Abstract
Copper (Cu) is an essential micronutrient required for normal growth and development of plants; however, at elevated concentrations in soil, copper is also generally considered to be one of the most toxic metals to plant cells due to its inhibitory effects against many physiological and biochemical processes. In spite of its potential physiological and economical significance, molecular mechanisms under Cu stress has so far been grossly overlooked in sorghum. To explore the molecular alterations that occur in response to copper stress, the present study was performed in ten-day-old Cu-exposed leaves of sorghum seedlings. The growth characteristics were markedly inhibited, and ionic alterations were prominently observed in the leaves when the seedlings were exposed to different concentrations (0, 100, and 150 µM) of CuSO4. Using two-dimensional gels with silver staining, 643 differentially expressed protein spots (≥1.5-fold) were identified as either significantly increased or reduced in abundance. Of these spots, a total of 24 protein spots (≥1.5-fold) from Cu-exposed sorghum leaves were successfully analyzed by MALDI-TOF-TOF mass spectrometry. Of the 24 differentially expressed proteins from Cu-exposed sorghum leaves, 13 proteins were up-regulated, and 11 proteins were down-regulated. The abundance of most identified protein species, which function in carbohydrate metabolism, stress defense and protein translation, was significantly enhanced, while that of another protein species involved in energy metabolism, photosynthesis and growth and development were severely reduced. The resulting differences in protein expression patterns together with related morpho-physiological processes suggested that these results could help to elucidate plant adaptation to Cu stress and provide insights into the molecular mechanisms of Cu responses in C4 plants.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Heavy metals are considered to be one of the most deleterious pollutants in the environment, posing a threat to plant growth and development (Hossain et al. 2012). Levels of heavy metals are rapidly increasing in the environment due to diverse anthropogenic activities, such as mining, frequent use of chemical fertilizers and a wide range of industrial activities (Ahsan et al. 2009). Copper (Cu) is an essential micronutrient that plays crucial roles in multiple biochemical processes, as well as in signaling of transcription and protein trafficking machinery at the cellular level (Yruela 2005). However, excess levels of Cu induce oxidative stress (Li et al. 2005) and may cause the accumulation of free radicals that are damaging to cellular components (Nagajyoti et al. 2010; Taddei et al. 2007).
At higher concentrations, Cu may interfere with many physiological processes such as photosynthesis, pigment synthesis, oxidative stress response, nitrogen and protein metabolism and mineral uptake. In addition, protein structures and membranes are severely interrupted due to the binding of Cu with sulfhydryl groups (Fernandes and Henriques 1991; Yruela 2009), which eventually leads to metabolic disturbance (Lou et al. 2004; Shen et al. 1998; Tewari et al. 2006).
To cope with metal toxicity as well as to control metal homeostasis and redox status, plants have developed multiple metal tolerance mechanisms within the cell including exclusion, compartmentalization, and chelation by organic ligands of organic acids, amino acids, proteins and peptides (Cobbett and Goldsbrough 2002; Hall 2002; Yruela 2009). Metal sensitivity and toxicity in plants are not only associated with elevated concentrations, but are also dependent on the developmental stage of the plants (Caspi et al. 1999).
Several investigations have postulated that plants respond to any type of stress condition via rapid alterations in gene expression and protein synthesis. Thereby, the identification and analysis of heavy metal genes and proteins as well as anti-oxidative enzymes have paved the way for the investigation of these mechanisms in plants (Ahsan et al. 2009; Clemens 2006; Cobbett and Goldsbrough 2002; Hall 2002; Utriainen et al. 1998). To this end, morpho-physiological analysis combined with molecular analysis has received considerable attention in the study of responses to heavy metal stress.
Phytoremediation is a cost-effective plant based approach that is used to avoid the danger of spreading contaminants. Consequently, this approach can immobilize pollutants and decrease soil or water pollution (Salt et al. 1998). Selection of appropriate plant species, i.e., metal-tolerant plants with high biomass production and known agronomic techniques, is a crucial requirement for effective phytoremediation. Notably, the above mentioned conditions are met by the most widely used energy crop, Sorghum bicolor (Soudek et al. 2014). Moreover, Sorghum bicolor is an important C4 crop due to its wide use as a food and feed. In addition, sorghum is a promising candidate for non-food uses, particularly as an energy crop (Barbanti et al. 2006; Meki et al. 2013) and for bioethanol production (Xin et al. 2008). Previous studies have demonstrated that sorghum plants are able to accumulate large quantities of heavy metal in the shoots (Epelde et al. 2009; Zhuang et al. 2009) and are highly tolerant to metal pollution (Angelova et al. 2011; Pinto et al. 2006).
However, very few studies have been conducted on sorghum response to heavy metals. In the past decade, there have been only a few studies dealing with the interactive effects of a combination of heavy metals on plants (Yang et al. 2004), which have prompted the development of bioremediation (Arora and Sharma 2009) and phytoremediation (Mendoza et al. 2006) strategies. Although phenotypic, physiological and biochemical analyses have been executed in different plant species and genotypes to explore the Cu stress response (Kasim 2006), Cu-induced alterations at the protein level have been overlooked.
Proteomics is one of the most powerful high-throughput molecular tools that is able to compare proteomes under different biotic and abiotic stress conditions. Using this approach, a number of studies have recently been conducted to identify Cu-responsive proteins in several plants and algal species (Ahsan et al. 2007; Bona et al. 2007; Chen et al. 2015; Cuypers et al. 2005; Ritter et al. 2010; Song et al. 2013, 2014; Zhang et al. 2009). More specifically, leaf proteome alterations in response to Cu have been investigated in the leaves of rice (Hajduch et al. 2001; Rakwal et al. 1999), the leaves and roots of wheat (Li et al. 2013), Elsholtzia splendens (Li et al. 2009), birch (Utriainen et al. 1998), and the seedlings of Arabidopsis (Smith et al. 2004).
To the best of our knowledge, proteome investigation has not been performed in response to Cu-stress in Sorghum bicolor, previously. In the present study, proteomic analysis combined with morpho-physiological investigation of sorghum seedlings exposed to Cu were executed to explore the Cu-responsive proteins in sorghum seedling leaves and their possible function in hydroponically grown sorghum seedlings. A two-dimensional proteome approach aided by mass spectrometry was employed to identify proteins regulated by Cu-stress that may provide new insight and a better understanding of the mechanisms of metal-stress tolerant plants.
Materials and methods
Plant material, growth conditions and copper (Cu) treatment
Seeds of Sorghum bicolor L. (BTX 623) were collected from the National Germplasm Resources of USDA-ARS, Plant Stress and Germplasm Development Unit, Lubbock, TX, USA. The seeds were surface-sterilized with 1 % (v/v) sodium hypochlorite for 15 min followed by washing with de-ionized water and germinated in controlled conditions (25 °C, 16 h day/8 h night, and 150 µmol.m−2.s−1 light intensity). After germination, the 5 days old seedlings were grown hydroponically in plastic dishes containing Hoagland solution (Hoagland and Arnon, 1950) for 5 days. The solution was aerated daily by air bubbling for 30 min and changed every 2 days. After 10 days, the seedlings were divided into three groups. Each group was irrigated with a Hoagland nutrient solution supplemented with 0, 100, and 150 µM CuSO4 and grown in the same controlled environment as described above. After Cu treatment for 5 days, the seedling leaves were harvested directly into liquid nitrogen and stored at −80 °C, and morpho-physiological and proteome analyses were performed. Three independent biological replicates were prepared for each sample.
Growth parameters
Growth parameters such as shoot length (cm), root length (cm), shoot fresh weight (g), and root fresh weight (g) were measured in the collected samples immediately following 5 days of Cu stress. Dry shoot and root weight (g) were determined after drying in a forced-air oven at 65 °C for 72 h (Barrs and Weatherley 1962). Three independent biological replicates with nine plants each were performed.
