Abstract
Rainfall patterns are predicted to change dramatically in many terrestrial landscapes, including drylands. The most limiting resources for plant growth in arid regions is nitrogen (N) as well as water. A natural aridity gradient provides appropriate candidate conditions for predicting the impacts of changes in rainfall on soil N dynamics. To comprehensively and mechanistically examine soil N dynamics, we focused on the steps of N transformation, their microbial drivers, and the determining soil properties. We divided the N transformation process into three steps: (i) organic matter degradation, (ii) N mineralization, and (iii) nitrification, which are driven primarily by fungi, prokaryotes, and ammonia oxidizers, respectively. Soil samples were collected from three black locust forests with mean annual precipitations ranging from 449 to 606 mm. Along the aridity gradient, all three steps changed while maintaining a balance. The degradation and mineralization steps varied with changes in the soil fungal and prokaryotic communities, respectively. The compositions of these communities were determined by soil substrate quality and quantity; saprotrophs and copiotrophs decreased along the aridity gradient. On the other hand, the abundance of ammonia-oxidizing bacteria, which correlated with the rate of nitrification, was likely determined by soil moisture. Therefore, if precipitation were to decrease, changes in the nitrification step might be the first mechanism to limit plant productivity in semi-arid forests. This limitation would extend to the other steps in the N cycling process via plant–soil feedback. Thus, N cycling dynamics are predicted to achieve new stable states suited to the changed precipitation regime.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Rainfall patterns are expected to change dramatically in the near future; drought events will likely increase in frequency, the areal extent of arid zones will expand, and floral compositions will change, especially in drylands (IPCC 2007). Water is usually the most limiting resource for plant growth in arid regions, but nitrogen (N) is a co-limiting factor for net primary production (NPP) (Austin and Sala 2002; Eskelinen and Harrison 2015). When drought periods are protracted, N is a greater limiting resource for NPP than water (Ren et al. 2017). Studies of N cycling responses to rainfall pattern changes are essential for a better understanding of soil dynamics in an era of rapid climate change. Many studies have examined the mechanism of soil N dynamics responses to rainfall (Landesman and Dighton 2010; Chen et al. 2013, 2017; Ladwig et al. 2015; Bowles et al. 2018). However, most have focused on only single parts of the N transformation process or have failed to provide information on microbial N drivers. A comprehensive study that includes steps of soil N transformation and relevant microbes will contribute significantly to a deeper understanding of the mechanism underlying the responses of soil N dynamics to rainfall pattern change.
The series of N transformation processes in the soil can be divided into three steps. The first step is degradation, which produces small-molecule organic N from large-molecule organic N, such as the organic matter in soil. In the second step, the small-molecule organic N compounds are mineralized and converted to ammonium N. The third step is nitrification, in which ammonium N is converted to nitrate N. Thus, soil N transformation is a step-by-step process; substrate quantity and quality can limit downstream transformations (Updegraff et al. 1995; Norman and Barrett 2014). Changes in an upstream step can affect the subsequent downstream step by acting as a bottleneck, i.e., a step that delays the progress of subsequent steps. For example, in cold ecosystems, the mineralization step is limited by temperature and can act as a bottleneck (Rustad et al. 2001; Schimel et al. 2004). The nitrification step can also act as a bottleneck when the pH is low (Ste-Marie and Paré 1999). Thus, each step may react differently to changes in rainfall patterns. Our understanding of the total soil N dynamics process will be improved by dividing the chain of events into separate steps that can be examined in detail.
The responses of soil N dynamics to rainfall pattern changes are mediated by soil microbes (Chen et al. 2017; Bowles et al. 2018). Microbes also fall into three categories of step drivers. Fungi and prokaryotes are the main drivers of the degradation and mineralization steps, respectively (Moore et al. 2003). In the nitrification step, Ammonia-oxidizing bacteria and archaea are important because they perform the rate-limiting process. (Kowalchuk and Stephen 2001; Isobe et al. 2015). Nitrite-oxidizing bacteria have recently received increasing attention, although concentrations of the nitrite N concentration was much smaller than those of ammonium and nitrate N (Burns et al. 1996; Smith et al. 1997; Xu et al. 2012; Maharjan and Venterea 2013; Venterea et al. 2015). A focus on microbial composition improves understanding of the mechanism of soil N dynamics because the abilities and strategies for N transformation vary among microbial taxa. For example, among fungi, saprotrophs and mycorrhizal species decompose organic matter primarily to obtain carbon (C) and N, respectively. Mycorrhizal fungi receive C from their host plants (Lindahl et al. 2007; Smith and Read 2010). The soil bacteria may be categorized as copiotrophs, which readily decompose labile compounds, and oligotrophs, which decompose recalcitrant compounds (Fierer et al. 2007). These two bacterial groups also differ in their ability to produce extracellular enzymes, including N-acetylglucosaminidase and chitinase, which decompose monomers and polymers, respectively (Zimmerman et al. 2013). Ammonia-oxidizing bacteria and archaea also have different oxidization capabilities: nitrification rates tend to correlate with ammonia-oxidizing bacterial abundance, but not with archaeal abundance, likely because ammonia-oxidizing bacteria are competitive at high ammonium concentrations (Di et al. 2009; Chen et al. 2013; Banning et al. 2015), although not in acidic soil (Prosser and Nicol 2008; Isobe et al. 2015).
Many studies have examined the ways in which soil physicochemical properties influence the microbes driving soil N processes. The low soil moisture, low organic matter content, and high pH in drylands limit fungal and bacterial growth (Frey et al. 1999; Maestre et al. 2015) and change community compositions by reducing the abundances of copiotrophic and ammonia-oxidizing bacteria (Adair and Schwartz 2008; Xiong et al. 2012). However, under such stressful conditions, microbial groups with high drought tolerance or enhanced abilities in recalcitrant nutrient decomposition flourish, as observed in Actinobacteria and Verrucomicrobia (Bouskill et al. 2013; Cederlund et al. 2014; Iwaoka et al. 2018). Thus, a comprehensive view of soil N dynamics will emerge from studies that link information on each step of soil N transformation, their drivers, the soil microbial community, and soil physicochemical properties. With a such a broad view, we can better predict how rainfall pattern shifts will alter the soil conditions that regulate microbial communities and the chain of N transformation processes.
Natural aridity gradients provide appropriate candidate conditions for studies aiming to predict the effects of rainfall pattern changes on soil N dynamics. Along an aridity gradient, soil moisture content changes with other soil properties, such as organic matter content, as a function of rainfall history, unlike in shorter-term artificial rainfall experiments. Soil N availability, N transformation rate, and microbial abundance tend to increase with rainfall toward the wetter end of an aridity gradient (Burke et al. 1997; Austin and Sala 2002; Adair and Schwartz 2008). However, this is not always the case, as mineralization rate, nitrification rate and N availability have been reported not to change with rainfall (Smith et al. 2002; Feral et al. 2003; Meier and Leuschner 2014). Contradictions among studies have arisen because the underlying mechanisms are not understood. In our study, we examined three black locust (Robinia pseudoacacia) forests occurring along a natural aridity gradient on the Loess Plateau in northeastern China. Differences in vegetation, including variability in canopy trees, strongly influence soil N dynamics (Finzi et al. 1998; Phillips et al. 2013); we therefore compared the same vegetation along the aridity gradient.
