Abstract
An increasing number of circRNAs have been found to be involved in the development of gastric cancer. However, the function of circ_0003789 in regulating gastric cancer progression is unclear. Here, we aimed to investigate the expression, function and molecular mechanism of circ_0003789 in gastric cancer pathogenesis. Circ_0003789, miR-429 and ZFP36 ring finger protein like 2 (ZFP36L2) mRNA were quantified by quantitative real-time polymerase chain reaction (qRT-PCR). Cell proliferation was illustrated by 5-Ethynyl-2′-deoxyuridine (Edu), cell counting kit-8 (CCK-8) and colony formation assays. Apoptosis was determined by flow cytometry. Protein level was detected by Western blotting assay. Xenograft assays were used for functional analysis of circ_0003789 in vivo. The relationship between miR-429 and circ_0003789 or ZFP36L2 was predicted by starbase3.0 online database and identified by dual luciferase reporter assay. The expression levels of circ_0003789 and ZFP36L2 were significantly upregulated in gastric cancer tissues and cells, while the expression of miR-429 was downregulated. Downregulation of circ_0003789 inhibited gastric cancer cell growth and invasion and promoted apoptosis in vitro. Circ_0003789 acted as a sponge of miR-429. Moreover, miR-429 silencing by miR-429 inhibitors attenuated the effects of circ_0003789 interference on cell growth, apoptosis and invasion. ZFP36L2 was targeted by miR-429, and the effects of miR-429 on cell growth, invasion and apoptosis were attenuated by ZFP36L2 overexpression. Circ_0003789 could enhance ZFP36L2 expression by interacting with miR-429. Silencing of circ_0003789 inhibited tumor growth in vivo. Circ_0003789 regulates tumor progression in gastric cancer through miR-429/ZFP36L2 axis. This finding implies that circ_0003789 may be a therapeutic target for gastric cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer, a common cancer of the digestive system, is the fifth most common malignant tumor and the fourth leading cause of cancer death (Sung et al. 2021). With the development of science and technology, the clinical diagnosis and treatment strategies of gastric cancer have been greatly improved. Nonetheless, the prognosis of some gastric cancer patients is still very poor because they were diagnosed at an advanced or metastatic stage (Allemani et al. 2015). Therefore, the molecular mechanisms underlying gastric carcinogenesis and gastric cancer progression still need to be explored.
Circular RNAs (circRNAs) are a unique class of RNA transcripts that are produced mainly by exons of the pre-mRNAs, which are characterized by the covalently closed-loop structure (Jeck and Sharpless 2014; Qu et al. 2015). CircRNAs have been found to be involved in regulating gene expression by functioning as miRNA sponges (Meng et al. 2017). CircRNAs have been implicated in almost all types of cancers, such as glioma (Ke et al. 2022), pancreatic cancer (Hu et al. 2022) and intrahepatic cholangiocarcinoma (Du et al. 2022). Importantly, the circRNA-miRNA-mRNA network is involved in a range of disease-related pathways and affects the progression and prognosis of cancer biology (He et al. 2022; Cui et al. 2018). As an example, circ_0042823 is reported to accelerate the development of laryngeal squamous cell carcinoma via regulating the miR-877-5p/FOXM1 axis (Wu et al. 2021). Circ-RanGAP1 can regulate VEGFA expression via sponging miR-877-3p to promote gastric cancer metastasis (Lu et al. 2020). Circ_0008673 sponges miR-153-3p to promote breast cancer progression by upregulating CFL2 expression (Zhang et al. 2022). However, many circRNAs are still poorly understood in gastric cancer.
Circ_0003789, located at chr2:122,514,815–122,520,660, is formed by back-splicing of exons 2–5 of TSN pre-mRNA, with a splice sequence length of 387 bp. Moreover, circ_0003789 is upregulated in gastric cancer, and circ_0003789 upregulation is positively associated with poor tumor differentiation, clinical advanced stage, and distal metastasis (Shao 2020). Nonetheless, the specific role and mechanism of circ_0003789 in gastric cancer progression is unclear.
In this paper, we sought to investigate the function of circ_0003789 in gastric cancer development. The current data demonstrated that circ_0003789 affects gastric cancer growth and apoptosis by regulating miR-429/ZFP36 ring finger protein like 2 (ZFP36L2) axis.