Analysis of relative water content
For the determination of water content during the copper stress period, leaves of seedlings were measured by a previously described method (Kim et al. 2005) with minor modifications. In brief, plants (three seedlings) were weighed (fresh weight, FW), saturated in water for 2 h, and then, their turgid weights (TWs) were calculated. The samples were then dried in an oven at 65 °C for 72 h and dry weights (DWs) were calculated. Statistical analysis was performed using Student’s t test, and p < 0.05 was considered statistically significant. The RWC was determined using the following formula:
Determination of copper content in sorghum leaves
Copper content in the leaf tissues was determined as previously described (Gong et al. 2003). In brief, the seedling leaves were collected, washed five times with Milli-Q water to remove superficially absorbed metal, and then dried immediately at 60 °C for 72 h. Then, the dried materials were ground into powder. The sample (500 mg) was digested in 5 ml of HNO3 (48 %, w/v) at 60 °C for 48 h. After dilution with Mili-Q water (1:20), copper in the solution was measured using inductively coupled plasma-optical emission spectrometry (ICP-OES; Optima 5300 V; Perkin-Elmer, Inc., Waltham, Massachusetts, USA). Copper concentrations in the tissues were calculated as mg per kg dry weight.
Measurement of ion concentration in sorghum leaves
The ion concentration observed in the seedling leaves of sorghum was determined as previously described (Oh et al. 2014b). In brief, a portion (500 mg) of the sorghum leaves was placed in a micro-Kjeldahl flask and then 5 ml H2SO4 was added. Filter paper, No. 6 or No. 7 was used to quantify Zn2+, Ca2+, Fe2+, and Mn2+. Concentrations of positive ions and copper in the leaves were determined by inductively coupled plasma-optical emission spectrometry (ICP-OES; Optima 5300 V; Perkin-Elmer, Inc., Waltham, Massachusetts, USA).
Protein extraction
Proteins from the leaves were extracted using the trichloroacetic acid (TCA)/acetone method as previously described (Li et al. 2013) with minor modifications. For two-dimensional gel electrophoresis (2-DE), three biological replicates were performed with two technical replicates each. For each sample, 500 mg of the frozen leaf tissues was ground with liquid nitrogen and homogenized in 10 mL of ice-cold 10 % trichloroacetic acid and 0.07 % 2-mercaptoethanol in acetone. The mixture was vortexed, and the suspension was sonicated for 10 min and then incubated for 1 h at −20 °C with regular shaking at 15 min intervals. After incubation, the suspension was centrifuged at 9000×g for 20 min at 4 °C. The supernatant was discarded, and the resulting pellet was washed twice with ice-cold acetone containing 0.07 % (v/v) 2-mercaptoethanol. The final pellet was vacuum-dried using a speed-vac concentrator (Hanil Science Medical, Modulspin 31, Seoul, South Korea) for 10 min and resuspended in lysis buffer containing 8 M urea, 2 M thiourea, 5 % (w/v) 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), and 2 mM tributylphosphine). After incubation at 25 °C for 1 h, the suspension was centrifuged at 20,000×g for 20 min at 25 °C, and then, the resulting supernatant was finally collected into a 1.5-ml tube as protein extract. The protein concentrations of the samples were determined using the Bradford assay (Bio Rad) (Bradford 1976) with a spectrophotometer (UV-1700 PharmaSpec; Shimadzu Corporation, Kyoto, Japan) at an absorbance of 595 nm with bovine serum albumin (BSA) as the standard.
Two-dimensional gel electrophoresis
Protein samples were purified using a 2D Cleanup Kit (GE Healthcare Biosciences Corp, 800 Centennial Ave, Piscataway, USA). Protein samples (100 µg) were directly loaded onto a focusing tray in a final volume of 150 µL rehydration buffer containing 8 M urea, 2 % CHAPS, 50 mM DTT, 0.2 % Bio-Lyte 3/10 ampholyte, and 0.001 % bromophenol blue (Bio-Rad, Hercules, CA, USA). Isoelectric focusing was performed using immobilized pH gradient strips (3–10 NL, 7 cm Bio-Rad, USA) that were actively rehydrated for 12 h at 50 v. The isoelectric focusing (IEF) was carried out using a Protean IEF cell (Bio- Rad) under the following conditions: 250 V for 15 min with a linear ramp; 4000 V for 1 h with a linear ramp; and finally, 4000 V at 12000 V/h with a rapid ramp at 20 °C. After IEF, the strips were incubated for 15 min in 2 ml of equilibration buffer I (6 M urea, 2 % w/v SDS, 0.375 M Tris- HCl pH (8.8), 20 % v/v glycerol and 2 % w/v DTT) and then equilibrated again for 15 min in the same buffer except that DTT was replaced with iodoacetamide (2.5 % w/v). For the second dimension, the equilibrated strips were transferred to 12 % SDS–polyacrylamide gels with 5 % stacking gels sealed in 1 % agarose. Separation was carried out at 25 V for the first 30 min, followed by 50 V until the bromophenol blue dye reached the bottom of the gel.
Protein visualization and gel-image analysis
Gels obtained from two dimensional electrophoresis were stained with a Plus One Silver Staining Kit (GE Healthcare Biosciences AB, Uppsala, Sweden). Triplicate gels were digitized with an image scanner (HP Scanjet G 4010) and then computationally analyzed using the Progenesis SameSpot software version 3.0 (Nonlinear Dynamics Ltd.). The intensity of all protein spots was normalized relative to the total abundance of all valid spots. After normalization and background subtraction, a matchset was created for both control gels (three replicates) and Cu-treated gels. Spots were considered reproducible if they were present or absent in all replicate gels for each treatment. To validate the automated spot detection and matching process, the images were edited manually, and streaks, speckles, and artifacts were removed. Spot patterns of all gels were matched to each other to quantify each spot after normalization using the local regression model available in Progenesis SameSpots. The average intensities of resolved spots were compared using quantitative, qualitative and statistical functions within the Progenesis SameSpots software. Significant changes between spots were determined using Student’s t-test for paired observations. Changes with a p value of <0.05 were considered statistically significant.
In-gel digestion
Selected protein spots were manually excised from silver stained 2-DE gels, and the gel slices were washed twice with double distilled water. The distilled water-washed gel slices were then destained with 100 mM sodium thiosulfate and 30 mM potassium ferricyanide (1:1). The sample was then vortexed for 10 min, washed with distilled water 3–5 times until the color was completely removed, squeezed for 10 min with 100 % acetonitrile (ACN) and dried by vacuum concentrator. After destaining, the proteins were reduced within the gel pieces with 10 mM dithiothreitol in 100 mM NH4HCO3 for 1 h at 60 °C followed by incubation for 40 min in the dark with 55 mM iodoacetamide in 100 mM NH4HCO3. Proteins were digested in 100 mM NH4HCO3 containing 7–8 µL (0.1 µg/µL) trypsin enzyme (Promega Corporation, Madison, WI 53711-5399, USA) and incubated at 37 °C for 16 h. The resulting tryptic peptides were extracted from the gel pieces with 5 % trifluoroacetic acid (TFA) in 50 % acetonitrile 3 times. The solution containing eluted peptide was concentrated up to drying by a vacuum concentrator, and the resulting extracts were analyzed by mass spectrometry.