We examined the process of soil N dynamics changes (mediated by shifts in the soil microbial community) along the aridity gradient. We aimed to use the data from this study to predict responses to rainfall pattern alterations in a time of global climate change. We studied all three steps of N transformation, the associated microbial communities, and soil physicochemical properties along the aridity gradient. Our fundamental hypothesis was that inhibition of a single step in the transformation process would cause a bottleneck in the soil N cycling process. Such bottlenecks would be key elements in the N limitation of forest productivity in arid ecosystems during future climate change.
Materials and methods
Study sites and soil sampling
We conducted the study in three black locust (Robinia pseudoacacia) forests spanning the southern to central sectors of the Loess Plateau. The three stands were located in mountainous areas of Shaanxi Province, China: (i) near Yongshou County (Huiping; 34°48N, 107°59E, 1395 m elevation), (ii) near Yan’an City (Mt. Gonglushan; 36°25N, 109°32E, 1320 m elevation), and (iii) near Ansai Country (Zhifanggou; 36°45N, 109°15E, 1175 m elevation), which we refer to hereafter as the Wet, Med (medium), and Dry sites, respectively. The vegetation type has been classified as a forest–steppe transitional zone; the mean annual rainfall decreases gradually from southeast to northwest (Tsunekawa 2014). The reference evaporation rate increases from south to north in our research region (Li et al. 2012). Annual rainfall and mean annual air temperature at the sites were as follows: 606 mm and 10.8 °C at the Wet site (Li et al. 2015), 514 mm and 10.2 °C at the Med site (Otsuki et al. 2005), and 449 mm and 8.8 °C at the Dry site (Qiu et al. 2012). The area has hot summers and cold winters; heavy precipitation occurs in summer. Although the precipitation gradient in this study, only 150 mm in mean annual precipitation, is smaller than those in other studies (Austin and Sala 2002; Feral et al. 2003; Adair and Schwartz 2008; Meier and Leuschner 2014), 150 mm is a critical difference in this area, the arid boundary of a forest. The rainfall amount at the Dry site was 74% of that at the Wet site; 30% exclusion of rainfall in semi-arid forests has been shown to reduce forest production (Ogaya and Penuelas 2007). Our gradient enables a comparison of the effects of precipitation under similar environmental conditions of mean annual temperature and soil type.
At all three sites, forest canopies were closed and occupied by > 90% black locust, with understory covers of shrubs and herbaceous species. Tree basal area, the amounts of organic layer and litter quality (C:N ratio) decreased along the aridity gradient (Table S1). In each forest, we established four plots (20 × 20 m), each > 30 m distant from all neighboring plots. All plots had similar slopes (flat land or gentle slopes near ridges) because topography considerably affects soil and N contents in this region (Tateno et al. 2017). Soil samples (0–10 cm deep) were taken from four locations in each plot. The sampling locations were ca. 10 m distant from one another. The four soil samples were combined into a single sample. Samples were taken in September 2015, and in April, June, and September 2016.
Measurement of soil physical and chemical properties
Soil water content was determined after samples were dried at 105 °C for > 3 days. Soil pH was measured with a pH meter (D-51; HORIBA, Kyoto, Japan) in a 2:5 soil/water suspension. After soil samples were ground, total C and N contents were measured using the dichromate oxidation and Kjeldahl methods, respectively. Soon after soil collection and incubation at 25 °C for > 6 days without freezing soil samples, we extracted dissolved C and N with 2 M KCl in a 1:10 soil/extractant ratio. The extracts were stored in a freezer until further analysis. The total dissolved organic C and N contents in the extracts were measured with a TOC-LCPH/CPN + TNM-L device (SHIMADZU, Kyoto, Japan) (Shimadzu 2013). We measured the amounts of ammonium and nitrate N in the extracts with a Bran + Luebbe AutoAnalyzer III device (BLTEC, Tokyo, Japan) using a colorimetric procedure. We also measured the amount of nitrite N in the extracts which was from soils collected on two occasions from the same sites. Soil dissolved organic N content, reportedly correlated with protein abundance that can be degraded by prokaryotes (Yu et al. 2002; Wang et al. 2013), was calculated as the difference between the concentrations of total dissolved N and inorganic N (i.e., ammonium + nitrate N). Net N mineralization and nitrification rates in the soil were calculated as the differences in inorganic N and nitrate N, respectively, before and after incubation. We report N content per kilogram of dry soil. Soil organic matter quality and dissolved organic matter quality were examined based on the total organic C:N and dissolved organic C:N ratios, respectively.
Soil DNA extraction and quantification by real-time quantitative PCR
DNA was extracted from 0.25-g soil samples using the MoBio Powersoil DNA Isolation Kit (MoBio, Carlsbad, CA, USA) according to the manufacturer’s instructions. The real-time quantitative polymerase chain reaction (qPCR) was run in a Light Cycler Nano thermal cycler device (Roche Diagnostics K.K., Mannheim, Germany) with the SYBR Green I intercalating dye. Fungal ITS, bacterial 16S rRNA, archaeal 16S rRNA, ammonia-oxidizing bacterial amoA, and ammonia-oxidizing archaeal amoA genes were targeted to estimate the population size of each soil microbial group. Fungal ITS abundances were determined using the universal primer sets ITS1F_KYO2/ITS2_KYO2 (Toju et al. 2012). The qPCR conditions for fungal ITS genes were as follows: initial denaturation at 95 °C for 10 min, followed by > 30 cycles at 95 °C for 1 min, 53 °C for 30 s, and 72 °C for 1 min (Fierer and Jackson 2005). The subsequent step in the analysis for 16S rRNA and amoA genes has been described in detail by Iwaoka et al. (2018).
Sequence analysis
We amplified the ITS and 16S rRNA genes in the DNA extracts using the primer sets ITS1F_KYO2/ITS2_KYO2 (Toju et al. 2012) and U519f (Suzuki and Giovannoni 1996)/U785r (Wang and Qian 2009), respectively, with the following reaction solution ratio: 44:1:1:4 Platinum PCR Super Mix High Fidelity (Thermo Fisher Scientific)/10 mM forward primer/10 mM reverse primer/tenfold-diluted DNA sample. The subsequent step in the analysis has been described in detail by Iwaoka et al. (2018). Sequencing was performed on an Ion Personal Genome Machine (PGM) (Thermo Fisher Scientific). The processing of the sequencing data for 16S rRNA genes was performed as described by Iwaoka et al. (2018). ITS sequences shorter than 160 bp in length were removed; ITSx was used to extract fungal ITS sequences (Bengtsson-palme et al. 2013). The remaining steps were the same as for 16S rRNA sequences. Sequence data were deposited in the Sequence Read Archive of the National Center for Biotechnology Information under accession number PRJDB7067.
