air drying according to the modified protocol of Carvalho and Saraiva (1993). Briefly, the digested roots were transferred to inclined slides, washed about 5 times with several drops of ice-cold fixative (methanol:acetic acid, 3:1) as the material was chopped, and dried with the aid of a hand pump. Finally, the slides were incubated in 45% acetic acid for 5 min and dried at 37 °C. Slides were stained in 0.1 μg/mL DAPI in 50% glycerol, selected under fluorescence microscopy, destained in ethanol:acetic acid (3:1) for 30 min, followed by absolute ethanol for 1 h, and stored at − 20 °C.
Obtaining and labelling probes
Twenty-three BACs previously mapped cytogenetically in P. leptostachyus and other species of the genus (Fonsêca et al. 2010; Bonifácio et al. 2012; Fonsêca and Pedrosa-Harand 2013) were selected for fluorescence in situ hybridization in P. macvaughii (Table 1). BAC DNA was extracted by the miniprep technique using the Plasmid Mini Kit (Qiagen). The probes were labelled with Cy3-dUTP (GE) or SpectrumGreen-dUTP (Vysis) by nick translation using the Nick Translation Mix kit (Roche). The bacteriophage SJ19.12, a marker for chromosome 10 (Fonsêca et al. 2010), as well as the 5S rDNA (D2, Pedrosa et al. 2002) and 35S rDNA (pTa71, Gerlach and Bedbrook 1979) were also used as probes and labelled with Cy3-dUTP or digoxigenin 11-dUTP (Roche).
Fluorescence in situ hybridization
FISHs were performed according to Fonsêca et al. (2010). The rehybridization of slides was performed according to Heslop-Harrison et al. (1992). The 35S rDNA probe was detected with antidigoxigenin produced in sheep and conjugated to FITC (Roche) and amplified with antisheep IgG produced in donkey and conjugated with FITC (Vector) in 1% BSA in PBS. For probes that generated additional dispersed signals, hybridization was performed using P. vulgaris genomic DNA, extracted according to the modified protocol of Weising et al. (2005) and fragmented in boiling water for 50 min (to obtain fragments less than 1 kb) as blocking at different concentrations (20–100×) depending on the BAC probe used.
Analysis of results
Metaphase cells were captured on a Leica DM5500B epifluorescence microscope by DFC345 FX capture system (Leica). The best metaphases were overlaid and adjusted for brightness and contrast in Adobe Photoshop CS6. Chromosomes were identified and numbered according to the orthology with P. vulgaris (Fonsêca et al. 2010). Chromosome sizes and approximate positions of markers along chromosomes are only schematically represented.
Results
Phaseolus macvaughii showed 2n = 20, as previously reported for this species (Mercado-Ruaro and Delgado-Salinas 1998) and also observed for P. leptostachyus (Fonsêca et al. 2016). In order to identify the chromosomes and the mechanisms involved in the formation of this dysploid karyotype, which has 20 instead of the 22 chromosomes observed in the rest of the genus, single copy clones for nine of its ten chromosomes were cytogenetically mapped and compared to the previous results of P. leptostachyus. First, five BACs and one bacteriophage from P. vulgaris chromosomes (Pvu) 6, 10 and 11 were hybridized to P. macvaughii chromosomes. These three chromosomes were involved in the nested chromosome fusion (NCF) that caused the dysploidy event and in the formation of the largest chromosome pair in P. leptostachyus (Table 1, Fonsêca et al. 2016). The hybridization with BAC 18B15 (Pvu6) labelled a small chromosome pair carrying the unique 35S site, identifying it as orthologous to Pvu6 and not involved in the formation of the largest chromosome pair (Fig. 1a). BAC 63H6 (Pvu10) evidenced a signal in the subterminal region of the long arm of the largest pair of P. macvaughii, the same arm in which the single 5S rDNA site is located (Fig. 1a–c). The other Pvu10 (5S rDNA, BAC 63H6 and SJ19.12) and Pvu11 (BACs 127J2 and 179N14) probes all hybridized to the largest chromosome pair in P. macvaughii, indicating the presence of the same NCF that placed the inverted Pvu10 in the centromeric region of Pvu11 and led to the formation of the largest chromosome pair (Fig. 1a–c). Thus, a single event caused the descending dysploidy in the ancestral of P. macvaughii and P. leptostachyus, although chromosome arm sizes vary between these species due to the additional translocation of Pv6 to the largest chromosome pair in P. leptostachyus only.