Protein identification by mass spectrometry
The protein spots were identified by matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometry (MALDI-TOF-TOF MS) as previously described (Weng et al. 2013) with minor modifications. In brief, the peptides were eluted with 1 µL matrix solution (α-cyano-4-hydroxy-cinnamic acid in 5 % TFA, 50 % acetonitrile) before application to the target plate. Samples were allowed to air-dry and analyzed by a 4700 MALDI-TOF-TOF analyzer (Applied Biosystems, Foster City, CA, USA). All mass spectra were acquired in reflection mode with 0-4000 m/z by a 4700 proteomics analyzer (Applied Bio-systems, Framingham, MA, USA). External calibration was performed using a standard peptide mixture of des-Arg bradykinin, angiotensin, Glu-fibrinopeptide B, adrenocorticotropic hormone (ACTH) clip 1-17, ACTH clip 18-39, and ACTH clip 7-38.
Bioinformatics analysis
The combined MS and MS/MS spectra were used to identify differentially regulated proteins using Mascot Generic File (MGF) with an in-house licensed MASCOT search engine (Mascot v. 2.4.0, Matrix Science, London, UK) against the viridiplantae within the uniprot database. In the MASCOT search, carbamidomethylation of cysteines was set as a fixed modification and the oxidation of methionines was set as a variable modification. MASCOT was used with the monoisotopic mass selected, a peptide mass tolerance of 100 ppm, and a fragment iron mass tolerance of 2 Da. The Trypsin was specified as the proteolytic enzyme with one potential missed cleavage. All proteins identified by high-scoring peptides were considered true matches, and at least two peptide matches. Protein hits were validated if the identification involved at least 10 top-ranking peptides with p < 0.05. When those peptides matched multiple members of a protein family, the presented protein was selected based on the highest score and the greatest number of matching peptides.
Results
Effects of Cu stress on the morphology of sorghum seedlings
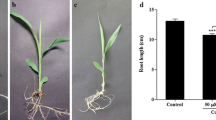
The present study focused on investigating the effects of Cu when sorghum seedling leaves were exposed to different concentrations (0 µM, 100 µM and 150 µM) of CuSO4 for 5 days. To explore the morphological alterations in sorghum seedlings under Cu stress, growth parameters such as shoot length, root length, fresh weight and relative water content (RWC) were investigated after 5 days of Cu stress. Overall plant growth was inhibited compared with control plants (Fig. 1).
Responses to copper stress on morphological alterations of sorghum seedlings (a Shoot length, b Root length, c Fresh weight, d Relative water content) under different concentrations of copper. Ten-day-old seedlings of sorghum (BTX 623) were exposed to different concentrations of copper (0 µM, 100 µM, 150 µM CuSO4) for 5 days. Three plants were randomly selected for measurement at each time interval for each replication, and the experiment was biologically replicated for three times. Each bar represents the average ± SE of three plants. Significant differences between the control and the copper-induced seedlings were determined by performing a one way analysis of variance (ANOVA) with Tukey’s all pairs of column comparison tests
After 5 days of Cu stress, the fresh weight of sorghum seedling leaves decreased (Fig. 1c) with increasing copper concentrations which correlates with the effects of copper on shoot (Fig. 1a) and root length (Fig. 1b). However, the results exhibited a significant variation in plant relative water content (RWC) between the two treatments compared with the controls (Fig. 1d).
From these results, it is concluded that copper has an adverse effect on growth characteristics as copper concentration increases, and the most significant growth inhibition was observed in plants treated with the highest concentrations of Cu2+ ions (150 μM) compared with control plants.
Copper accumulation level and ionic changes in leaves of sorghum
After 5 days of copper treatments, the copper content was measured in sorghum seedling leaves. Copper content increased remarkably with higher concentrations of Cu, and the highest accumulation level of Cu was observed with the concentration of 150 µM CuSO4 (Fig. 2). The results obtained from accumulated leaves suggested that the accumulation of Cu is dose dependant. In the present study, the concentrations of Zn2+, Ca2+, Fe2+ and Mn2+ interacting ions were elucidated in sorghum leaves to investigate the ionic imbalance. The interacting ion concentrations decreased when the sorghum leaves were exposed to Cu2+ (Fig. 3a–d).
Leaf morphology and copper accumulation level in sorghum leaf. a Control. b 100 µM CuSO4, c 150 µM CuSO4, and d. Cu content under in response to Cu treatment. Ten-day-old sorghum seedlings (BTX 623) were exposed to different concentrations of copper (0 µM, 100 µM, 150 µM CuSO4) for 5 days. Values (mean ± SD) were determined with 3 independent experiments (n = 3)
Ionic alterations observed in sorghum leaf a Zn2+, b Ca2+, c Fe2+, d Mn2+ mg/g DW under copper stress. Ten-day-old sorghum seedlings (BTX 623) were exposed to different concentrations of copper (0 µM, 100 µM, 150 µM CuSO4) for 5 days. Values (mean ± SD) were determined with 3 independent experiments (n = 3). Significant differences between the control and the Cu-induced seedlings were determined by performing a one-way analysis of variance (ANOVA) with Tukey’s all pairs of column comparison test
Protein expression patterns in sorghum seedling leaves in response to copper
To investigate how many and which proteins are altered in the leaf of sorghum under copper stress, 10-day-old seedlings were exposed to different concentrations of copper (0, 100, and 150 µmol CuSO4) for 5 days and subjected to two-dimensional electrophoresis. Total soluble proteins were extracted from the leaves of control and Cu-treated (100 and 150 µmol CuSO4) sorghum seedlings, and the representative silver staining gel images from the control and Cu-treated groups are shown in Fig. 4. Copper-responsive proteome maps produced from 2-DE gels showed high reproducibility among the three independent extractions, and more than 643 protein spots were reproducibly detected on the silver staining gel using Progenesis SameSpot software. Twenty-four (24) protein spots were differentially altered, and their abundance was significantly responsive to copper treatment, with more than a 1.5-fold change in intensity. A total of 13 proteins showed increased expression, and 11 proteins showed decreased expression in treated samples compared with the control samples. The representative proteins that could be distinguished visually are enlarged in Fig. 5. Clustering analysis was performed to categorize the identified proteins that showed differential expression profiles in response to Cu stress (Fig. 6). The differentially expressed proteins identified in the present work are summarized in Table 1.
Representative gel images from 2-DE analysis of Sorghum bicolor leaves exposed to 0, 100 and 150 µM CuSO4 in comparison to untreated controls. Leaf tissues were fractionated with TCA-acetone precipitation method as described in “Materials and Methods” section. Proteins were extracted from the Cu-exposed 15-day-old seedling leaves. For iso-electric focusing, 100 µg of proteins was loaded onto pH 3-10 NL IPG strips (7 cm). SDS-PAGE was performed on 12 % gel, and the proteins were separated using 2-DE and then stained with silver staining. The differentially expressed protein spots (>1.5-fold difference) are indicated with different colored arrows on the 2-D gel map and are statistically significant to a level of 95 % per group (Student’s t test) using biological and analytical replicates (n = 3). The MW of each protein was determined by standard markers
Cluster analysis of copper-responsive leaf proteins. Ten-day-old sorghum seedlings were exposed to copper stress for 5 days. Samples from non-treated control plants were collected on the same days as treated plants. Differences in the intensity of proteins in both control and copper treated plants are shown in the form of clusters. Any intensity showing statistical significance (p < 0.05) was considered to be positive. The protein spot number is indicated on the right side of the cluster. The clusters were determined using Genesis software (ver. 1.7.6)
Functional classification of identified proteins
To increase our understanding of the roles of proteins involved in the Cu stress response, the identified proteins were categorized into different groups. The 24 differentially expressed protein spots were analyzed using MALDI-TOF/TOF MS. Identified proteins were annotated and classified into functional categories. Gene ontology categories were assigned to all 24 proteins according to their molecular function, cellular component and biological processes (Fig. 7a–c).