Materials and Methods
Specimen Collection
Sixty specimens including surgically resected gastric cancer tissues and corresponding paracancerous tissues were collected from gastric cancer patients at Xianning Central hospital. The tissue specimens were rapidly frozen in liquid nitrogen. The gastric cancer tissues were confirmed by pathological examination. The patients had not undergone surgery or radiotherapy before surgery, and all patients signed the written informed consent. The current study was reviewed and approved by the ethics committee of Xianning Central hospital.
Cell Culture and Transfection
Gastric cancer cell lines (AGS and SNU-1) and human normal gastric mucosal cells (GES-1) were commercially provided by EK-Bioscience (Shanghai, China) and cultured in RPMI-1640 medium (EK-Bioscience) containing 10% fetal bovine serum in standard conditions (5% CO2, 37 °C). Cells at logarithmic growth stage were taken for experiments.
For cell transfection, AGS and SNU-1 cells were adjusted to a density of 1.0 × 105 cells/mL. Transient transfection of cell lines was done according to the product instructions of Liposome Lipofectamine™ 6000 (Beyotime, Shanghai, China). Small interfering RNA (siRNA) against circ_0003789 (si-circ_0003789) and negative control siRNA (si-NC), miR-429 mimics (miR-429) and negative mimics control (miR-NC), miR-429 inhibitors (anti-miR-429) and inhibitors control (anti-miR-NC), and ZFP36L2 overexpression plasmid (ZFP36L2) and negative control plasmid (pcDNA) were transfected into gastric cancer cells.
Quantitative Reverse Transcription-PCR (qRT-PCR)
Total RNA was extracted with TRIzol reagent. Synthesis of first strand cDNA was conducted using AMV Reverse Transcriptase (Solarbio, Beijing, China), and then SYBR Premix Ex Taq II (TaKaRa, Dalian, China) was used for qRT-PCR assay. For relative expression, U6 (miRNA) or GAPDH (for circRNA/mRNA) was used as the internal reference, and data were analyzed by the 2−△△Ct method. The primers were synthesized by RiBoBio (Guangzhou, China) and were shown as below: circ_0003789 Forward: 5′-TGAGCCAGATCGGGAGAAAG-3′, Reverse: 5′-CTGATGGACCCCTTGCAGTA-3′; ZFP36L2 Forward: 5′-ATCAACTCCACGCGCTACAA-3′, Reverse: 5′-GGCAGAAGCCGATGGTATGA-3′; GAPDH Forward: 5′-GACAGTCAGCCGCATCTTCT-3′, Reverse: 5′-GCGCCCAATACGACCAAATC-3′; miR-429 Forward: 5′-GTATGAGTAATACTGTCTGGTAA-3′, Reverse: 5′-CTCAACTGGTGTCGTGGAG-3′; U6 Forward: 5′-CTCGCTTCGGCAGCACA-3′, Reverse: 5′-AACGCTTCACGAATTTGCGT-3′.
RNase R Treatment Assay
The extracted total RNA was incubated without or with RNase R (Seebio, Shanghai, China) for 30 min. Finally, circ_0003789 and GAPDH mRNA expression were determined by qRT-PCR.
5-Ethynyl-2′-Deoxyuridine (Edu) Staining
The EdU kit (RiboBio) was used to test DNA synthesis. Cells were seeded in 24-well plates. After transfection for 48 h, EdU solution (50 μM) was added into per well, followed by incubation for 2 h. After being fixed with paraformaldehyde (4%), cells were permeated using Triton-X-100 (0.5%). After being stained with Apollo reaction in the dark for 20 min, cells were then stained with DAPI for 10 min, followed by observation using a fluorescence microscope (Leica, Wetzlar, Germany).
Cell Counting Kit-8 (CCK-8) Assay
The cells were grown in a 96-well plate for 48 h. After that, CCK-8 solution (10 μL, Solarbio) was added to each well at the different time points and incubation was done for 2 h. The absorbance values at 450 nm were collected by a microplate reader. All experiments were repeated for 3 times. The average values were detected and the proliferation curves were plotted.