Fluorescence in situ hybridization in mitotic metaphases of P. macvaughii (Pma, a–b, d–f) and P. leptostachyus (Ple, g–h), showing the rearrangements involving chromosomes Pvu1, 2, 6, 10 and 11 (schematically represented in c and i). BACs (a–b, d–f and g–h), bacteriophage (b) and rDNA (a and g) are indicated on the upper side of each cell in the respective colours. Subtelomeric BACs in Pvu are between parenthesis. In a–b, rearrangements in Pvu10 and Pvu11, but not Pvu6, originated the largest chromosome pair in P. macvaughii. In d–f, rearrangements involving chromosomes Pvu1 and Pvu2 in P. macvaughii. In g–h, rearrangements involving Pvu2 and Pvu6 in P. leptostachyus. Note that 35S rDNA is highly decondensed in (g). Chromosomes were counterstained with DAPI and visualized in grey. Bar in (h) correspond to 5 μm
In addition, three other rearrangements were observed on P. macvaughii chromosomes when compared to P. vulgaris. The first change involved chromosomes 1 and 2. BAC 221F15 (Pvu1) hybridized at the proximal region of the long arm, and BAC 257 L12 (Pvu1) showed a signal at the end of the short arm of the same chromosome (Table 1, Fig. 1d), whereas in P. vulgaris these BACs are in opposite arms, short and long arms, respectively, and in P. leptostachyus, these BACs are at different chromosomes. BACs from Pvu2 hybridized to two chromosomal pairs of P. macvaughii. BACs 127F19 and 225P10 hybridized at the end of the long arm of chromosome 1, in the same arm where BAC 221F15 from Pvu1 was mapped (Fig. 1e–f), revealing a translocation of the terminal portion of the long arm of Pvu2 to the short arm of Pvu1 in P. macvaughii. To verify whether this translocation was a shared rearrangement between the two species in the Leptostachyus group, the BACs 127F19 and 225P10 were hybridized in P. leptostachyus. However, these two BACs of Pvu2 showed signals on the long arm of chromosome 6, identified by the 35S DNAr site (Fig. 1g), while BAC 221F15 (Pv1) showed a signal on another chromosome (Fig. 1h–i). Thus, the translocations involving chromosome Pv2 are distinct between P. macvaughii (1/2) and P. leptostachyus (6/2).
For Pvu3, there was a pericentric inversion in P. macvaughii, revealed by the hybridization of BAC 77J14 in the proximal region of the short arm of the chromosome, opposite to BAC 91K16 (Fig. 2a–b), instead of both BACs present in the long arm, as in P. vulgaris (Table 1). The other BACs of this chromosome showed to be collinear in relation to P. vulgaris, while in P. leptostachyus BAC 77J14 is in a different chromosome (Fig. 2b–d). For Pvu4, BACs 221J10 and 190C15 were in opposite arms when compared to P. vulgaris, suggesting a putative pericentric inversion. In P. leptostachyus these BACs are at different chromosomes (Fig. 2e-g).