In our analyses of the fungal and prokaryotic community structures, we standardized the read numbers to 10031 and 8726, respectively, using random pick-up based on the minimum read number. We searched for fungal trophic modes from the fungal taxonomy and predicted specific functions based on the 16S rRNA genes using the FUNGuild database and the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) pipeline, respectively (Langille et al. 2013; Nguyen et al. 2016). To perform PICRUSt searches, the sequences were re-picked at a 97% identity level with reference to the Greengenes database (version 13_05). We eliminated reads not corresponding to the reference database; the OTU table was normalized by gene copy number, and the metagenome functional profiles were predicted using PICRUSt (ver. 1.1.1) to generate a table of Kyoto Encyclopedia of Gene and Genomes (KEGG) orthologs (KOs). We selected the genes necessary for coding extracellular enzymes responsible for N compound degradation in soil, i.e., cellulase (EC 3.2.1.4), N-acetylglucosaminidase (EC 3.2.1.52), and chitinase (EC 3.2.1.14), based on the KOs (Isobe et al. 2018).
Statistical analyses
We used one-way analysis of variance (ANOVA) with the sampling date (September 2015/April 2016/June 2016/September 2016) as a random effect to test for significant differences among the sites in soil physicochemical properties, total and dissolved organic C, N contents and C:N ratios, inorganic N content and net transformation rate, microbial abundance, relative abundances of fungal and prokaryotic groups, and predicted gene abundances. We chose a significance level of P < 0.05 for all tests. Prior to ANOVA, we tested for normality and homogeneity of variance in the data using the Kolmogorov–Smirnov and median Levene’s tests, respectively. The car package in R software (version 3.1.2; R Development Core Team 2014) was used to perform Levene’s test (Fox et al. 2014). When the P values for these tests were < 0.05, data were square-root or reciprocally transformed prior to ANOVA analysis. Significant pairwise differences between sites were identified by Tukey–Kramer multiple comparison tests, or Steel–Dwass multiple comparison tests when the homogeneity of variance was violated. Correlations were tested using the Pearson procedure or by a generalized linear mixed model with site and sampling date as random effects.
Non-metric multidimensional scaling (NMDS) analysis of community structure dissimilarity based on the Bray–Curtis index was performed using the metaMDS function in the vegan package (Oksanen et al. 2016) of R software. The envfit function in the vegan package was used to identify significant correlations between (i) soil properties and the relative abundances of the fungal trophic groups, saprotrophic fungal orders, and the predicted abundances of genes for N-degrading enzymes and (ii) the NMDS values of points as vectors on the NMDS ordination plot. Permutational multivariate analysis of variance (PerMANOVA) was performed to test the significance of the effect of site on the microbial community, with sampling date as a random effect, using the Adonis function in the R vegan package.
Results
Soil physicochemical properties and organic matter quality and quantity
Soil water content was significantly higher at the Wet site than at the other two sites (Table 1). Soil pH also differed significantly among sites and was lowest at the Wet site. Soil total N content differed significantly among sites and was highest at the Wet site, followed by the Med site. Total C:N ratios did not differ significantly among sites. The dissolved organic C:N ratio differed significantly among sites, and was lowest at the Wet site. For soil water content, pH, and dissolved organic C:N ratio, the values for the Med site were more similar to those in the Dry site, but for total N content, the Med site was more similar to the Wet site.
Extractable N dynamics in soils
Dissolved organic N, ammonium N, nitrate N, and total inorganic N contents differed significantly among sites (Fig. 1). The Wet site had significantly higher dissolved organic N and ammonium N contents than did the Dry site. The Wet site had significantly higher nitrate N and total inorganic N contents than did the Med and Dry sites. The net N mineralization rate in the soil did not differ significantly among sites (Table 1). The net nitrification rate differed significantly among sites, and was highest at the Wet site. Nitrite N was generally undetectable, with a maximum of 0.008 ppm (< 0.1 mg kg−1).
Extractable N contents in the soils. Values are means ± SDs. Significant differences among sites, identified by one-way ANOVA with the sampling date as a random effect, are indicated in the upper right text panel. *P < 0.05, ***P < 0.001. Results of Tukey–Kramer or Steel–Dwass tests are shown inside the bars; different lowercase letters indicate significant pairwise differences between sites for each parameter
Soil fungal communities
Fungal ITS gene abundance in the soil did not differ significantly among sites (Table S2). However, the PerMANOVA showed that soil fungal community structure was influenced significantly by site (F = 11.8, partial R2 = 0.34, P < 0.001, Table S3), and the fungal communities at the three sites along the aridity gradient were separated on axis 1 of the NMDS ordination (Fig. 2a).
NMDS based on a fungal and b prokaryotic community dissimilarities. Blue, green, and red symbols represent the Wet, Med (medium), and Dry sites, respectively. Circles, triangles, squares, and diamonds represent samples collected in September 2015, April 2016, June 2016, and September 2016, respectively. Only significant vectors are plotted on the ordination. Black vectors represent soil properties. Dark gray vectors represent relative abundances of trophic groups or predicted gene abundances. WC water content, TN total N content, TC total C content, C:N total C:N ratio; NO3 nitrate N content, DON dissolved organic N content, DOC:DON dissolved organic C:N ratio
The relative abundances of saprotrophic and pathotrophic fungi differed significantly among sites (Fig. 3, Table S4). The relative abundance of saprotrophic fungi was highest and second highest at the Wet and Med sites, respectively. The relative abundance of pathotrophic fungi was highest at the Dry and Med sites. Saprotrophic fungal community composition also changed along the aridity gradient, with an apparent reduction in the relative abundance of the order Mortierellales as dryness increased (Fig. S1; Table S4). In our NMDS ordination plot, the vectors for the relative abundances of saprotrophic fungi and Mortierellales were more highly correlated with the vectors for soil total N content than with those for water content and pH, and they were also strongly negatively correlated with dissolved organic C:N ratio (Fig. 2a).
Relative abundances of fungal trophic groups based on FUNGuild database trophic modes. The values in the bar graphs are the averages. The FUNGuild algorithm frequently outputs multiple trophic modes for single OTUs (e.g., saprotroph–symbiotroph); in these cases, the OTUs were placed in the “Others” category
Soil prokaryotic communities
The soil 16S rRNA gene abundances of bacteria and archaea did not differ significantly among sites (Table S2), but the PerMANOVA showed that the soil prokaryotic community structure differed significantly by site along the aridity gradient (F = 14.9, partial R2 = 0.36, P < 0.001, Table S3). The NMDS also identified clear differences in prokaryotic community structures among sites (Fig. 2b).