Colony Formation Assay
The indicated treated cells were placed in 6-well plates, and the medium was renewed every 3 days during incubation. Cells were cultured for approximately 12 days, stained with crystal violet (0.5%), and counted.
Flow Cytometry Analysis
After being resuspended in 500 μL binding buffer, the cells were dyed with Annexin V-FITC and PI (Sangon, Shanghai, China) for 15 min in a dark condition, followed by detection of the apoptosis rate using flow cytometer (BD Biosciences, San Jose, CA, USA).
Transwell Invasion Assay
Transfected cells were plated in Matrigel-coated chamber (8 µm pores, Corning, Corning, NY, USA) in non-serum medium and translocated toward 10%-serum growth media in the lower chamber for 24 h. Crystal violet (0.2%, Solarbio) was used to stain cells before the count of cells on the undersurface under a 100X microscope (Olympus, Tokyo, Japan).
Western Blot Analysis
Proteins were extracted according to the instructions of RIPA lysis buffer (Beyotime), and protein quantification was performed with the BCA kit (Beyotime). Ten micrograms of proteins was subjected to SDS-PAGE, followed by transferring onto the PVDF membranes using electro-transferring. The proteins were blocked with skim milk (5%, Beyotime) for 1–2 h, and the corresponding primary antibodies were added overnight at 4 °C: anti-Ki67 (ab197547, 1:1500), anti-PCNA (ab18197, 1:1000), anti-Bax (ab104156, 1:1500), anti-Bcl-2 (ab196495, 1:2000), anti-ZFP36L2 (ab70775, 1:1000), and anti-β-actin (ab8227, 1:2000), and followed by the probing with HRP-marked secondary antibody (ab205718, 1:2000) for 2 h. At last, the blots were developed with ECL reagent (Abcam, Cambridge, MA, USA) in a dark room to determine the grayscale values of each protein band. The antibodies used in this experiment were commercially provided by Abcam.
Dual-Luciferase Reporter Assay
The fragments of circ_0003789 or ZFP36L2 3′UTR containing the miR-429 binding sequence region or miss-matched seed region were synthesized and individually cloned into the psiCHECK2 vector (YouBia, Changsha, China) to produce the wild-type and mutant luciferase vectors (circ_0003789-WT, circ_0003789-MUT, ZFP36L2 3′UTR-WT, and ZFP36L2 3′UTR-MUT). The corresponding reporter vectors were cotransfected into gastric cancer cells with miR-429 mimics/miR-NC using Lipofectamine™ 6000. After 48 h, a dual-luciferase reporter assay kit (Yeasen, Shanghai, China) was employed for measuring the luciferase activity.
Xenograft Tumor Assay
The lentivirus-mediated (RiboBio) short hairpin RNA (shRNA) against circ_0003789 (sh-circ_0003789) or matched control (sh-NC) was used for stable transfection in AGS cells. Approximately 4 × 106 AGS cells (control or circ_0003789-downregulating) were injected into BALB/c nude mice (N = 12). All mice were fed in a pathogen-free environment. The volume of tumors was measured every week. The mice were sacrificed 5 weeks later, and tumor tissues were removed. The animal experiments were approved by the ethics committee of Xianning Central hospital.
Statistical Analysis
GraphPad Prism 7.0 software was used for statistical analysis, and the data were expressed as means ± standard deviation. A Student’s t test and one-way ANOVA were used for comparison between two groups and multiple groups, respectively. Expression correlation was analyzed by Pearson’s correlation coefficient analysis. The significance was determined when P < 0.05.
Results
Circ_0003789 was Elevated in Gastric Cancer
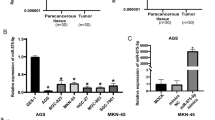
We firstly explored whether circ_0003789 was dysregulated in gastric cancer by qRT-PCR. The level of circ_0003789 was higher in gastric cancer than that in adjacent normal tissues (Fig. 1A). Similarly, the expression of circ_0003789 was elevated in gastric cancer cells (AGS and SNU-1) compared with GES-1 cells (Fig. 1B). In addition, RNase R treatment assay was used to determine the stability of circ_0003789, and the results showed that circ_0003789 was resistant to RNase R digestion (Fig. 1C, D). These data suggested that circ_0003789 might play a key role in gastric cancer progression.