Protein encoding gene function of 24 differentially expressed proteins from leaves of Sorghum bicolor. Frequency distribution of identified proteins within functional categories was based on molecular functions (a), cellular components (b), and biological processes (c). Classifications were made using iProClass databases, and assignment by function based on gene ontology
Based on molecular function, the proteins were classified into 9 groups. Among all the identified proteins, the molecular functional group of nucleic acid binding accounted for the largest number of differentially expressed proteins followed by oxidoreductase activity, ATP binding and catalytic activity (Fig. 7a). Regarding cellular component localization, the identified proteins were classified into 5 categories. Most of the proteins were localized in the membrane followed by plastids and cytoplasm (Fig. 7b).
Based on biological function, the identified proteins were categorized into the following 10 biological process categories: protein translation and synthesis [tRNA synthase-like protein (spot 116), thymidine kinase (spot 237), putative uncharacterized protein Sb09g019170 (spot 287), uncharacterized protein (spot 330)]; carbohydrate metabolism [glyceraldehyde-3-phosphate dehydrogenase (spot 261), glyceraldehyde-3-phosphate dehydrogenase (fragment) (spot 264), glyceraldehyde-3-phosphate dehydrogenase (spot 265), glyceraldehyde-3-phosphate dehydrogenase 1, cytosolic (spot 268)]; stress and defense-related proteins [thaumatin-like protein (spot 253), rossmann-fold NAD(P)(+)-binding proteins (spot 246), putative uncharacterized protein Sb06g034150 (spot 256), zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 (spot 262)]; growth and development [maturase K (spot 296)]; energy metabolism [ATP synthase beta-subunit (spot 297), aspartate aminotransferase (spot 385)]; photosynthesis [Fructose-bisphosphate aldolase (spot 286)]; oxidation–reduction processes [Alcohol dehydrogenase (spot 260)]; sucrose biosynthesis [sucrose phosphate synthase (spot 233)]; transport [uncharacterized protein (spot 182), AT3G59320 protein (spot 234)]; and unknown functions [putative uncharacterized protein (spot 129), transferase, transferring glycosyl groups, putative (spot 217), RNA-dependent DNA polymerase (spot 258), Os03g0127000 protein (spot 377)]. Among the identified proteins, carbohydrate metabolism (17 %), stress defense (17 %) and protein translation and synthesis (17 %) accounted for the largest number of differentially expressed proteins (Fig. 7c). In addition, proteins related to energy metabolism (8 %), transport (8 %) and photosynthesis (4 %) also constituted larger proportions of the differential proteins.
Discussion
The current study was carried out to investigate the morphological, physiological and proteomic changes in sorghum seedling leaves in response to copper stress. Recently, copper has been frequently considered a deleterious heavy metal associated with different types of morphological, physiological (Adrees et al. 2015) and proteomic alterations due to its toxic effects and molecular tolerance in plants (Chen et al. 2015; Li et al. 2013).
Morphological responses and water content of sorghum seedlings under Cu stress
Elevated concentrations of Cu may adversely affect plant growth and metabolism and can interfere physiological and biochemical activities, leading to oxidative damage to all organisms including plants (Ahsan et al. 2012). In the present study, a dose-dependent inhibition was observed for shoot and root length and fresh and dry weights of leaves in sorghum seedlings. Similar reports have previously described that a significant decrease in the growth parameters occurred in Solanum nigrum L. as Cu availability increased in the nutrient solution at elevated concentrations (Fidalgo et al. 2013). The morphological alterations observed in the present study were also similar to those of an earlier study (Li et al. 2013) in which the growth parameters were severely reduced in response to Cu when wheat plants were exposed to Cu. Moreover, a similar response has been reported in previous studies of other species in response to Cu stress (Aly and Mohamed 2012; Gori et al. 1998; Li et al. 2009).
Copper severely affects plant water relation through the decrease of water absorption and transport, which is thereby reflected in the water status of plants under stress condition (Kaya et al. 2007). In the present study, RWC was significantly reduced in sorghum seedling leaves when exposed to different concentrations of Cu. Consequently, Cu reduced cell wall elasticity, resulting in lower water stress resistance by plants under Cu stress, causing turgor loss at higher water contents.
Abiotic stress has an adverse effect on water content. Relative water content has been shown to decline in rice seedling leaves under salt stress (Kim et al. 2005) and in the leaves of the alfalfa when the plants were exposed to high temperature (Li et al. 2013).
Taken together, the morphological results revealed that Cu stress significantly affects water content and decreased the growth and development of sorghum seedlings in a dose-dependent manner.
Alteration of Cu accumulation level and ionic homeostasis in response to Cu
In the present study, the accumulation level of Cu in shoots of control plants and Cu-exposed plants showed that the Cu level was significantly higher in treated plants than in controls. However, the extent of Cu translocation in shoots has been shown to be drastically low (Ducic and Polle 2005; Liao et al. 2000) as indicated by the Cu content accumulated in roots. The accumulation level of Cu increased remarkably when the plants were exposed to Cu stress. Notably, under Cu stress, the plants appeared to readily uptake Cu in hydroponic systems, and comparatively more Cu accumulation occurred in roots than in leaves that has been reported in several prior studies (Li et al. 2009, 2013).
The metal ions were quantitated to investigate the differential interactions of divalent metal ions with proteins and how these interactions potentially modulate protein function. In the present study, the positive ion concentrations of Fe2+, Zn2+, Ca2+ and Mn2+ were elucidated using ICP-OES in sorghum seedling leaves. The concentrations of Fe2+, Zn2+, Ca2+ and Mn2+ were decreased when the plants were exposed to Cu stress. However, the entry of Ca2+ into the cytosol triggers catabolic processes by activation of phospholipases. This leads to liberation of linolenic or linoleic acids supplying substrates for lipoxygenase or free radical processes directly induced by Cu ions (Maksymiec 1998).
The Cu/Fe interaction is one of the most interesting and complicated within plants. These two essential metals depend on each other for proper cellular metabolism (Harris 1994); although, it is very difficult to determine precisely where in the organism the two metals interact or whether there is a third component somehow controlling the two. Previous results have suggested that the reduced chlorophyll content in plant leaves grown in high Cu concentrations makes leaves more susceptible to photoinhibition as a consequence of a Cu-induced Fe deficiency (Patsikka et al. 2002). A previous study has investigated potential mechanisms for the nutrient balances within plant cells. However, the results of that investigation concluded that high translocation of Cu to shoots has an adverse effect on plant growth and development (Wissuwa et al. 2006).
Changes in protein expression patterns responsive to Cu stress in sorghum seedling leaves
To study plant-metal stress interactions, several attempts have demonstrated the utility of proteomics research in plants to explore novel stress-responsive proteins (Ahsan et al. 2009). In the present study, a total of 24 protein species were identified using a 2-DE method that exhibited altered abundance in Cu-treated sorghum seedling leaves compared with untreated seedlings (Control). Among the 24 identified proteins, some proteins have been well characterized according to their response to heavy metals or other stresses such as glyceraldehyde-3-phosphate dehydrogenase, thaumatin-like protein, rossmann-fold NAD(P)(+)-binding proteins, ATP synthase beta-subunit, alcohol dehydrogenase, while others, such as AT3G59320 protein, maturase K, RNA-dependent DNA polymerase, Os03g0127000 protein have not been studied well with respect to their roles in plant stress response.
The proteins identified in this investigation are mainly associated with protein translation and synthesis (Spots no. 116, 237, 287, 330), carbohydrate metabolism (Spots no. 261, 264, 265, 268), stress and defense (Spots no. 246, 253, 256, 262), growth and development (Spot no. 296), energy metabolism (Spots no. 297, 385), photosynthesis (Spot no. 286), oxidation–reduction processes (Spot no. 260), sucrose biosynthesis (Spot no. 233), transport (Spots no. 182, 234) and unknown functions (Spots no. 129, 217, 258, 377) (Table 1). Protein spots with unknown functions were not identified, most likely because genomic sequencing is insufficient to attain any sustainable hypothesis for this grass species. Some of the identified Cu-responsive protein species associated with primary biological processes are discussed below.