The predicted relative abundances of genes associated with N-acetylglucosaminidase and chitinase differed significantly among sites and were highest at the Wet and Med sites, respectively (Fig. 4). However, the predicted relative abundances of genes associated with cellulase did not differ among sites. The relative abundances of the dominant copiotrophic bacteria, Betaproteobacteria, Gammaproteobacteria, and Bacteroidetes and its classes (Cleveland et al. 2007; Fierer et al. 2007), were significantly different between sites (Fig. S2, Table S5) and were highest in the Wet site. In contrast, the relative abundance of the second-ranking Actinobacteria significantly differed among sites and was highest in the Med site. In our NMDS, the vectors of predicted abundances of genes associated with N-acetylglucosaminidase pointed in directions consistent with the vector for dissolved organic N content and inorganic N content (Fig. 2b).
Predicted abundances of genes associated with soil N decomposition (cellulase, chitinase, and N-acetylglucosaminidase) enumerated using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) pipeline procedures. Values are means ± SDs. Significant differences among sites, identified by one-way ANOVA with the sampling date as a random effect, are indicated in the upper right corner (***P < 0.001). Different lowercase letters indicate significant pairwise differences between sites (Tukey–Kramer or Steel–Dwass test)
Soil ammonia-oxidizing communities
Abundances of amoA genes in the soil differed significantly among sites for bacteria, but not for archaea (Fig. 5). The ammonia-oxidizing bacterial abundances were significantly positively and negatively correlated with soil moisture and pH, respectively (Table 2). Ammonia-oxidizing archaea abundance was not correlated with soil moisture, pH, or ammonium N content.
Logged values of soil bacterial and archaeal amoA gene abundances. Values are means ± SDs. Significantly different values among sites, identified by one-way ANOVA with the sampling date as a random effect, are indicated in the upper right sector of the figure (*P < 0.05, ***P < 0.001). Different lowercase letters above the bars indicate significant pairwise differences between sites (Tukey–Kramer or Steel–Dwass test)
Discussion
Soil N dynamics changes along the aridity gradient
Each of the soil extractable N contents, i.e., dissolved organic N, ammonium N, and nitrate N, was reduced with increasing aridity (Fig. 1), which was contrary to our expectation that the content of a specific form of N would remain constant or increase with increasing aridity due to the bottleneck step. Furthermore, nitrite N did not accumulate in the study sites, as expected. The change in net nitrification rate was consistent with the decrease in nitrate N content (Table 1). The net N mineralization rate by laboratory incubation did not differ significantly among sites, although the ammonium N and total inorganic N contents decreased with increasing aridity. N mineralization may be activated after a rainfall event, as suggested in previous studies in which high microbial activity was observed after the re-wetting of dry soil (Franzluebbers et al. 1996; Mikha et al. 2005). Therefore, N mineralization activity in the field may have been higher after rainfall, and rainfall frequency increases from the southeast to northwest on the Loess Plateau (Wan et al. 2014); therefore, the Wet site likely had more frequent rainfall and higher N mineralization activity in the field. According to the pattern of soil extractable N contents, all N degradation steps were affected by the aridity gradient. Substrate accumulates when a bottleneck develops in the stepwise degradation process, but we did not detect accumulated forms of N, suggesting that changes maintained a balance, with no evidence of a continuous bottleneck along the aridity gradient.
Degradation of organic matter by the fungal community
Fungal abundance did not decrease with increasing aridity (Table S2), although dissolved organic N content which was primarily produced by fungal community decreased with increasing aridity. However, fungal community composition changed with the aridity level (Figs. 2a, 3). The relative abundances of saprotrophic fungi, especially that of the dominant order Mortierellales, decreased markedly with increasing aridity (Figs. 2a, 3, S1). As saprotrophs obtain C energy from decomposition and Mortierella-dominated communities have high C-degrading potential (e.g., phenol-oxidase production) (Phillips et al. 2014), the decreases in these groups should reduce the degradation rate and contribute to the increase in the C:N ratio of dissolved organic matter. Pathotrophs, which alternatively increased in abundance along the aridity gradient (Fig. 3), acquire energy from host cells (Nguyen et al. 2016); therefore, the group cannot contribute to decomposition.
Reductions in the abundances of saprotrophs such as Mortierellales with increasing aridity may be caused by reductions in the content of soil organic matter: we found a strong positive correlation between (i) saprotroph and Mortierellales relative abundances and (ii) soil total N contents (Fig. 2a). This finding is consistent with those of other studies, in which increases in saprotrophic fungi and taxa in Mortierellales with addition of organic matter have been observed (Hanson et al. 2008; Kao-Kniffin and Balser 2008; Chigineva et al. 2009), and is further supported by the apparent decrease in the amount of plant input and soil organic matter content with increasing aridity (Table S1). Moreover, fungal communities are reportedly resilient to changes in water content or pH (Rousk et al. 2010; Barnard et al. 2013; de Vries et al. 2018). Thus, the decrease in organic matter content along the aridity gradient likely reduced saprotrophic fungi, particularly Mortierellales, thereby decreasing the dissolved organic N content and increasing the dissolved organic C:N ratio. In contrast, it has been reported that soil moisture content determines fungal decomposition activity which may control soil organic matter content (Hawkes et al. 2011; Cregger et al. 2012). However, if fungal degradation decreased at our study sites due to soil dryness, then soil organic matter content would have increased, not decreased, with increasing aridity. Therefore, organic matter content was a limiting factor of fungal degrading ability on the aridity gradient in the current study.
N mineralization by the soil prokaryotic community
Prokaryotic abundance did not decrease with increasing aridity (Table S2), although ammonium N content, which was produced mainly by prokaryotic community, decreased with increasing aridity. However, community composition changed with aridity, which was also the case for the degradation step (Figs. 2b, S2). The Wet site had the highest N-acetylglucosaminidase production potential for labile compound decomposition, whereas the Med site had the highest chitinase production potential for recalcitrant compound decomposition (Fig. 4). Consistently, the relative abundances of copiotrophs such as the Betaproteobacteria, Gammaproteobacteria, and Bacteroidetes were highest in the Wet site (Fig. S2; Table S5). And that of Actinobacteria, which are strongly competitive in the utilization of recalcitrant compounds such as chitin, was highest at Med site (Bastian et al. 2009; Bell et al. 2013; Cretoiu et al. 2013). Active decomposition of labile compounds by abundant copiotrophs at the Wet site and decomposition of recalcitrant compounds by Actinobacteria at the Med site were key elements in the N mineralization step gradient along the aridity gradient.