Circ_0003789 was elevated in gastric cancer. A Circ_0003789 expression in gastric cancer tissues and adjacent normal tissues was detected by qRT-PCR. B Circ_0003789 expression in gastric cancer cell lines was measured by qRT-PCR. C, D Circ_0007841 expression was analyzed in AGS and SNU-1 cells with RNase R treatment. *P < 0.05
Circ_0003789 Knockdown Suppressed the Progression of Gastric Cancer In Vitro
To verify the function of circ_0003789 in gastric cancer, we silenced circ_0003789 expression in AGS and SNU-1 cells by transfection of si-circ_0003789. qRT-PCR verified that transfection of si-circ_0003789 resulted in a significant decrease in circ_0003789 expression, indicating that circ_0003789 was successfully disrupted (Fig. 2A). Edu and CCK8 results showed that silencing of circ_0003789 inhibited the proliferative capacity of gastric cancer cells (Fig. 2B–D). Also, knockdown of circ_0003789 reduced the number of colonies in AGS and SNU-1 cells compared to control group (Fig. 2E). Ki67 is a DNA binding protein and is considered to be one of the best clinical markers for cell proliferation (Yang et al. 2018). PCNA is known as a molecular marker of proliferation (Wang 2014). The protein levels of Ki67 and PCNA were reduced by knockdown of circ_0003789 (Fig. 2F). The effect of si-circ_0003789 on cell apoptosis was detected by flow cytometry. Apoptosis of gastric cancer cells was significantly increased by si-circ_0003789 compared to the si-NC group (Fig. 2G). Western blot assay was used to detect the expression levels of pro-apoptotic protein Bax and anti-apoptotic protein Bcl-2 in AGS and SNU-1 cells. Circ_0003789 silencing resulted in a significant upregulation in Bax expression and a significant downregulation in Bcl-2 expression, reinforcing that circ_0003789 interference promoted apoptosis in gastric cancer cells (Fig. 2H). Additionally, cell invasion was hindered by circ_0003789 interference (Fig. 2I). Overall, interfering circ_0003789 suppressed the malignant phenotypes of gastric cancer cells.
Circ_0003789 knockdown suppressed the progression of gastric cancer cells in vitro. A The knockdown efficiency of si-circ_0003789 was detected by qRT-PCR in AGS and SNU-1 cells. B–D Cell proliferation was evaluated by Edu and CCK-8 staining assays in gastric cancer cells transfected with si-circ_0003789. E Number of colonies was examined by colony formation assay in gastric cancer cells transfected with si-circ_0003789. F Ki67 and PCNA protein levels were measured by Western blotting in gastric cancer cells transfected with si-circ_0003789. G Apoptosis was determined by flow cytometry in gastric cancer cells transfected with si-circ_0003789. H Bax and Bcl-2 protein levels were detected by Western blotting in gastric cancer cells transfected with si-circ_0003789. I Cell invasion was detected by transwell assay. *P < 0.05
Circ_0003789 Acted as a Sponge of miR-429
To explore the potential mechanism of circ_0003789 in regulating the malignant phenotypes of gastric cancer, we predicted its targeted miRNAs. Among these candidates, we selected five miRNAs that play essential roles in gastric cancer development and found that miR-429 was preferentially enriched by biotinylated circ_0003789 probe (Supplementary Fig. 1). StarBase3.0 database predicted that miR-429 was partially complementary bound to circ_0003789 sequence (Fig. 3A). To verify the targeting relationship between circ_0003789 and miR-429, we carried out luciferase assays. The overexpression efficiency of miR-429 mimics was first examined by qRT-PCR, and results showed that the expression of miR-429 was dramatically increased by transfection of miR-429 mimics in gastric cancer cells (Fig. 3B). Dual luciferase reporter assays showed that miR-429 overexpression significantly inhibited the luciferase activity of circ_0003789-WT group but had no effect on luciferase activity of the circ_0003789-MUT group (Fig. 3C, D). Moreover, miR-429 expression was significantly reduced in gastric cancer tissues compared to adjacent normal tissues (Fig. 3E). Similarly, miR-429 expression was significantly decreased in gastric cancer cells (Fig. 3F). In addition, Pearson's correlation analysis showed that the expression of circ_0003789 was negatively correlated with miR-429 expression in gastric cancer tissues (Fig. 3G). The above results indicated that circ_0003789 acted as a sponge for miR-429.