Fluorescence in situ hybridization in mitotic metaphases of P. macvaughii (Pma, a–c, e) and P. leptostachyus (Ple, f), showing rearrangements involving chromosomes Pvu3 (a–c) and Pvu4 (e–f), schematically represented in d and g, respectively. The BACs used are indicated on the upper side of each cell in the respective colours. Chromosomes were counterstained with DAPI and visualized in grey. Red boxes on chromosomes indicate putative inversion events. Bar in (f) correspond to 5 μm
Chromosomes Pvu7 and Pvu8 did not show any rearrangement in P. macvaughii, since BACs 22I21, 33M20 and 86I17, as well as BACs 169G16 and 177I19, respectively, were syntenic in Pma7 and Pma8 (Table 1, Fig. 3a-d). Thus, it was possible to identify in P. macvaughii, in addition to chromosome 6, two other pairs of conserved chromosomes to P. vulgaris. Phaseolus macvaughii chromosome 9 is metacentric, with the presence of BACs 163I7 and 224I16 in the long and short arm, respectively (Fig. 3e and g). This differs from P. vulgaris, which has an acrocentric chromosome 9 carrying a 35S rDNA site on the short arm and both BACs on the long arm, as well as from P. leptostachyus, with BACs 163I7 and 224I16 on different chromosomes (Fig. 3f-g). The results of the cytogenetic mapping in P. macvaughii are summarized and compared to P. leptostachyus and P. vulgaris in Fig. 4.
Fluorescence in situ hybridization in mitotic metaphases of P. macvaughii (Pma, a, c, e) and P. leptostachyus (Ple, partial, f) showing conservation of synteny for chromosomes Pvu7 (a–b), Pvu8 (c–d), and Pvu9 (e, g) in P. macvaughii when compared to P. vulgaris. Difference between Pma9 and Pvu9 is attributed to rearrangement in P. vulgaris, but Ple9 shows synteny break (f). The BACs used are indicated on the upper side of each cell in the respective colours. Chromosomes were counterstained with DAPI and visualized in grey. Bar in f correspond to 5 μm
Schematic representation of P. macvaughii chromosomes compared to P. vulgaris (Fonsêca et al. 2010) and P. leptostachyus (Fonsêca et al. 2016 and present data). Subtelomeric BACs in Pvu are between parenthesis. Chromosomal rearrangements are represented above each branch in red by the abbreviations: NCF (nested chromosome fusion), Inv (inversion), Pe (pericentric), Pa (paracentric) and Tr (translocation). The numbers indicate the chromosome pairs involved. Red boxes on chromosomes indicate putative inversion events. Phylogenetic relationships between species according to Delgado-Salinas et al. (2006)
Discussion
In this work, nine of the ten chromosome pairs of P. macvaughii could be mapped and compared to P. vulgaris (Fonsêca et al. 2010) and P. leptostachyus (Fonsêca et al. 2016 and present work). We demonstrated that the descending dysploidy that originated the karyotype with 2n = 20 in the Leptostachyus group was a single event resulting from the centric insertion of all or a large part of Pvu10 in Pvu11 (Fonsêca et al. 2016). However, unlike in P. leptostachyus (Fonsêca et al. 2016), chromosome 6 was not involved in the formation of the largest pair in P. macvaughii, and, therefore, the dysploidy in the group is associated to a single NCF (Fig. 4). In the cotton tribe (Gossypieae), a dysploidy in the clade that includes Kokia Lewton and Gossypioides Skovst. ex J.B.Hutch. (n = 12) occurred after divergence of this branch from Gossypium (n = 13). This reduction of one chromosome pair resulted from several structural rearrangements involving three chromosome pairs (Udall et al. 2019). A scenario of multiple rearrangements is also proposed to explain the dysploid reduction observed in tribe Boechereae Al-Shehbaz, Beilstein and E.A. Kellogg, from Brassicaceae (n = 8 → n = 7; Mandáková et al. 2020).
Despite the collinearity of sequences along chromosome Ple/Pma 10/11, all BACs located in the long arm in P. macvaughii are in the short arm of P. leptostachyus, and vice versa. It is possible that the additional translocation of part of chromosome 6 to chromosome 10/11 of P. leptostachyus (Fonsêca et al. 2016) resulted in a slight change in the length of the chromosome arms, transforming the short arm of the largest ancestral pair, conserved in P. macvaughii, into the long arm in P. leptostachyus. Additionally, it is possible that quantitative changes in the pericentromeric heterochromatin of this chromosome, after the separation of both species, also contributed to this change in arm ratio. Differences in centromere position for chromosomes 6, 8 and 10 were observed between Vigna aconitifolia (Jacq.) Marechal. and V. unguiculata (L.) Walp. without detected changes in collinearity and may be related to variation in the 35S rDNA block size or other repetitive sequences (Oliveira et al. 2020).