The Wet site had the highest dissolved organic N content and the lowest dissolved organic C:N ratio; such organic matters would be suitable substrates for N-acetylglucosaminidase. The Med site would contain relatively more high-molecular-weight organic compounds such as chitin, the substrate of chitinase, based on that the Med site had relatively low dissolved organic N contents and a relatively high dissolved organic C:N ratio (similar to the Dry site), but relatively high total C and N contents (similar to the Wet site; Table 1). Increasing pH and decreasing moisture with increasing aridity may reduce N-acetylglucosaminidase production potential by limiting the growth of copiotrophs such as Betaproteobacteria (Lauber et al. 2009; Lennon et al. 2012; Armstrong et al. 2016). However, Actinobacteria are resilient to low moisture and alkaline soil (Lauber et al. 2009; Placella et al. 2012; Barnard et al. 2013). The increase in Actinobacteria in the Med site are difficult to explain without considering organic matter quality and quantity, because the Med site had medium moisture and pH. Our findings agree with previous observations that the response of bacterial communities to soil water conditions is mediated by organic matter type and availability (de Vries et al. 2012; Placella et al. 2012; Göransson et al. 2013).
Nitrification
The gradient in nitrification was likely related to the activities of the ammonia oxidizer communities. Ammonia-oxidizing bacterial abundance decreased along the aridity gradient, but the abundances of ammonia-oxidizing archaea did not differ among sites (Fig. 5), as observed in previous studies (Adair and Schwartz 2008; Chen et al. 2013). The frequently observed positive relationship between the net nitrification rate and ammonia-oxidizing bacterial abundance (Di et al. 2009; Chen et al. 2013; Banning et al. 2015) suggests that nitrification is driven mainly by ammonia-oxidizing bacteria at this study site. However, we did not test differences in archaeal versus bacterial ammonia oxidation activity through addition of octyne as previously described (Taylor et al. 2013).
Ammonia-oxidizing bacterial abundance was significantly positively correlated with soil moisture and negatively correlated with pH, but the abundance of these microbes was not related to ammonium N content (Table 2). The positive correlation between the abundance of this group of prokaryotes and moisture content was consistent with the findings of previous studies (Gleeson et al. 2010; Placella and Firestone 2013; Marcos et al. 2016), but the negative correlation with pH was not (Hu et al. 2013). Ammonium N content has been shown to determine ammonia-oxidizing bacterial abundance with a tenfold gap in the ammonium N content (Verhamme et al. 2011; Wang et al. 2011). Hence, the determining factor in ammonia-oxidizing bacterial abundance at this study site was probably soil moisture and not substrate ammonium N, unlike in upstream steps. Soil moisture obviously declined along the aridity gradient, thereby reducing ammonia-oxidizing bacterial abundance and creating the nitrification step gradient.
Predicting shifts in N transformation with changing rainfall pattern
In summary, our findings suggest that substrate quality and quantity strongly affect the degradation and N mineralization steps by changing fungal and prokaryotic community compositions. Although soil moisture or pH may contribute to changes in the fungal and prokaryotic community, it is difficult to explain the changes observed in these communities without substantial effects of their substrate quality and quantity. However, in the nitrification step, soil moisture appeared to be more important than substrate for the ammonia-oxidizing bacteria, although ammonia-oxidizing archaeal population was resilient to changes in soil moisture. Hence, as rainfall patterns change, N degradation and mineralization will proceed well unless the soil organic matter content changes, but the nitrification rate will slow as moisture decreases. Substrate and soil moisture reportedly determine N mineralization and nitrification rates (Saetre and Stark 2005; Ford et al. 2007; Marcos et al. 2016), but substrate has a greater effect on N mineralization than does moisture, which has a greater effect on nitrification (Yahdjian and Sala 2010; Hu et al. 2015), as demonstrated in this study.
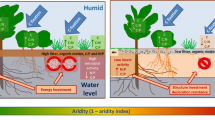
Furthermore, the vulnerability of nitrification to soil moisture depletion resulting from climate change may control whole-forest N cycling. A decrease in N availability will limit N uptake, NPP and litter production (Norby et al. 2010; Ren et al. 2017; Tateno et al. 2017), consequently reducing soil organic matter content (Fig. 6). This reduction in soil organic matter content imposes substrate limitation on the fungal community, which will limit the whole N dynamics sequence through mineralization (Fig. 6). Thus, precipitation decreases may limit whole-forest N cycling, beginning with nitrification limitation, via the plant–soil feedback process. In this study, we observed no accumulation of specific N forms, perhaps because limitation in the nitrification step extended to the other steps; hence, the N cycling process was maintained in particular stable states, each suited to the precipitation at the different sites. Thus, we found balanced changes in extractable content of all N forms along the aridity gradient. The nitrification step is likely to become a temporary bottleneck step with increasing aridity. However, we expect the whole N cycling process to achieve a new stable state suited to the decreased precipitation pattern under future climate change.