Circ_0003789 acted as a sponge of miR-429. A Predictive target sites of circ_0003789 and miR-429 were provided by StarBase3.0. B The level of miR-429 was examined by qRT-PCR in AGS and SNU-1 cells transfected with miR-NC or miR-429. C, D The associated relationship between circ_0003789 and miR-429 was illustrated by dual-luciferase reporter assay. E, F miR-429 expression in gastric cancer tissues and cell lines was detected by qRT-PCR. G Correlation between circ_0003789 and miR-429 expression was analyzed in gastric cancer tissues. *P < 0.05
Circ_0003789 Knockdown Suppressed the Progression of Gastric Cancer by Targeting miR-429
To investigate the function of circ_0003789/miR-429 axis in gastric cancer progression, si-circ_0003789 and miR-429 inhibitors were co-transfected into AGS and SNU-1 cells. qRT-PCR verified that miR-429 inhibitors significantly reduced miR-429 expression in cells (Fig. 4A). MiR-429 inhibitors attenuated the inhibitorsy effect of circ_0003789 silencing on gastric cancer cell proliferation, colony formation, Ki67 and PCNA protein levels (Fig. 4B–F). In addition, the promotion effect of circ_0003789 interference on apoptosis was restored by transfection of miR-429 inhibitors (Fig. 4G). Western blot results showed that the impact of circ_0003789 knockdown on Bax and Bcl-2 levels was reversed by miR-429 inhibitors in AGS and SNU-1 cells (Fig. 4H). Additionally, reduction of miR-429 abated circ_0003789 interference-mediated invasion suppression in gastric cancer cells (Fig. 4I). Taken together, circ_0003789 knockdown suppressed the malignant phenotypes of gastric cancer cells by targeting miR-429.
Circ_0003789 knockdown suppressed the progression of gastric cancer cells by targeting miR-429. A The effect of miR-429 inhibitors on miR-429 expression was presented by qRT-PCR. B–H AGS and SNU-1 cells were transfected with si-NC, si-circ_0003789, si-circ_0003789 + anti-miR-NC, or si-circ_0003789 + anti-miR-429. B–D Cell proliferation was evaluated by Edu and CCK-8 staining assays. E Number of colonies was examined by colony formation assay. F Ki67 and PCNA expression levels were measured by Western blotting. G Cell apoptosis was determined via flow cytometry. H Bax and Bcl-2 expression was detected via Western blotting. I Cell invasion was detected by transwell assay. *P < 0.05
MiR-429 Directly Targeted ZFP36L2
Next, we identified the downstream effectors of miR-429 in regulating growth and apoptosis of gastric cancer cells. StarBase3.0 database showed that miR-429 possessed the binding site of ZFP36L2 mRNA (Fig. 5A). Dual luciferase reporter assays showed that increased miR-429 expression significantly inhibited the luciferase activity of ZFP36L2 3'UTR-WT but had no effect on the luciferase activity of ZFP36L2 3′UTR-MUT (Fig. 5B, C). ZFP36L2 mRNA and protein levels were significantly decreased in gastric cancer cells transfected with miR-429 mimics, while they were significantly increased in gastric cancer cells transfected with miR-429 inhibitors (Fig. 5D, E). In addition, ZFP36L2 mRNA and protein levels were significantly increased in gastric cancer tissues compared to adjacent normal tissues (Fig. 5F, G). Similarly, ZFP36L2 mRNA and protein levels were significantly increased in gastric cancer cells (Fig. 5H, I). Pearson’s correlation analysis showed a negative expression correlation between miR-429 and ZFP36L2 in gastric cancer tissues (Fig. 5J). The above results suggested that miR-429 directly targeted ZFP36L2.