Chromosome 6 has a 35S rDNA site in P. macvaughii, as all previously analysed Phaseolus species (Pedrosa-Harand et al. 2006; Bonifácio et al. 2012; Fonsêca and Pedrosa-Harand 2013; Fonsêca et al. 2016), reinforcing this terminal 35S site on the short arm as a plesiomorphic character. Similarly, the 5S rDNA site was conserved in chromosome 10, which correspond to the largest chromosome pair (10/11) in P. macvaughii and P. leptostachyus (Fonsêca et al. 2016). However, the two species of Leptostachyus group share with P. lunatus a putative pericentric inversion on chromosome 10 that placed the 5S rDNA site at the short arm (Bonifácio et al. 2012). This event probably occurred before the separation of the Leptostachyus and Lunatus groups and, thus, is not related to the dysploidy or to the other rearrangements in Leptostachyus.
Excepted for the NCF that gave rise to the dysploid karyotype, none of the detected rearrangements in P. macvaughii and P. leptostachyus was shared within this group. Chromosome 2 was involved in translocations both in P. macvaughii and P. leptostachyus, but while in P. macvaughii the translocation was with chromosome 1, in P. leptostachyus, it was with chromosome 6 (Fonsêca et al. 2016). Furthermore, P. leptostachyus showed exclusive translocations involving at least chromosomes 1, 2, 3, 4, 6, 7 and 9 (Fonsêca et al. 2016). Similarly, while P. macvaughii chromosome 3 showed a pericentric inversion, Ple3 showed a translocation and a paracentric inversion (Fonsêca et al. 2016; Fig. 4). Therefore, multiple, independent events occurred after the dysploidy and the separation of the two species. In Ricotia L. (Brassicaceae), species with n = 14 were the result of independent events of dysploidy, and part of one n = 14 group went through further rearrangements resulting in n = 13 (Mandáková et al. 2018).
In this study, we demonstrated that a single NCF gave rise to the 2n = 20 karyotype in the ancestral of the Leptostachyus group. After this event, further species-specific rearrangements occurred in each lineage (Fig. 4). All these events occurred in the last 2.5 million years, during or after species separation (Delgado-Salinas et al. 2006). Chromosome rearrangements may contribute to species isolation, as observed in Drosophila (Fuller et al. 2019), Lepidoptera (de Vos et al. 2020) and rodents (Capilla et al. 2016). In plants, rearrangements also contributed to speciation in wild emmer wheat (Wang et al. 2020) and reproductive isolation in Carex L. (Cyperaceae; Escudero et al. 2016). They are also believed to constitute key evolutionary innovation underlying the diversification of Boechereae (Mandáková et al. 2020). The presence of exclusive rearrangements for the two species after the NCF suggests not a single moment of major genomic restructuring, but a high rate of karyotype evolution, with successive and independent rearrangements, in a relatively short period since the origin of the group. The investigation of the third species, P. micranthus, may reveal further rearrangements, shedding light to the chromosome evolution after the dysploidy event in this group. This future work may benefit from the recently developed oligonucleotide painting probes for chromosomes 2 and 3 of P. vulgaris (Martins, Lívia do Vale et al., unpublished results). In the absence of a diploidization process, since polyploidy did not occur in the genus Phaseolus (Schmutz et al. 2014), the cause for this accelerated rate of chromosome evolution in the Leptostachyus group remains unknown.
Data availability
Not applicable.