Conceptual N cycling model based on results from this study. Black solid arrows and black thick dashed arrows refer to the effects observed in this study and the predicted effects, respectively. Aridity determines the nitrification step, which is controlled by soil moisture and ammonia oxidizers. The nitrification step would affect N uptake and net primary production (NPP), consequently controlling the degradation and mineralization steps by changing their substrates and drivers. Thin black dashed arrows indicate other possible effects that were not strongly supported in this study
References
Adair KL, Schwartz E (2008) Evidence that ammonia-oxidizing archaea are more abundant than ammonia-oxidizing bacteria in semiarid soils of northern Arizona, USA. Microb Ecol 56:420–426. https://doi.org/10.1007/s00248-007-9360-9
Armstrong A, Valverde A, Ramond J et al (2016) Temporal dynamics of hot desert microbial communities reveal structural and functional responses to water input. Nat Publ Gr. https://doi.org/10.1038/srep34434
Austin AT, Sala OE (2002) Carbon and nitrogen dynamics across a natural precipitation gradient in Patagonia, Argentina. J Veg Sci 13:351–360. https://doi.org/10.1111/j.1654-1103.2002.tb02059.x
Banning NC, Maccarone LD, Fisk LM, Murphy DV (2015) Ammonia-oxidising bacteria not archaea dominate nitrification activity in semi-arid agricultural soil. Sci Rep. https://doi.org/10.1038/srep11146
Barnard RL, Osborne CA, Firestone MK (2013) Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J 7:2229–2241. https://doi.org/10.1038/ismej.2013.104
Bastian F, Bouziri L, Nicolardot B, Ranjard L (2009) Impact of wheat straw decomposition on successional patterns of soil microbial community structure. Soil Biol Biochem 41:262–275. https://doi.org/10.1016/j.soilbio.2008.10.024
Bell TH, Yergeau E, Maynard C et al (2013) Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J 7:1200–1210. https://doi.org/10.1038/ismej.2013.1
Bengtsson-Palme J, Ryberg M, Hartmann M et al (2013) Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4:914–919. https://doi.org/10.1111/2041-210X.12073
Bouskill NJ, Lim HC, Borglin S et al (2013) Pre-exposure to drought increases the resistance of tropical forest soil bacterial communities to extended drought. ISME J 7:384–394. https://doi.org/10.1038/ismej.2012.113
Bowles TM, Jackson LE, Cavagnaro TR (2018) Mycorrhizal fungi enhance plant nutrient acquisition and modulate nitrogen loss with variable water regimes. Glob Chang Biol 24:e171–e182. https://doi.org/10.1111/gcb.13884
Burke IC, Lauenroth WK, Parton WJ, Parton WJ (1997) Regional and temporal variation in net primary production and nitrogen mineralization in grasslands. Ecology 78:1330–1340. https://doi.org/10.2307/2266128
Burns LC, Stevens RJ, Laughlin RJ (1996) Production of nitrite in soil by simultaneous nitrification and denitrification. Soil Biol Biochem 28:609–616. https://doi.org/10.1016/0038-0717(95)00175-1
Cederlund H, Wessén E, Enwall K et al (2014) Soil carbon quality and nitrogen fertilization structure bacterial communities with predictable responses of major bacterial phyla. Appl Soil Ecol 84:62–68. https://doi.org/10.1016/j.apsoil.2014.06.003
Chen Y, Xu Z, Hu H et al (2013) Responses of ammonia-oxidizing bacteria and archaea to nitrogen fertilization and precipitation increment in a typical temperate steppe in Inner Mongolia. Appl Soil Ecol 68:36–45. https://doi.org/10.1016/j.apsoil.2013.03.006
Chen J, Xiao G, Kuzyakov Y et al (2017) Soil nitrogen transformation responses to seasonal precipitation changes are regulated by changes in functional microbial abundance in a subtropical forest. Biogeosciences 14:2513–2525. https://doi.org/10.5194/bg-14-2513-2017
Chigineva NI, Aleksandrova AV, Tiunov AV (2009) The addition of labile carbon alters litter fungal communities and decreases litter decomposition rates. Appl Soil Ecol 42:264–270. https://doi.org/10.1016/j.apsoil.2009.05.001
Cleveland CC, Nemergut DR, Schmidt SK, Townsend AR (2007) Increases in soil respiration following labile carbon additions linked to rapid shifts in soil microbial community composition. Biogeochemistry 82:229–240. https://doi.org/10.1007/s10533-006-9065-z
Cregger MA, Schadt CW, McDowell NG et al (2012) Response of the soil microbial community to changes in precipitation in a semiarid ecosystem. Appl Environ Microbiol 78:8587–8594. https://doi.org/10.1128/AEM.02050-12
Cretoiu MS, Korthals GW, Visser JHM, Van Elsas JD (2013) Chitin amendment increases soil suppressiveness toward plant pathogens and modulates the actinobacterial and oxalobacteraceal communities in an experimental agricultural field. Appl Environ Microbiol 79:5291–5301. https://doi.org/10.1128/AEM.01361-13
de Vries FT, Liiri ME, Bjørnlund L et al (2012) Legacy effects of drought on plant growth and the soil food web. Oecologia 170:821–833. https://doi.org/10.1007/s00442-012-2331-y
de Vries FT, Griffiths RI, Bailey M et al (2018) Soil bacterial networks are less stable under drought than fungal networks. Nat Commun. https://doi.org/10.1038/s41467-018-05516-7
Di HJ, Cameron KC, Shen JP et al (2009) Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2:621–624. https://doi.org/10.1038/ngeo613
Eskelinen A, Harrison SP (2015) Resource colimitation governs plant community responses to altered precipitation. Proc Natl Acad Sci 112:13009–13014. https://doi.org/10.1073/pnas.1508170112
Feral CJW, Epstein HE, Otter L et al (2003) Carbon and nitrogen in the soil-plant system along rainfall and land-use gradients in southern Africa. J Arid Environ 54:327–343. https://doi.org/10.1006/jare.2002.1091
Fierer N, Jackson J (2005) Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol 71:4117. https://doi.org/10.1128/AEM.71.7.4117
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. https://doi.org/10.1890/05-1839
Finzi AC, Canham CD, Van Breemen N (1998) Canopy tree soil interactions within temperate forests: species effects on pH and cations. Ecol Appl 8:447–454. https://doi.org/10.2307/2641084
Ford DJ, Cookson WR, Adams MA, Grierson PF (2007) Role of soil drying in nitrogen mineralization and microbial community function in semi-arid grasslands of north-west Australia. Soil Biol Biochem 39:1557–1569. https://doi.org/10.1016/j.soilbio.2007.01.014
Fox J, Weisberg S, Adler D et al (2014) Package ‘car’ (Version 2.1-3)
Franzluebbers AJ, Haney RL, Hons FM, Zuberer DA (1996) Determination of microbial biomass and nitrogen mineralization following rewetting dried soil. Soil Sci Soc Am J 60:1133–1139
Frey SD, Elliott ET, Paustian K (1999) Bacterial and fungal abundance and biomass in conventional and no-tillage agroecosystems along two climatic gradients. Soil Biol Biochem 31:573–585. https://doi.org/10.1016/S0038-0717(98)00161-8