MiR-429 directly bound to ZFP36L2. A The putative binding sites between miR-429 and ZFP36L2 were predicted by StarBase3.0 online database. B, C The interaction between miR-429 and ZFP36L2 was detected by luciferase reporter assay. D, E The impacts of miR-429 mimics or inhibitors on the mRNA and protein expression of ZFP36L2 were determined by qRT-PCR and Western blot, respectively. F–I ZFP36L2 mRNA and protein levels in gastric cancer tissues and cell lines were detected by qRT-PCR and Western blot assays. J Pearson’s correlation coefficient was used for correlation analysis between miR-429 and ZFP36L2. *P < 0.05
ZFP36L2 Could Reverse the Effects of miR-429 on Growth and Apoptosis of Gastric Cancer Cells
To investigate the function of miR-429/ZFP36L2 axis in gastric cancer progression, miR-429 mimics and ZFP36L2 overexpression plasmids were co-transfected into AGS and SNU-1 cells. The transfection efficiency of ZFP36L2 overexpression plasmid in gastric cancer cells was verified by qRT-PCR and Western blot (Fig. 6A, B). Overexpression of miR-429 inhibited AGS and SNU-1 cell proliferation, colony number, Ki67 and PCNA protein levels, which were reversed by ZFP36L2 re-expression (Fig. 6C–G). In addition, miR-429 mimics significantly promoted apoptosis, and the effect was restored by ZFP36L2 overexpression (Fig. 6H). Meanwhile, miR-429 mimics-mediated increase in Bax expression and decrease in Bcl-2 expression were counteracted by overexpression of ZFP36L2 (Fig. 6I). Furthermore, miR-429 mimics significantly impeded cell invasion, while ZFP36L2 re-expression abrogated this effect (Fig. 6J). The above results suggested that overexpression of miR-429 inhibited the development of gastric cancer by targeting ZFP36L2.
ZFP36L2 could reverse the effects of miR-429 on the biological functions in gastric cancer cells. A, B The overexpression efficiency of ZFP36L2 was determined by qRT-PCR and western blot assays. C–I AGS and SNU-1 cells were transfected with miR-NC, miR-429, miR-429 + pcDNA, or miR-429 + ZFP36L2. C–E Cell proliferation was evaluated by Edu and CCK-8 staining assays. F Number of colonies was examined by colony formation assay. G Ki67 and PCNA protein levels were measured by Western blotting assay. H Apoptosis was determined via flow cytometry. I Bax and Bcl-2 protein levels were detected via Western blotting assay. J Cell invasion was detected by transwell assay. *P < 0.05
Circ_0003789 Sponged miR-429 to Regulate ZFP36L2 Expression in Gastric Cancer Cells
We have demonstrated that circ_0003789 acts as a sponge for miR-429 and miR-429 directly targets ZFP36L2. Therefore, we next further explored whether circ_0003789 regulates ZFP36L2 expression by sponging miR-429. Pearson’s correlation analysis showed that in gastric cancer tissues, circ_0003789 expression was positively correlated with ZFP36L2 expression (Fig. 7A). Silencing of circ_0003789 reduced ZFP36L2 mRNA and protein levels, which were reversed by the addition of miR-429 inhibitors (Fig. 7B, C). This result suggested that circ_0003789 regulated ZFP36L2 expression by sponging miR-429.
Circ_0003789 sponged miR-429 to regulate ZFP36L2 expression in gastric cancer cells. A The linear association between ZFP36L2 and circ_0003789 levels in gastric cancer tissues was analyzed. B, C The qRT-PCR and Western blotting were adopted to analyze the ZFP36L2 mRNA and protein levels in gastric cancer cells transfected with si-NC, si-circ_0003789, si-circ_0003789 + anti-miR-NC or si-circ_0003789 + anti-miR-429. *P < 0.05
Circ_0003789 Knockdown Inhibited the Growth of Xenograft Tumors In Vivo
To determine whether circ_0003789 silencing has an inhibitory effect on tumor growth, we established a xenograft tumor model in nude mice. The results showed that tumor volume and weight were significantly suppressed in the sh-circ_0003789 group compared to the sh-NC group (Fig. 8A, B). In addition, qRT-PCR results showed that circ_0003789 silencing resulted in a significant decrease in circ_0003789 level and an increase in miR-429 expression (Fig. 8C, D). Knockdown of circ_0003789 decreased ZFP36L2 mRNA and protein levels in tumor tissues (Fig. 8E, F). Taken together, knockdown of circ_0003789 could inhibit tumor growth in vivo by upregulating miR-429 and downregulating ZFP36L2.