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- Chr:
-
Chromosome
- DAPI:
-
4,6-Diamidino-2-phenylindole
- FISH:
-
Fluorescence in situ hybridization
- NCF:
-
Nested Chromosome Fusion
- Mya:
-
Million years ago
- rDNA:
-
Ribosomal DNA
- Ple :
-
Phaseolus leptostachyus
- Pma :
-
Phaseolus macvaughii
- Pvu :
-
Phaseolus vulgaris
References
Bonifácio EM, Fonsêca A, Almeida C, dos Santos KGB, Pedrosa-Harand A (2012) Comparative cytogenetic mapping between the lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.). Theor Appl Genet 124:1513–1520. https://doi.org/10.1007/s00122-012-1806-x
Broughton WJ, Hérnadez G, Blair MW, Beebe SE, Gepts P, Vanderleyden J (2003) Beans (Phaseolus spp.)—model food legumes. Plant Soil 252:55–128. https://doi.org/10.1023/A:1024146710611
Capilla L, Sánchez-Guillén RA, Farré M, Paytuví-Gallart A, Malinverni R, Ventura J, Larkin DM, Ruiz-Herrera A (2016) Mammalian comparative genomics reveals genetic and epigenetic features associated with genome reshuffling in Rodentia. Genome Biol Evol 8:3703–3717. https://doi.org/10.1093/gbe/evw276
Carvalho CR, Saraiva LS (1993) An air drying technique for maize chromosomes without enzymatic maceration. Biotech Histochem 68:142–145. https://doi.org/10.3109/10520299309104684
Cusimano N, Souza A, Renner SS (2012) Maximum likelihood inference implies a high, not a low, ancestral haploid chromosome number in Araceae, with a critique of the bias introduced by ‘x’. Ann Bot 109:681–692. https://doi.org/10.1093/aob/mcr302
de Vos JM, Hannah A, Livio B, Kay L (2020) Speciation through chromosomal fusion and fission in Lepidoptera. Philos Trans R Soc B 375:20190539. https://doi.org/10.1098/rstb.2019.0539
Delgado-Salinas A, Bibler R, Lavin M (2006) Phylogeny of the genus Phaseolus (Leguminosae): a recent diversification in an ancient landscape. Syst Bot 31:779–791. https://doi.org/10.1600/036364406779695960
Escudero M, Hahn M, Brown BH, Lueders K, Hipp AL (2016) Chromosomal rearrangements in holocentric organisms lead to reproductive isolation by hybrid dysfunction: the correlation between karyotype rearrangements and germination rates in sedges. Am J Bot 103:1529–1536. https://doi.org/10.3732/ajb.1600051
Fonsêca A, Pedrosa-Harand A (2013) Karyotype stability in the genus Phaseolus evidenced by the comparative mapping of the wild species Phaseolus microcarpus. Genome 56:335–343. https://doi.org/10.1139/gen-2013-0025
Fonsêca A, Ferreira J, dos Santos TRB, Mosiolek M, Bellucci E, Kami J, Gepts P, Geffroy V, Schweizer D, dos Santos KGB, Pedrosa-Harand A (2010) Cytogenetic map of common bean (Phaseolus vulgaris L.). Chromosom Res 18:487–502. https://doi.org/10.1007/s10577-010-9129-8
Fonsêca A, Ferraz ME, Pedrosa-Harand A (2016) Speeding up chromosome evolution in Phaseolus: multiple rearrangements associated with a one-step descending dysploidy. Chromosoma 125:413–421. https://doi.org/10.1007/s00412-015-0548-3
Fuller ZL, Koury SA, Phadnis N, Schaeffer SW (2019) How chromosomal rearrangements shape adaptation and speciation: case studies in Drosophila pseudoobscura and its sibling species Drosophila persimilis. Mol Ecol 28:1283–1301. https://doi.org/10.1111/mec.14923
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885. https://doi.org/10.1093/nar/7.7.1869
Guerra M (2000) Chromosome number variation and evolution in monocots. In: Wilson KL, Morrison DA (eds) Monocots - Systematics and Evolution. CSIRO, Melbourne, pp 127–136
Guerra M (2008) Chromosome numbers in plant cytotaxonomy: concepts and implications. Cytogenet Genome Res 120:339–350. https://doi.org/10.1159/000121083
Guerra M, Souza MJ (2002) Como observarcromossomos: Um Guia de Técnica sem Citogenética Vegetal, Animal e Humana. 1a edição, Ribeirão Preto, FUNPEC, pp 23