Gleeson DB, Müller C, Banerjee S et al (2010) Response of ammonia oxidizing archaea and bacteria to changing water filled pore space. Soil Biol Biochem 42:1888–1891. https://doi.org/10.1016/j.soilbio.2010.06.020
Göransson H, Godbold DL, Jones DL, Rousk J (2013) Bacterial growth and respiration responses upon rewetting dry forest soils: impact of drought-legacy. Soil Biol Biochem 57:477–486. https://doi.org/10.1016/j.soilbio.2012.08.031
Hanson CA, Allison SD, Bradford MA et al (2008) Fungal taxa target different carbon sources in forest soil. Ecosystems 11:1157–1167. https://doi.org/10.1007/s10021-008-9186-4
Hawkes CV, Kivlin SN, Rocca JD et al (2011) Fungal community responses to precipitation. Glob Chang Biol 17:1637–1645. https://doi.org/10.1111/j.1365-2486.2010.02327.x
Hu H, Zhang L, Dai Y et al (2013) pH-dependent distribution of soil ammonia oxidizers across a large geographical scale as revealed by high-throughput pyrosequencing. J Soils Sediments 13:1439–1449. https://doi.org/10.1007/s11368-013-0726-y
Hu H, Macdonald CA, Trivedi P et al (2015) Water addition regulates the metabolic activity of ammonia oxidizers responding to environmental perturbations in dry subhumid ecosystems. Environ Microbiol 17:444–461. https://doi.org/10.1111/1462-2920.12481
IPCC (2007) Climate change 2007: synthesis report. Contribution of working groups I, II and III to the fourth assessment report of the Intergovernmental Panel on Climate Change
Isobe K, Ohte N, Oda T et al (2015) Microbial regulation of nitrogen dynamics along the hillslope of a natural forest. Front Environ Sci 2:1–8. https://doi.org/10.3389/fenvs.2014.00063
Isobe K, Oka H, Watanabe T et al (2018) High soil microbial activity in the winter season enhances nitrogen cycling in a cool-temperate deciduous forest. Soil Biol Biochem 124:90–100. https://doi.org/10.1016/j.soilbio.2018.05.028
Iwaoka C, Imada S, Taniguchi T et al (2018) The impacts of soil fertility and salinity on soil nitrogen dynamics mediated by the soil microbial community beneath the halophytic shrub Tamarisk. Microb Ecol 75:985–996. https://doi.org/10.1007/s00248-017-1090-z
Kao-Kniffin J, Balser TC (2008) Soil fertility and the impact of exotic invasion on microbial communities in Hawaiian forests. Microb Ecol 56:55–63. https://doi.org/10.1007/s00248-007-9323-1
Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol 55:485–529. https://doi.org/10.1146/annurev.micro.55.1.485
Ladwig LM, Sinsabaugh RL, Collins SL, Thomey ML (2015) Soil enzyme responses to varying rainfall regimes in Chihuahuan Desert soils. Ecosphere 6:1–10. https://doi.org/10.1890/ES14-00258.1
Landesman WJ, Dighton J (2010) Response of soil microbial communities and the production of plant-available nitrogen to a two-year rainfall manipulation in the New Jersey Pinelands. Soil Biol Biochem 42:1751–1758. https://doi.org/10.1016/j.soilbio.2010.06.012
Langille M, Zaneveld J, Caporaso JG et al (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. https://doi.org/10.1038/nbt.2676
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120. https://doi.org/10.1128/AEM.00335-09
Lennon JT, Aanderud ZT, Lehmkuhl BK, Schoolmaster DR (2012) Mapping the niche space of soil microorganisms using taxonomy and traits. Ecology 93:1867–1879. https://doi.org/10.1890/11-1745.1
Li Z, Zheng FL, Liu WZ (2012) Spatiotemporal characteristics of reference evapotranspiration during 1961-2009 and its projected changes during 2011-2099 on the Loess Plateau of China. Agric For Meteorol. https://doi.org/10.1016/j.agrformet.2011.10.019
Li T, Ren B, Wang D, Liu G (2015) Spatial variation in the storages and age-related dynamics of forest carbon sequestration in different climate zones-evidence from black locust plantations on the loess plateau of China. PLoS One. https://doi.org/10.1371/journal.pone.0121862
Lindahl BD, Ihrmark K, Bogberg J et al (2007) Spatial separation of litter decomposition and mycorrhizal nitrogen uptake in a boreal forest. New Phytol 173:611–620. https://doi.org/10.1111/j.1469-8137.2006.01936.x
Maestre FT, Delgado-Baquerizo M, Jeffries TC et al (2015) Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc Natl Acad Sci 112:15684–15689. https://doi.org/10.1073/pnas.1516684112
Maharjan B, Venterea RT (2013) Nitrite intensity explains N management effects on N2O emissions in maize. Soil Biol Biochem 66:229–238. https://doi.org/10.1016/j.soilbio.2013.07.015
Marcos MS, Bertiller MB, Cisneros HS, Olivera NL (2016) Nitrification and ammonia-oxidizing bacteria shift in response to soil moisture and plant litter quality in arid soils from the Patagonian Monte. Pedobiologia (Jena) 59:1–10. https://doi.org/10.1016/j.pedobi.2015.11.002
Meier IC, Leuschner C (2014) Nutrient dynamics along a precipitation gradient in European beech forests. Biogeochemistry 120:51–69. https://doi.org/10.1007/s10533-014-9981-2
Mikha MM, Rice CW, Milliken GA (2005) Carbon and nitrogen mineralization as affected by drying and wetting cycles. Soil Biol Biochem 37:339–347. https://doi.org/10.1016/j.soilbio.2004.08.003
Moore JC, McCann K, Setälä H, De Ruiter PC (2003) Top-down is bottom-up: does predation in the rhizosphere regulate aboveground dynamics? Ecology 84:846–857. https://doi.org/10.1890/0012-9658(2003)084%5b0846:TIBDPI%5d2.0.CO;2
Nguyen NH, Song Z, Bates ST et al (2016) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248. https://doi.org/10.1016/j.funeco.2015.06.006
Norby RJ, Warren JM, Iversen CM et al (2010) CO2 enhancement of forest productivity constrained by limited nitrogen availability. Proc Natl Acad Sci 107:19368–19373. https://doi.org/10.1073/pnas.1006463107
Norman JS, Barrett JE (2014) Substrate and nutrient limitation of ammonia-oxidizing bacteria and archaea in temperate forest soil. Soil Biol Biochem 69:141–146. https://doi.org/10.1016/j.soilbio.2013.11.003
Ogaya R, Penuelas J (2007) Tree growth, mortality, and above-ground biomass accumulation in a holm oak forest under a five-year experimental field drought. Plant Ecol 189:291–299. https://doi.org/10.1007/s11258-006-9184-6
Oksanen AJ, Blanchet FG, Friendly M et al (2016) Package ‘vegan’ (Version 2.4-0)
Otsuki K, Yamanaka N, Du S et al (2005) Seasonal changes of forest ecosystem in an artificial forest of Robinia pseudoacacia in the Loess Plateau in China. J Agric Meteorol 60:613–616. https://doi.org/10.2480/agrmet.613
Phillips RP, Brzostek E, Midgley MG (2013) The mycorrhizal-associated nutrient economy: a new framework for predicting carbon—nutrient couplings in temperate forests. New Phytol 199:41–51. https://doi.org/10.1111/nph.12221
Phillips LA, Ward V, Jones MD (2014) Ectomycorrhizal fungi contribute to soil organic matter cycling in sub-boreal forests. ISME J 8:699–713. https://doi.org/10.1038/ismej.2013.195
Placella SA, Firestone MK (2013) Transcriptional response of nitrifying communities to wetting of dry soil. Appl Environ Microbiol 79:3294–3302. https://doi.org/10.1128/AEM.00404-13