Discussion
With the advances in high-throughput sequencing and biology, many circRNAs have been shown to be aberrantly expressed in cancer, playing an integral oncogenic or tumor-suppressing role in tumorigenesis and tumor progression (Li et al. 2020; Wang et al. 2021). For example, circ_0046264 acts as a potential biomarker for lung cancer diagnosis and prognosis, and its overexpression promotes lung cancer cell proliferation and invasive ability (Liu et al. 2020). Circ-ERBB2 promotes the Warburg effect and triple-negative breast cancer development via miR-136-5p/PDK4 pathway (Huang et al. 2021). Circ_0005273 increases pancreatic cancer cell proliferative and migratory abilities by activating KLF12 and predicts lymphatic metastasis, distant metastasis and prognosis in pancreatic cancer patients (Hou and Li 2020). Consistent with previous work (Shao et al. 2017), our data indicated that circ_0003789 expression was upregulated in gastric cancer. Functional studies showed that silencing of circ_0003789 repressed cell growth and increased apoptosis in vitro, as well as hindered tumor growth in vivo. Furthermore, we revealed that the circ_0003789/miR-429/ZFP36L2 axis was associated with gastric cancer development.
Previous reports have shown that miR-429 acts as a tumor-suppressing miRNA to participate in cancer cell biological behaviors, such as apoptosis, stemness, proliferation and metastasis (Liu and Shen 2020; Wang et al. 2019). For example, miR-429 is down-regulated and impedes cell proliferation and metastasis in ovarian cancer (Xu et al. 2021), oral squamous cell cancer (Sun et al. 2021) and breast cancer (Zhang et al. 2020). Here, miR-429 was also found to be lowly expressed in gastric cancer cells and tissues. In addition, miR-429 mimics inhibited cell growth and induced apoptosis of gastric cancer cells. Furthermore, miR-429 level was found to be negatively correlated with circ_0003789 level in gastric cancer, and miR-429 inhibition attenuated the effects of circ_0003789 silencing on cell growth and apoptosis. These results indicated that miR-429 was involved in circ_0003789 silencing-mediated antitumor effects.
Next, ZFP36L2 was validated as a downstream target for miR-429. ZFP36L2 has established a potent oncogenic role in various cancers, such as pancreatic dual adenocarcinoma and acute myeloid leukemia (Yonemori et al. 2017; Liu et al. 2018). Moreover, ZFP36L2 has been implicated in gastric cancer pathogenesis (Xing et al. 2019). Our data also suggested that miR-429 regulated gastric cancer progression by targeting ZFP36L2. Importantly, our data established that circ_0003789 could modulate the expression of ZFP36L2 via sponging miR-429.
In summary, our study confirmed that circ_0003789 regulated gastric cancer cell growth and apoptosis. Moreover, circ_0032821 mediated gastric cancer progression via the regulation of miR-429/ZFP36L2 axis. This present research may uncover a potential biomarker and target for gastric cancer diagnosis and treatment.
Data Availability
Not applicable.
References
Allemani C et al (2015) Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). The Lancet 385(9972):977–1010
Cui X et al (2018) Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer 17(1):123
Du J et al (2022) CircNFIB inhibits tumor growth and metastasis through suppressing MEK1/ERK signaling in intrahepatic cholangiocarcinoma. Mol Cancer 21(1):18
He C et al (2022) Graph convolutional network approach to discovering disease-related circRNA-miRNA-mRNA axes. Methods 198:45–55
Hou YS, Li X (2020) Circ_0005273 induces the aggravation of pancreatic cancer by targeting KLF12. Eur Rev Med Pharmacol Sci 24(22):11578–11586
Hu C et al (2022) circFARP1 enables cancer-associated fibroblasts to promote gemcitabine resistance in pancreatic cancer via the LIF/STAT3 axis. Mol Cancer 21(1):24
Huang Y et al (2021) Circular RNA circ-ERBB2 elevates the warburg effect and facilitates triple-negative breast cancer growth by the MicroRNA 136–5p/pyruvate dehydrogenase kinase 4 axis. Mol Cell Biol 41(10):e0060920
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32(5):453–461