Protein involved in carbohydrate metabolism
Carbohydrate metabolism is thought to be one of the most critical pathways in plants that regulate sugar synthesis and transformation as well as carbon partitioning (Hrazdina and Jensen 1992). The proteins identified in the present study showed significant changes in abundance after acute Cu stress, and a large portion of the proteins (4 proteins) were associated with carbohydrate metabolism. The abundance of glyceraldehyde-3-phosphate dehydrogenase (Table 1, spot 261, 265), glyceraldehyde-3-phosphate dehydrogenase (fragment) (Table 1, spot 264), and glyceraldehyde-3-phosphate dehydrogenase 1, cytosolic (Table 1, spot 268) was markedly increased in the leaves of Cu-stressed sorghum seedlings. A key role of metal species in biological organisms are acting as cofactors of diverse enzymes (Andreini et al. 2007). The identified proteins with common metal binding properties, such as the enzymes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) have been known to function with metal ions as their cofactors (Mounaji et al. 2003). GAPDH has been well documented in several previous studies both in plants and algal species (Contreras et al. 2010; Li et al. 2009; Liu et al. 2014; Ritter et al. 2010; Song et al. 2014; Wang et al. 2014; Zou et al. 2015), in which this enzyme was found to be up-regulated by Cu stress. It has been shown that GAPDH in plants remains an active enzyme required to maintain energy and reduce power and control the generation of H2O2 under oxidative stress conditions (Contreras et al. 2010). Taken together, the over-expression of GAPDH plays an important role in assisting Sorghum bicolor to attenuate the negative effects of oxidative stress caused by Cu.
Proteins involved in stress and defense
Cu, as a redox-active metal can catalyze the formation of hydroxyl radicals via a Haber–Weiss or Fenton-like reaction to produce ROS that induce oxidative stress in cells and may cause abnormalities in a wide range of cellular macromolecules including lipids, proteins, and DNA, consequently, leading to cell death (Mattie and Freedman 2004). Stress-related proteins have been observed to play an important role in plant resistance to Cu stress (Ahsan et al. 2007; Li et al. 2009, 2013; Zou et al. 2015).
Four stress-related proteins, named thaumatin-like protein (Table 1, spot 253), rossmann-fold NAD(P)(+)-binding proteins (Table 1, spot 246), putative uncharacterized protein Sb06g034150 (Table 1, spot 256), zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 (Table 1, spot 262), were differentially regulated by excessive Cu treatment (Fig. 4, Table 1). Thaumatin-like protein (TLP), is known as a pathogen-related proteins that plays a crucial role in plant defense (Van Loon et al. 2006) and protects plants from biotic and abiotic stress. In the present study, TLP was up-regulated by Cu stress. TLPs, are known to be induced not only by several pathogens but also by a wide array of abiotic stress factors such as heavy metals (Kieffer et al. 2008). Though the linkage between TLPs and Cu stress has not been clearly defined, a significant increase in TLPs was noticed in other heavy metal stress studies, such as in the leaves of Lupinus albus (Alves et al. 2006) in response to boron deficiency and in grapevine leaves in response to manganese (Yao et al. 2012). Zinc finger proteins were identified in the present study as stress-associated proteins. This protein was also increased in rice roots in response to copper stress (Song et al. 2013). Taken together, these findings suggest that the proteins involved in resistance to stress helped Sorghum bicolor to tolerate high levels of Cu.
Protein involved in translation and synthesis
Four of the identified proteins, such as tRNA synthase-like protein (Table 1, spot 116), thymidine kinase (Table 1, spot 237), putative uncharacterized protein Sb09g019170 (Table 1, spot 287) and uncharacterized protein (Table 1, spot 330), showed a decrease in abundance at low concentrations of Cu but significantly increased at elevated concentrations of Cu. The tRNA synthetases, better known as aminoacyl tRNA synthetases, play an important role in translation during protein synthesis. The primary function of aminoacyl-tRNA synthetases is to couple tRNAs to corresponding amino acids participating in protein translation. The up-regulation of translatetion-related proteins such as tRNA synthetases has been observed in studies of endoplasmic reticulum stress responses in rice seeds (Qian et al. 2015). Aminoacyl-tRNA synthetases have been found in marine alga and were up-regulated in response to Cu stress (Contreras et al. 2010).
Proteins involved in growth and development
Excess Cu induces oxidative stress that may interrupt several metabolic processes through the over-expression of stress proteins, thereby maintaining cellular homeostasis (Ahsan et al. 2007). In the present study, maturase K (Table 1, spot 296) was up-regulated when the plants were subjected to elevated concentrations of Cu. Maturase K (mat K), has been observed in other studies (Liu et al. 2014) wherein mat K was shown to potentially play a role in plant growth and cell division as well as in protecting metabolic processes against oxidative stress. It has also been found that mat K protein has an adverse effect on growth and development (Li et al. 2015), and this protein was found to be increased in response to heavy metal stress. The results obtained from the present study suggest that successful growth and development of the plants are drastically affected in response to Cu exposure.
Protein involved in energy metabolism
ATP synthetase decreases with increasing stress (Tezara et al. 1999), and it also acts as an essential metabolite in the cell wall associated with energy conversion (Chivasa et al. 2005). Energy metabolism was significantly altered in response to Cu stress as revealed by the altered expression of ATP synthase beta-subunit (Table 1, spot 297) and aspartate aminotransferase (Table 1, spot 385). In the present study, reductions in the abundance of ATP synthase beta-subunit protein, involved in energy metabolism, were observed in response to Cu stress. ATP synthase beta-subunit down-regulation at the protein level has also been observed in Elsholtzia splendens roots and leaves (Li et al. 2009) and in the marine brown algae Sargassum fusiforme (Zou et al. 2015). This reduction in protein abundance was thought to most likely impose significant restrictions on the TCA cycle and electron transport, leading to inhibition of ATP synthesis (Sweetlove et al. 2002). Moreover, the down-regulation of this protein has revealed that energy is diverted to fuel those metabolic pathways, and this protein may have been involved in the detoxification process when algae were subjected to chronic Cu stress (Zou et al. 2015). The results from the present study suggest that energy metabolism was inhibited under oxidative stress caused by elevated Cu stress.
Protein involved in photosynthesis
Several reports have demonstrated that photosynthesis is severely affected by Cu stress (Han et al. 2008; Ritter et al. 2010; Zou et al. 2015). A reduction in the proteins associated with the Calvin cycle and photosynthetic electron transport chain has been were reported in plants exposed to heavy metals (Himelblau and Amasino 2000). In this study, fructose-bisphosphate aldolase (Table 1, spot 286), that involved in photosynthesis, was considerably down-regulated after exposure to Cu stress. The down-regulation of this protein has often been observed in other studies of the effects of cadmium stress on plants (Li et al. 2015; Zhao et al. 2011). The results of the present study indicated that the efficiency of fructose-bisphosphate aldolase decreased under elevated Cu stress, thereby reducing of photosynthesis.
Protein involved in oxidation–reduction process
The role of alcohol dehydrogenase in the tolerance of crops to abiotic stress (Liao and Lin 2001; Oh et al. 2014a) has already been identified. In the present study, alcohol dehydrogenase (Table 1, spot 260) was up-regulated in response to Cu stress. However, several studies have demonstrated that alcohol dehydrogenases have been found to be increased in various fungi under oxidative stress (Bro et al. 2003; Vallino et al. 2005; Wang et al. 2006).