Placella SA, Brodie EL, Firestone MK (2012) Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc Natl Acad Sci 109:10931–10936. https://doi.org/10.1073/pnas.1204306109
Prosser JI, Nicol GW (2008) Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 10:2931–2941. https://doi.org/10.1111/j.1462-2920.2008.01775.x
Qiu L, Zheng F, Yin R (2012) SWAT-based runoff and sediment simulation in a small watershed, the loessial hilly-gullied region of China: capabilities and challenges. Int J Sediment Res 27:226–234. https://doi.org/10.1016/S1001-6279(12)60030-4
Ren H, Xu Z, Isbell F et al (2017) Exacerbated nitrogen limitation ends transient stimulation of grassland productivity by increased precipitation. Ecol Monogr 87:457–469. https://doi.org/10.1002/ecm.1262
Rousk J, Bååth E, Brookes PC et al (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351. https://doi.org/10.1038/ismej.2010.58
Rustad LE, Campbell JL, Marion GM et al (2001) A meta-analysis of the response of soil respiration, net nitrogen mineralization, and aboveground plant growth to experimental ecosystem warming. Oecologia 126:543–562. https://doi.org/10.1007/s004420000544
Saetre P, Stark JM (2005) Microbial dynamics and carbon and nitrogen cycling following re-wetting of soils beneath two semi-arid plant species. Oecologia 142:247–260. https://doi.org/10.1007/s00442-004-1718-9
Schimel JP, Bilbrough C, Welker JM (2004) Increased snow depth affects microbial activity and nitrogen mineralization in two Arctic tundra communities. Soil Biol Biochem 36:217–227. https://doi.org/10.1016/j.soilbio.2003.09.008
Shimadzu (2013) 680°C combusion catalytic oxidation method measurement principles
Smith SE, Read DJ (2010) Mycorrhizal symbiosis. Academic Press, London
Smith RV, Doyle RM, Burns LC, Stevens RJ (1997) A model for nitrite accumulation in soils. Soil Biol Biochem 29:1241–1247. https://doi.org/10.1016/S0038-0717(97)00028-X
Smith JL, Halvorson JJ, Bolton H (2002) Soil properties and microbial activity across a 500 m elevation gradient in a semi-arid environment. Soil Biol Biochem 34:1749–1757. https://doi.org/10.1016/S0038-0717(02)00162-1
Ste-Marie C, Paré D (1999) Soil, pH and N availability effects on net nitrification in the forest floors of a range of boreal forest stands. Soil Biol Biochem 31:1579–1589. https://doi.org/10.1016/S0038-0717(99)00086-3
Suzuki MT, Giovannoni SJ (1996) Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes. Appl Environ Microbiol 62:2–8
Tateno R, Taniguchi T, Zhang J et al (2017) Net primary production, nitrogen cycling, biomass allocation, and resource use efficiency along a topographical soil water and nitrogen gradient in a semi-arid forest near an arid boundary. Plant Soil 420:209–222. https://doi.org/10.1007/s11104-017-3390-y
Taylor AE, Vajrala N, Giguere AT et al (2013) Use of aliphatic n-alkynes to discriminate soil nitrification activities of ammonia-oxidizing thaumarchaea and bacteria. Appl Environ Microbiol 79(21):6544–6551. https://doi.org/10.1128/AEM.01928-13
Toju H, Tanabe AS, Yamamoto S, Sato H (2012) High-coverage ITS primers for the DNA-based identification of Ascomycetes and Basidiomycetes in environmental samples. PLoS ONE. https://doi.org/10.1371/journal.pone.0040863
Tsunekawa A (2014) Knowledge and technology to save drylands: solutions to desertification, land degradation and drought. Maruzen Publishing (in Japanese)
Updegraff K, Pastor J, Bridgham SD, Johnston CA (1995) Environmental and substrate controls over carbon and nitrogen mineralization in northern wetlands. Ecol Appl 5:151–163. https://doi.org/10.2307/1942060
Venterea RT, Clough TJ, Coulter JA et al (2015) Ammonium sorption and ammonia inhibition of nitrite-oxidizing bacteria explain contrasting soil N2O production. Sci Rep. https://doi.org/10.1038/srep12153
Verhamme DT, Prosser JI, Nicol GW (2011) Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J 5:1067–1071. https://doi.org/10.1038/ismej.2010.191
Wan L, Zhang XP, Ma Q et al (2014) Spatiotemporal characteristics of precipitation and extreme events on the Loess Plateau of China between 1957 and 2009. Hydol Proccess 28:4971–4983. https://doi.org/10.1002/hyp.9951
Wang Y, Qian P (2009) Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE. https://doi.org/10.1371/journal.pone.0007401
Wang S, Wang Y, Feng X (2011) Quantitative analyses of ammonia-oxidizing Archaea and bacteria in the sediments of four nitrogen-rich wetlands in China. Appl Microbiol Biotechnol 90:779–787. https://doi.org/10.1007/s00253-011-3090-0
Wang X, Ye J, Perez PG et al (2013) The impact of organic farming on the soluble organic nitrogen pool in horticultural soil under open field and greenhouse conditions: a case study. Soil Sci Plant Nutr 59:237–248. https://doi.org/10.1080/00380768.2013.770722
Xiong J, Liu Y, Lin X et al (2012) Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ Microbiol 14:2457–2466. https://doi.org/10.1111/j.1462-2920.2012.02799.x
Xu Z, Wan S, Ren H et al (2012) Effects of water and nitrogen addition on species turnover in temperate grasslands in northern China. PLoS ONE. https://doi.org/10.1371/journal.pone.0039762
Yahdjian L, Sala OE (2010) Size of precipitation pulses controls nitrogen transformation and losses in an arid Patagonian ecosystem. Ecosystems 13:575–585. https://doi.org/10.1007/s10021-010-9341-6
Yu Z, Kraus TEC, Dahlgren RA et al (2002) Contribution of amino compounds to dissolved organic nitrogen in forest soils. Biogeochemistry 61:173–198. https://doi.org/10.1023/A:1020221528515
Zimmerman AE, Martiny AC, Allison SD (2013) Microdiversity of extracellular enzyme genes among sequenced prokaryotic genomes. ISME J 7:1187–1199. https://doi.org/10.1038/ismej.2012.176
Acknowledgements
We greatly thank to members of the Institute of Soil and Water Conservation of Chinese Academy of Science (CAS), the Arid Land Research Center (ALRC) of Tottori University, and the Field Science, Education and Research Center (FSERC) of Kyoto University, for cooperation and logistics in both of field survey and laboratory analysis. We also greatly thank Dr. Kazuo Isobe, Dr. Takahito Yoshioka, Dr. Naoko Tokuchi and Dr. Kazuya Kobayashi for helpful comments. This study was financially supported in part by JSPS-KAKENHI (Grant No.15H05113), Grant-in-Aid for JSPS Research Fellow (Grant No. 17J07686) and Fund of Joint Research Program of Arid Land Research Center, Tottori University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Steven Perakis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tatsumi, C., Taniguchi, T., Du, S. et al. The steps in the soil nitrogen transformation process vary along an aridity gradient via changes in the microbial community. Biogeochemistry 144, 15–29 (2019). https://doi.org/10.1007/s10533-019-00569-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10533-019-00569-2