Ke Y et al (2022) Hsa_circ_0076931 suppresses malignant biological properties, down-regulates miR-6760-3p through direct binding, and up-regulates CCBE1 in glioma. Biosci Rep 42(1):BSR20211895
Li J et al (2020) Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer 6(4):319–336
Liu X, Shen Z (2020) LncRNA TMPO-AS1 aggravates the development of hepatocellular carcinoma via miR-429/GOT1 axis. Am J Med Sci 360(6):711–720
Liu J et al (2018) ZFP36L2, a novel AML1 target gene, induces AML cells apoptosis and inhibits cell proliferation. Leuk Res 68:15–21
Liu ZH et al (2020) Correlations of hsa_circ_0046264 expression with onset, pathological stage and chemotherapy resistance of lung cancer. Eur Rev Med Pharmacol Sci 24(18):9511–9521
Lu J et al (2020) Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett 471:38–48
Meng S et al (2017) CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 16(1):94
Qu S et al (2015) Circular RNA: a new star of noncoding RNAs. Cancer Lett 365(2):141–148
Shao Y et al (2017) Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med 6(6):1173–1180
Shao Z et al (2020) Circ_0003789 Facilitates gastric cancer progression by inducing the epithelial-mesenchymal transition through the Wnt/beta-catenin signaling pathway. Cancer Biother Radiopharm
Sun B et al (2021) LncRNA LINC01303 promotes the progression of oral squamous cell carcinomas via the miR-429/ZEB1/EMT axis. J Oncol 2021:7974012
Sung H et al (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249
Wang SC (2014) PCNA: a silent housekeeper or a potential therapeutic target? Trends Pharmacol Sci 35(4):178–186
Wang Z et al (2019) miR-429 suppresses cell proliferation, migration and invasion in nasopharyngeal carcinoma by downregulation of TLN1. Cancer Cell Int 19:115
Wang Y, Chen H, Wei X (2021) Circ_0007142 downregulates miR-874-3p-mediated GDPD5 on colorectal cancer cells. Eur J Clin Invest 51(7):e13541
Wu T et al (2021) Hsa_circ_0042823 accelerates cancer progression via miR-877-5p/FOXM1 axis in laryngeal squamous cell carcinoma. Ann Med 53(1):960–970
Xing R et al (2019) Whole-genome sequencing reveals novel tandem-duplication hotspots and a prognostic mutational signature in gastric cancer. Nat Commun 10(1):2037
Xu H, Wang L, Jiang X (2021) Silencing of lncRNA DLEU1 inhibits tumorigenesis of ovarian cancer via regulating miR-429/TFAP2A axis. Mol Cell Biochem 476(2):1051–1061
Yang C et al (2018) Ki67 targeted strategies for cancer therapy. Clin Transl Oncol 20(5):570–575
Yonemori K et al (2017) ZFP36L2 promotes cancer cell aggressiveness and is regulated by antitumor microRNA-375 in pancreatic ductal adenocarcinoma. Cancer Sci 108(1):124–135
Zhang L et al (2020) MiR-429 suppresses proliferation and invasion of breast cancer via inhibiting the Wnt/beta-catenin signaling pathway. Thorac Cancer 11(11):3126–3138
Zhang L et al (2022) Circ_0008673 regulates breast cancer malignancy by miR-153-3p/CFL2 axis. Arch Gynecol Obstet 305(1):223–232
Acknowledgements
None.
Funding
None.
Author information
Authors and Affiliations
Contributions
YJ designed and performed the research; NC and SZ analyzed the data; LW and YJ wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Coflict of interest
The authors declare no competing interests.
Ethical Approval and Consent to Participate
Written informed consents were obtained from all participants and this study was permitted by the Ethics Committee of Xianning Central hospital, The First Affiliated Hospital of Hubei University of Science and Technology.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10528_2023_10535_MOESM1_ESM.tif
Supplementary file1 Supplementary Figure 1. Screen of miR-429. (A and B) The qRT-PCR was used to detect the enrichment levels of several miRNAs following the incubation of circ_0003789 probe or control probe. *P < 0.05 (TIF 324 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wan, L., Jia, Y., Chen, N. et al. Circ_0003789 Knockdown Inhibits Tumor Progression by miR-429/ZFP36L2 Axis in Gastric Cancer. Biochem Genet 62, 2504–2521 (2024). https://doi.org/10.1007/s10528-023-10535-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10535-1