Heslop-Harrison JS, Harrison GE, Leitch IJ (1992) Reprobing of DNA: DNA in situ hybridization preparations. Trends Genet 8:372–373. https://doi.org/10.1016/0168-9525(92)90287-E
Hoagland DR, Arnon DI (1950) The water culture method for growing plants without soils. Berkeley, California Agric Exp Station 347:29–32
Idziak D, Hazuka I, Poliwczak B, Wiszynska A, Wolny E, Hasterok R (2014) Insight into the karyotype evolution of Brachypodium species using comparative chromosome barcoding. PLoS One 9:e93503. https://doi.org/10.1371/journal.pone.0093503
International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768. https://doi.org/10.1038/nature08747
Khandelwal S (1990) Chromosome evolution in the genus Ophioglossum L. Bot J Linn Soc 102:205–217. https://doi.org/10.1111/j.1095-8339.1990.tb01876.x
Li J, Yuan J, Zhao Y, Meng F, Liu C, Zhang Z, Guo H, Xie Y, Hou Y, Li X, Wang X (2019) Reconstruction of evolutionary trajectories of coffee chromosomes. BMC Genomics 20:180. https://doi.org/10.1186/s12864-019-5566-8
Luo MC, Deal KR, Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT, Coleman-Derr D, Conley EJ, Crossman CC, Dubcovsky J, Gill BS, Gu YQ, Hadam J, Heo HY, Huo N, Lazo G, Ma Y, Matthews DE, McGuire PE, Morrell PL, Qualset CO, Renfro J, Tabanao D, Talbert LE, Tian C, Toleno DM, Warburton ML, You FM, Zhang W, Dvorak J (2009) Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc Natl Acad Sci USA 106:15780–15785. https://doi.org/10.1073/pnas.0908195106
Lysak MA, Pecinka A, Schubert I (2003) Recent progress in chromosome painting of Arabidopsis and related species. Chromosom Res 11:195–204. https://doi.org/10.1023/A:1022879608152
Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci U S A 103:5224–5229. https://doi.org/10.1073/pnas.0510791103
Mandáková T, Guo X, Özüdoğru B, Mummenhoff K, Lysak MA (2018) Hybridization-facilitated genome merger and repeated chromosome fusion after 8 million years. Plant J 96:748–760. https://doi.org/10.1111/tpj.14065
Mandáková T, Hloušková P, Windham MD, Mitchell-Olds T, Ashby K, Price B, Carman J, Lysak MA (2020) Chromosomal evolution and Apomixis in the cruciferous tribe Boechereae. Front Plant Sci 11:514. https://doi.org/10.3389/fpls.2020.00514
Mas de Xaxars G, Garnatje T, Pellicer J, Siljak-Yakovlev S, Vallès J, Garcia S (2015) Impact of dysploidy and polyploidy on the diversification of high mountain Artemisia (Asteraceae) and allies. Alp Bot 126:35–48. https://doi.org/10.1007/s00035-015-0159-x
Mercado-Ruaro P, Delgado-Salinas A (1998) Karyotypic studies on species of Phaseolus (Fabaceae: Phaseolinae). Am J Bot 85:1–9. https://doi.org/10.2307/2446547
Oliveira ARS, Martins LD, Bustamante FO, Muñoz-Amatriaín M, Close T, da Costa AF, Benko-Iseppon AM, Pedrosa-Harand A, Brasileiro-Vidal AC (2020) Breaks of macrosynteny and collinearity among moth bean (Vigna aconitifolia), cowpea (V. unguiculata), and common bean (Phaseolus vulgaris). Chromosom Res. https://doi.org/10.1007/s10577-020-09635-0
Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A (2002) Chromosomal map of the model legume Lotus japonicus. Genetics 161:1661–1672
Pedrosa-Harand A, de Almeida CC, Mosiolek M, Blair MW, Schweizer D, Guerra M (2006) Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. Theor Appl Genet 112:924–933. https://doi.org/10.1007/s00122-005-0196-8
Schmutz J, McClean PE, Mamidi S et al (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46:707–713. https://doi.org/10.1038/ng.3008
Schubert I, Lysak MA (2011) Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet 27:207–216. https://doi.org/10.1016/j.tig.2011.03.004