Proteins with miscellaneous functions
In our investigation, many proteins were identified with unknown functions of which three proteins were up-regulated (spot 217, spot 258 and spot 377) and one was down-regulated (spot 129) in response to Cu treatment. However, two proteins that are associated with transport activity, such as uncharacterized protein (Table 1, spot 182) and AT3G59320 protein (Table 1, spot 234), were up-regulated, and one protein named sucrose phosphate synthase (Table 1, spot 233) was identified that is involved in sucrose biosynthesis. In the present study, this protein was down-regulated in response to Cu stress. In addition, sucrose phosphate synthase showed no involvement in the response to Cu stress; however, this protein had an adverse effect on abiotic stress (Yin et al. 2014).
Although the availability of the full genome sequence of sorghum (Paterson et al. 2009) makes it a reasonable C4 plant, most sorghum gene products remain experimentally uncharacterized and are submitted as hypothetical proteins. By definition, hypothetical proteins are proteins predicted from genome sequences but whose existence has not been experimentally proven at the protein level (Lubec et al. 2005). However, the present results are consistent with other findings that have been reported recently for sorghum (Ngara et al. 2012; Swami et al. 2011).
To this end, we suggest that the role of these proteins could not be stated here during Cu stress because their function either may not be clear or unable to elucidate their role to attain any sustainable hypothesis for their implication in responses to Cu stress.
Conclusion
Excess Cu is toxic to plants, producing a wide range of adverse effects and, thereby, seriously limiting crop quality and production. This is the first proteome investigation of the extensive changes in sorghum seedling leaves in response to Cu stress. The present study sheds light on the copper stress-generated morpho-physiological alterations in sorghum seedlings and their correlation with the leaf proteome. The morphological results revealed that Cu stress significantly inhibited the growth of sorghum seedlings in a dose-dependent manner. Notably, Cu uptake increased remarkably with elevated concentrations of Cu and, thereby, had an adverse effect on ionic balances when the plants were exposed to Cu treatment. Our proteomic analysis identified 24 protein species in the leaves of Cu-exposed sorghum seedlings that exhibited significant changes in abundance. These protein species were particularly involved in carbohydrate metabolism, stress defense, energy metabolism, oxidation–reduction processes and photosynthesis. Finally, the obtained results suggest that these findings may help to provide new insights into the further investigations of Cu tolerance mechanisms and phytoremediation purposes in C4 plants using genetic and proteomic approaches.
References
Adrees M et al (2015) The effect of excess copper on growth and physiology of important food crops: a review. Environ Sci Pollut Res 22(11):8148–8162. doi:10.1007/s11356-015-4496-5
Ahsan N et al (2007) Excess copper induced physiological and proteomic changes in germinating rice seeds. Chemosphere 67:1182–1193. doi:10.1016/j.chemosphere.2006.10.075
Ahsan N, Renaut J, Komatsu S (2009) Recent developments in the application of proteomics to the analysis of plant responses to heavy metals. Proteomics 9:2602–2621. doi:10.1002/pmic.200800935
Ahsan N, Nakamura T, Komatsu S (2012) Differential responses of microsomal proteins and metabolites in two contrasting cadmium (Cd)-accumulating soybean cultivars under Cd stress. Amino Acids 42:317–327. doi:10.1007/s00726-010-0809-7
Alves M, Francisco R, Martins I, Ricardo C (2006) Analysis of Lupinus albus leaf apoplastic proteins in response to boron deficiency. Plant Soil 279:1–11. doi:10.1007/s11104-005-3154-y
Aly AA, Mohamed AA (2012) The impact of copper ion on growth, thiol compounds and lipid peroxidation in two maize cultivars (Zea mays L.) grown in vitro. Aust J Crop Sci 6:541–549
Andreini C, Banci L, Bertini I, Rosato A (2007) Occurrence of copper proteins through the three domains of life: a bioinformatic approach. J Proteome Res 7:209–216. doi:10.1021/pr070480u
Angelova V, Ivanova R, Delibaltova V, Ivanov K (2011) Use of sorghum crops for in situ phytoremediation of polluted soils. J Agric Sci Technol A 1:693–702. doi:10.1016/j.plaphy.2007.03.018
Arora K, Sharma S (2009) Toxic metal (Cd) removal from soil by AM fungi inoculated sorghum. Asian J Exp Sci 23:341–348
Barbanti L, Grandi S, Vecchi A, Venturi G (2006) Sweet and fibre sorghum (Sorghum bicolor (L.) Moench), energy crops in the frame of environmental protection from excessive nitrogen loads. Eur J Agron 25:30–39. doi:10.1016/j.eja.2006.03.001
Barrs H, Weatherley P (1962) A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust J Biol Sci 15:413–428. doi:10.1071/BI9620413
Bona E, Marsano F, Cavaletto M, Berta G (2007) Proteomic characterization of copper stress response in Cannabis sativa roots. Proteomics 7:1121–1130. doi:10.1002/pmic.200600712
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi:10.1016/0003-2697(76)90527-3
Bro C, Regenberg B, Lagniel G, Labarre J, Montero-Lomeli M, Nielsen J (2003) Transcriptional, proteomic, and metabolic responses to lithium in galactose-grown yeast cells. J Biol Chem 278:32141–32149. doi:10.1074/jbc.M304478200
Caspi V, Droppa M, Horváth G, Malkin S, Marder JB, Raskin VI (1999) The effect of copper on chlorophyll organization during greening of barley leaves. Photosynth Res 62:165–174. doi:10.1023/A:1006397714430
Chen C, Song Y, Zhuang K, Li L, Xia Y, Shen Z (2015) Proteomic analysis of copper-binding proteins in excess copper-stressed roots of two rice (Oryza sativa L.) varieties with different Cu tolerances. PLoS ONE 10(4):e0125367. doi:10.1371/journal.pone.0125367
Chivasa S, Ndimba BK, Simon WJ, Lindsey K, Slabas AR (2005) Extracellular ATP functions as an endogenous external metabolite regulating plant cell viability. Plant Cell 17:3019–3034. doi:10.1105/tpc.105.036806
Clemens S (2006) Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88:1707–1719. doi:10.1016/j.biochi.2006.07.003
Cobbett C, Goldsbrough P (2002) Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol 53:159–182. doi:10.1146/annurev.arplant.53.100301.135154
Contreras L, Moenne A, Gaillard F, Potin P, Correa JA (2010) Proteomic analysis and identification of copper stress-regulated proteins in the marine alga Scytosiphon gracilis (Phaeophyceae). Aquat Toxicol 96:85–89. doi:10.1016/j.aquatox.2009.10.007
Cuypers A, Koistinen KM, Kokko H, Karenlampi S, Auriola S, Vangronsveld J (2005) Analysis of bean (Phaseolus vulgaris L.) proteins affected by copper stress. J Plant Physiol 162:383–392. doi:10.1016/j.jplph.2004.07.018
Ducic T, Polle A (2005) Transport and detoxification of manganese and copper in plants. Braz J Plant Physiol 17:103–112. doi:10.1007/s12011-012-9532-4