Schubert I, Vu GTH (2016) Genome stability and evolution: attempting a holistic view. Trends Plant Sci 21:749–757. https://doi.org/10.1016/j.tplants.2016.06.003
Udall JA, Long E, Ramaraj T, Conover JL, Yuan D, Grover CE, Gong L, Arick MA II, Masonbrink RE, Peterson DG, Wendel JF (2019) The genome sequence of Gossypioides kirkii illustrates a descending dysploidy in plants. Front Plant Sci 10:1541. https://doi.org/10.3389/fpls.2019.01541
Vanzela AL, Guerra M, Luceno M (1996) Rhynchospora tenuis link (Cyperaceae), a species with the lowest number of holocentric chromosomes. Cytobios 88:219–228
Wang H, Yin H, Jiao C, Fang X, Wang G, Li G, Ni F, Li P, Su P, Ge W, Lyu Z, Xu S, Yang Y, Hao Y, Cheng X, Zhao J, Liu C, Xu F, Ma X, Sun S, Zhao Y, Bao Y, Liu C, Zhang J, Pavlicek T, Li A, Yang Z, Nevo E, Kong L (2020) Sympatric speciation of wild emmer wheat driven by ecology and chromosomal rearrangements. PNAS 117:5955–5963. https://doi.org/10.1073/pnas.1920415117
Weising K, Nybom H, Wolff K, Kahl G (2005) CTAB protocol I. In: DNA fingerprinting in plants : principles, methods, and applications, 2nd edn. Taylor & Francis Group, pp 100–102
Winterfeld G, Ley A, Hoffmann MH, Paule J, Röser M (2020) Dysploidy and polyploidy trigger strong variation of chromosome numbers in the prayer-plant family (Marantaceae). Plant Syst Evol 306:36. https://doi.org/10.1007/s00606-020-01663-x
Yogeeswaran K, Frary A, York T, Amenta A, Lesser AH, Nasrallah JB, Tanksley SD, Nasrallah ME (2005) Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res 15:505–515. https://doi.org/10.1101/gr.3436305
Acknowledgements
The authors thank Embrapa Arroz e Feijão (Santo Antônio de Goiás, Brazil) and the International Center of Tropical Agriculture – CIAT (Cali, Colombia), for seed supply; Paul Gepts (University of California, Davis, USA) for supplying the BAC clones from P. vulgaris; and Valérie Geffroy (Université Paris-Sud, Orsay Cedex, France) for the bacteriophage SJ19.12 from P. vulgaris. The authors also thank Gustavo Souza for help with image editing, as well as Tiago Ribeiro and Thiago Henrique do Nascimento for critical reading of the manuscript. We are also thankful to the National Council for Scientific and Technological Development – CNPq and CAPES (Coordenação de Pessoal de Nível Superior: Finance Code001) Brazil, for financial support.
Funding
The present study received financial support and fellowships from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) and CAPES (Coordenação de Pessoal de Nível Superior: Finance Code 001).
Author information
Authors and Affiliations
Contributions
MEF: performed experiments, organized the figures, and drafted the manuscript. AF: analysed and discussed the data. APH: designed and supervised the research and corrected the manuscript. All authors read, discussed, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors approved the final version of the manuscript.
Code availability
Not applicable.
Additional information
Responsible Editor: Jiming Jiang
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key message
After a nested chromosome fusion (NCF) and consequent dysploidy, successive and independent rearrangements differentiated the chromosome complements of two species of the Leptostachyus group in a relatively short period of time.
Rights and permissions
About this article
Cite this article
Ferraz, M.E., Fonsêca, A. & Pedrosa-Harand, A. Multiple and independent rearrangements revealed by comparative cytogenetic mapping in the dysploid Leptostachyus group (Phaseolus L., Leguminosae). Chromosome Res 28, 395–405 (2020). https://doi.org/10.1007/s10577-020-09644-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-020-09644-z