Epelde L, Mijangos I, Becerril JM, Garbisu C (2009) Soil microbial community as bioindicator of the recovery of soil functioning derived from metal phytoextraction with sorghum. Soil Biol Biochem 41:1788–1794. doi:10.1016/j.soilbio.2008.04.001
Fernandes J, Henriques F (1991) Biochemical, physiological, and structural effects of excess copper in plants. Bot Rev 57:246–273. doi:10.1007/BF02858564
Fidalgo F, Azenha M, Silva AF, Sousa A, Santiago A, Ferraz P, Teixeira J (2013) Copper-induced stress in Solanum nigrum L. and antioxidant defense system responses. Food Energy Sec 2:70–80. doi:10.1002/fes3.20
Gong J-M, Lee DA, Schroeder JI (2003) Long-distance root-to-shoot transport of phytochelatins and cadmium in Arabidopsis. Proc Natl Acad Sci 100:10118–10123
Gori P, Schiff S, Santandrea G, Bennici A (1998) Response of shape in vitro cultures of shape Nicotiana tabacum L. to copper stress and selection of plants from Cu-tolerant callus. Plant Cell Tissue Organ Cult 53:161–169. doi:10.1023/A:1006048031956
Hajduch M, Rakwal R, Agrawal GK, Yonekura M, Pretova A (2001) High-resolution two-dimensional electrophoresis separation of proteins from metal-stressed rice (Oryza sativa L.) leaves: Drastic reductions/fragmentation of ribulose-1, 5-bisphosphate carboxylase/oxygenase and induction of stress-related proteins. Electrophoresis 22:2824–2831. doi:10.1002/1522-2683(200108)
Hall J (2002) Cellular mechanisms for heavy metal detoxification and tolerance. J Exp Bot 53:1–11. doi:10.1093/jexbot/53.366.1
Han T, Kang S-H, Park J-S, Lee H-K, Brown MT (2008) Physiological responses of Ulva pertusa and U. armoricana to copper exposure. Aquat Toxicol 86:176–184. doi:10.1016/j.aquatox.2007.10.016
Harris ED (1994) Iron–Copper interactions: some new revelations. Nutr Rev 52:311–315. doi:10.1111/j.1753-4887.1994.tb01462.x
Himelblau E, Amasino RM (2000) Delivering copper within plant cells. Curr Opin Plant Biol 3:205–210. doi:10.1016/S1369-5266(00)80066-7
Hossain MA, Piyatida P, da Silva JAT, Fujita M (2012) Molecular mechanism of heavy metal toxicity and tolerance in plants: central role of glutathione in detoxification of reactive oxygen species and methylglyoxal and in heavy metal chelation. J Bot. doi:10.1155/2012/872875
Hrazdina G, Jensen RA (1992) Spatial organization of enzymes in plant metabolic pathways. Annu Rev Plant Biol 43:241–267. doi:10.1146/annurev
Kasim W (2006) Changes induced by copper and cadmium stress in the anatomy and grain yield of Sorghum bicolor (L.) Moench. Int J Agric Biol 8:123–128
Kaya C, Tuna AL, Ashraf M, Altunlu H (2007) Improved salt tolerance of melon (Cucumis melo L.) by the addition of proline and potassium nitrate. Environ Exp Bot 60:397–403. doi:10.1016/j.envexpbot.2006.12.008
Kieffer P et al (2008) Combining proteomics and metabolite analyses to unravel cadmium stress-response in poplar leaves. J Proteome Res 8:400–417. doi:10.1021/pr800561r
Kim DW et al (2005) A hydroponic rice seedling culture model system for investigating proteome of salt stress in rice leaf. Electrophoresis 26:4521–4539. doi:10.1002/elps.200500334
Li W, Khan MA, Yamaguchi S, Kamiya Y (2005) Effects of heavy metals on seed germination and early seedling growth of Arabidopsis thaliana. Plant Growth Regul 46:45–50. doi:10.1007/s10725-005-6324-2
Li F, Shi J, Shen C, Chen G, Hu S, Chen Y (2009) Proteomic characterization of copper stress response in Elsholtzia splendens roots and leaves. Plant Mol Biol 71:251–263. doi:10.1007/s11103-009-9521-y
Li G, Peng X, Xuan H, Wei L, Yang Y, Guo T, Kang G (2013) Proteomic analysis of leaves and roots of common wheat (Triticum aestivum L.) under copper-stress conditions. J Proteome Res 12:4846–4861. doi:10.1021/pr4008283
Li X, Zhou Y, Yang Y, Yang S, Sun X, Yang Y (2015) Physiological and proteomics analyses reveal the mechanism of Eichhornia crassipes tolerance to high-concentration cadmium stress compared with Pistia stratiotes. PLoS ONE 10(4):e0124304. doi:10.1371/journal.pone.0124304
Liao C-T, Lin C-H (2001) Physiological adaptation of crop plants to flooding stress. Proc Natl Sci Counc Repub China B 25:148–157
Liao M, Hedley M, Woolley D, Brooks R, Nichols M (2000) Copper uptake and translocation in chicory (Cichorium intybus L. cv Grasslands Puna) and tomato (Lycopersicon esculentum Mill. cv Rondy) plants grown in NFT system. II. The role of nicotianamine and histidine in xylem sap copper transport. Plant Soil 223:245–254. doi:10.1023/A:1004843505053
Liu T, Shen C, Wang Y, Huang C, Shi J (2014) New insights into regulation of proteome and polysaccharide in cell wall of Elsholtzia splendens in response to copper stress. PLoS ONE 9(10):e109573. doi:10.1371/journal.pone.0109573
Lou L-Q, Shen Z-G, Li X-D (2004) The copper tolerance mechanisms of Elsholtzia haichowensis, a plant from copper-enriched soils. Environ Exp Bot 51:111–120. doi:10.1016/j.envexpbot.2003.08.002
Lubec G, Afjehi-Sadat L, Yang J-W, John JPP (2005) Searching for hypothetical proteins: theory and practice based upon original data and literature. Prog Neurobiol 77:90–127. doi:10.1016/j.pneurobio.2005.10.001
Maksymiec W (1998) Effect of copper on cellular processes in higher plants. Photosynthetica 34:321–342. doi:10.1023/A:1006818815528
Mattie MD, Freedman JH (2004) Copper-inducible transcription: regulation by metal-and oxidative stressresponsive pathways. Am J Physiology-Cell Physiol 286:C293–C301. doi:10.1152/ajpcell.00293.2003
Meki MN, Snider JL, Kiniry JR, Raper RL, Rocateli AC (2013) Energy sorghum biomass harvest thresholds and tillage effects on soil organic carbon and bulk density. Ind Crops Prod 43:172–182. doi:10.1016/j.indcrop.2012.07.033
Mendoza J, Garrido T, Castillo G, San Martin N (2006) Metal availability and uptake by sorghum plants grown in soils amended with sludge from different treatments. Chemosphere 65:2304–2312. doi:10.1016/j.chemosphere.2006.05.012
Mounaji K, Vlassi M, Erraiss N-E, Wegnez M, Serrano A, Soukri A (2003) In vitro effect of metal ions on the activity of two amphibian glyceraldehyde-3-phosphate dehydrogenases: potential metal binding sites. Comp Biochem Physiol B 135:241–254. doi:10.1016/S1096-4959(03)00051-4
Nagajyoti P, Lee K, Sreekanth T (2010) Heavy metals, occurrence and toxicity for plants: a review. Environ Chem Lett 8:199–216. doi:10.1007/s10311-010-0297-8
Ngara R, Ndimba R, Borch-Jensen J, Jensen ON, Ndimba B (2012) Identification and profiling of salinity stress-responsive proteins in Sorghum bicolor seedlings. J Proteomics 75:4139–4150. doi:10.1016/j.jprot.2012.05.038
Oh M, Nanjo Y, Komatsu S (2014a) Gel-free proteomic analysis of soybean root proteins affected by calcium under flooding stress. Front Plant Sci. doi:10.3389/fpls.2014.00559
Oh MW et al (2014b) Proteome analysis of roots of wheat seedlings under aluminum stress. Mol Biol Rep 41:671–681. doi:10.1007/s11033-013-2905-8
Paterson AH et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556. doi:10.1038/nature07723
Patsikka E, Kairavuo M, Sersen F, Aro E-M, Tyystjarvi E (2002) Excess copper predisposes photosystem II to photoinhibition in vivo by outcompeting iron and causing decrease in leaf chlorophyll. Plant Physiol 129:1359–1367. doi:10.1104/pp.004788
Pinto A, de Varennes A, Goncalves M, Mota A (2006) Sorghum detoxification mechanisms. J Plant Nutr 29:1229–1242. doi:10.1080/01904160600767450
Qian D, Tian L, Qu L (2015) Proteomic analysis of endoplasmic reticulum stress responses in rice seeds. Sci Rep. doi:10.1038/srep14255
Rakwal R, Agrawal GK, Yonekura M (1999) Separation of proteins from stressed rice (Oryza sativa L.) leaf tissues by two-dimensional polyacrylamide gel electrophoresis: induction of pathogenesis-related and cellular protectant proteins by jasmonic acid, UV irradiation and copper chloride. Electrophoresis 20:3472–3478. doi:10.1002/(SICI)1522
Ritter A et al (2010) Copper stress proteomics highlights local adaptation of two strains of the model brown alga Ectocarpus siliculosus. Proteomics 10:2074–2088. doi:10.1002/pmic.200900004
Salt DE, Smith R, Raskin I (1998) Phytoremediation. Annu Rev Plant Biol 49:643–668. doi:10.1146/annurev.arplant.49.1.643
Shen Z, Zhang F, Zhang F (1998) Toxicity of copper and zinc in seedlings of mung bean and inducing accumulation of polyamine. J Plant Nutr 21:1153–1162. doi:10.1080/01904169809365474
Smith AP, DeRidder BP, Guo W-J, Seeley EH, Regnier FE, Goldsbrough PB (2004) Proteomic analysis of Arabidopsis glutathione S-transferases from benoxacor-and copper-treated seedlings. J Biol Chem 279:26098–26104. doi:10.1074/jbc.M402807200
Song Y, Cui J, Zhang H, Wang G, Zhao F-J, Shen Z (2013) Proteomic analysis of copper stress responses in the roots of two rice (Oryza sativa L.) varieties differing in Cu tolerance. Plant Soil 366:647–658. doi:10.1007/s11104-012-1458-2
Song Y, Zhang H, Chen C, Wang G, Zhuang K, Cui J, Shen Z (2014) Proteomic analysis of copper-binding proteins in excess copper-stressed rice roots by immobilized metal affinity chromatography and two-dimensional electrophoresis. Biometals 27:265–276. doi:10.1007/s10534-014-9707-x
Soudek P, Petrova S, Vankova R, Song J, Vanek T (2014) Accumulation of heavy metals using Sorghum sp. Chemosphere 104:15–24. doi:10.1016/j.chemosphere.2013.09.079
Swami AK, Alam SI, Sengupta N, Sarin R (2011) Differential proteomic analysis of salt stress response in Sorghum bicolor leaves. Environ Exp Bot 71:321–328. doi:10.1016/j.envexpbot.2010.12.017
Sweetlove L, Heazlewood J, Herald V, Holtzapffel R, Day D, Leaver C, Millar A (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32:891–904. doi:10.1046/j.1365-313X.2002.01474.x
Taddei S, Bernardi R, Salvini M, Pugliesi C, Durante M (2007) Effect of copper on callus growth and gene expression of in vitro-cultured pith explants of Nicotiana glauca. Plant Biosyst 141:194–203. doi:10.1080/11263500701401521
Tewari RK, Kumar P, Sharma PN (2006) Antioxidant responses to enhanced generation of superoxide anion radical and hydrogen peroxide in the copper-stressed mulberry plants. Planta 223:1145–1153. doi:10.1007/s00425-005-0160-5
Tezara W, Mitchell V, Driscoll S, Lawlor D (1999) Water stress inhibits plant photosynthesis by decreasing coupling factor and ATP. Nature 401:914–917. doi:10.1038/44842
Utriainen M, Kokko H, Auriola S, Sarrazin O, Karenlampi S (1998) PR-10 protein is induced by copper stress in roots and leaves of a Cu/Zn tolerant clone of birch, Betula pendula. Plant Cell Environ 21:821–828. doi:10.1046/j.1365-3040.1998.00326.x
Vallino M, Drogo V, Perotto S (2005) Gene expression of the ericoid mycorrhizal fungus Oidiodendron maius in the presence of high zinc concentrations. Mycorrhiza 15:333–344. doi:10.1007/s00572-004-0335-0
Van Loon LC, Rep M, Pieterse C (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162. doi:10.1146/annurev.phyto.44.070505.143425
Wang Y et al (2006) Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic Biol Med 40:1201–1209. doi:10.1016/j.freeradbiomed.2005.11.019
Wang Y, Li H, Qiu Y, Li N, Sun W, Shan Z (2014) Identification of copper-binding proteins in soybean seeds by immobilized metal affinity chromatography and mass spectrometry. Plant Biosystems-An Int J Dealing Aspects of Plant Biol 148:88–95. doi:10.1080/11263504.2013.770807
Weng Z-X et al (2013) Proteomic and physiological analyses reveal detoxification and antioxidation induced by Cd stress in Kandelia candel roots. Trees 27:583–595. doi:10.1007/s00468-012-0811-7
Wissuwa M, Ismail AM, Yanagihara S (2006) Effects of zinc deficiency on rice growth and genetic factors contributing to tolerance. Plant Physiol 142:731–741. doi:10.1104/pp.106.085225
Xin Z, Wang ML, Barkley NA, Burow G, Franks C, Pederson G, Burke J (2008) Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol 8:103. doi:10.1186/1471-2229-8-103
Yang X, Long X, Ye H, He Z, Calvert D, Stoffella P (2004) Cadmium tolerance and hyperaccumulation in a new Zn-hyperaccumulating plant species (Sedum alfredii Hance). Plant Soil 259:181–189. doi:10.1023/B:PLSO.0000020956.24027.f2
Yao YA et al (2012) Proteomic analysis of Mn-induced resistance to powdery mildew in grapevine. J Exp Bot 63:5155–5170. doi:10.1093/jxb/ers175
Yin X, Sakata K, Komatsu S (2014) Phosphoproteomics reveals the effect of ethylene in soybean root under flooding stress. J Proteome Res 13:5618–5634. doi:10.1021/pr500621c
Yruela I (2005) Copper in plants. Braz J Plant Physiol 17:145–156. doi:10.1590/S1677-04202005000100012
Yruela I (2009) Copper in plants: acquisition, transport and interactions. Funct Plant Biol 36:409–430. doi:10.1071/FP08288
Zhang H, Lian C, Shen Z (2009) Proteomic identification of small, copper-responsive proteins in germinating embryos of Oryza sativa. Ann Bot 103:923–930. doi:10.1093/aob/mcp012
Zhao L et al (2011) Cd-induced changes in leaf proteome of the hyperaccumulator plant Phytolacca americana. Chemosphere 85:56–66. doi:10.1016/j.chemosphere.2011.06.029
Zhuang P, Wensheng S, Zhian L, Bin L, Jintian L, Jingsong S (2009) Removal of metals by sorghum plants from contaminated land. J Environ Sci 21:1432–1437. doi:10.1016/S1001-0742(08)62436-5
Zou H-X, Pang Q-Y, Zhang A-Q, Lin L-D, Li N, Yan X-F (2015) Excess copper induced proteomic changes in the marine brown algae Sargassum fusiforme. Ecotoxicol Environ Saf 111:271–280. doi:10.1016/j.ecoenv.2014.10.028
Author information
Authors and Affiliations
Corresponding author
Additional information
Swapan Kumar Roy and Seong-Woo Cho have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Roy, S.K., Kwon, S.J., Cho, SW. et al. Leaf proteome characterization in the context of physiological and morphological changes in response to copper stress in sorghum. Biometals 29, 495–513 (2016). https://doi.org/10.1007/s10534-016-9932-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-016-9932-6