Abstract
Circular RNAs (circRNAs) are a class of novel RNAs, and aberrant expression of circRNAs has been implicated in human diseases, including gastric cancer (GC). This study aimed to identify the mechanism of circRNA_0043691 in regulating the progression of GC. GSE141977 was downloaded from Gene Expression Omibus (http://www.ncbi.nlm.nih.gov/geo/). Differentially expressed circRNAs were obtained by R software. The expression levels of circRNA_0043691 in GC tissue and normal tissue were identified by quantitative real-time polymerase chain reaction (qRT-PCR). Knockdown of circRNA_0043691 was then constructed and verified by qRT-PCR. Cell viability, migration, and invasion capacity were determined by Cell Counting Kit-8 assay, Transwell migration, and invasion, respectively. Next, knockdown of miR-873-3p was constructed and co-cultured with circRNA_0043691 knockdown to identify whether knockdown of miR-873-3p could attenuate the circRNA_0043691 knockdown on GC cells proliferation, migration, and invasion. The relationship between miR-873-3p and circRNA_0043691 or GART was predicted by bioinformatics tools and verified by dual-luciferase reporter. A total of 211 circRNAs were significantly differentially expressed, including 143 remarkably downregulated circRNAs and 68 significantly upregulated circRNAs. CircRNA_0043691 was upregulated in GC tissue. Knockdown of circRNA_0043691 decreased cell viability, migration, and invasion in GC cells. CircRNA_0043691 has potential putative binding sites with miR-873-3p. Moreover, CircRNA_0043691 positively regulated GART expression by sponging miR-873-3p. Furthermore, knockdown of miR-591 could partially attenuate the si-circRNA_0043691 on the GART expression. GART was upregulated in GC tissue and knockdown of GART could inhibit GC cells proliferation and invasion. Knockdown of circRNA_0043691 delayed the progression of GC via modulating the miR-873-3p-GART axis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Gastric cancer (GC), a prevalent and malignant tumor of the stomach, is the third leading cause of cancer-related mortality (Tan 2019; Orman and Cayci 2019). The progression of GC is a multistep and multifactorial process involving genetic variation, geographic location, life style, and genetic background (Vohlonen et al. 2017; Venerito et al. 2016). Treatment strategies for GC mainly include surgery, radiation, and chemotherapy. However, the survival rate remains unsatisfactory due to the pathogenesis of GC that remains poorly understood (Re 2018). Therefore, a deeper understanding of the mechanisms involved in GC progression is urgently needed (Correa 2013; Ang and Fock 2014).
Circular RNAs (circRNAs) are a novel competing endogenous noncoding RNA (ncRNAs) that participated into cancer development, progression, and metastasis (Feng et al. 2019). Previous research studies have demonstrated that several circRNAs are dysregulated in GC, including CircRNA_100269 (Zhang et al. 2017a), circ-HuR (Yang et al. 2019), CircRNA_0043691 (Yu et al. 2020), and circNHSL1 (Zhu et al. 2019a). Owing to the development of high-throughput sequencing, many more circRNAs are involved in the progression of oncogenesis by regulating microRNAs (miRNAs). Zhang et al. (2017a) found that circRNA_100269 is downregulated in GC and suppresses tumor cell growth by targeting miR-630. Yang et al. (2019) found that circ-HuR suppresses HuR expression and GC progression by inhibiting CNBP transactivation. Yu et al. (2020) revealed that circRNA_0043691 sponges miR-873-3p to promote proliferation and metastasis of GC by upregulating GART expression. Zhu et al. (2019a) found that circular RNA circNHSL1 promotes GC progression through the miR-1306-3p/SIX1/vimentin axis.
The Gene Expression Omnibus (GEO) dataset was available from the public (Toro-Domínguez et al. 2019). In this study, we firstly performed bioinformatic analysis of GSE141977 and found that circRNA_0043691 was upregulated in GC tissue. However, the molecular function of circRNA_0043691 in GC remains largely unknown.
MicroRNAs have emerged as key post-transcriptional regulators of gene expression, directly involved in human cancers (Zhang et al. 2019a). Many previous studies suggested that miRNAs may be identified as sponge and finally regulate mRNA expression (Qi et al. 2018). CircRNAs sharing target sites for one or more common miRNAs compete with each other for the limited pool of cellular miRNAs and thus affect the competing RNA’s level, a phenomenon known as Competing endogenous RNAs (ceRNA) effect (Zhang et al. 2019b). Deng et al. (2021) revealed that circTP63 enhances estrogen receptor-positive breast cancer progression and malignant behaviors through the miR-873-3p/FOXM1 axis. Zhu et al. (2019b) found that miR-873-5p inhibits the progression of colon cancer via repression of tumor suppressor candidate 3/AKT signaling. However, the role of miR-873-3p in GC cancer was unknown.
To evaluate the functional roles of circRNA_0043691 in GC progression, we performed loss-of-function and gain-of-function assays. Our findings may give insight into understanding the mechanism of GC pathogenesis and provide new biomarkers for clinical diagnosis.
Materials and methods
Bioinformatic analysis
GSE141977 was downloaded from Gene Expression Omibus (http://www.ncbi.nlm.nih.gov/geo/). This gene expression data set contained 3 gene expression profiles of gastric cancer (GC) and 3 gene expression profiles of adjacent normal tissue (NT), all of which were included in the analysis. The raw data were normalized using Limma package from Bioconductor as described previously (Wu et al. 2021). Differential expression analysis was conducted using the R limma package. The threshold cutoffs to identify significant differentially expressed circRNAs between GC and NT were False Discovery Rate (FDR)-adjusted P value (q value) < 0.05 and absolute log-fold change (log FC) > 1. A heatmap was generated using the “pheatmap” package of the R software. Using a bioinformatics database (Circular RNA Interactome, https://circinteractome.nia.nih.gov/), we found several target miRNAs that had potential binding sites with circRNA_0043691. The commonly targeted miRNAs were obtained between Circular RNA Interactome, TargetScan, and miRanda using a Venn diagram overlap analysis (Bardou et al. 2014). Target genes of miR-873-3p were predicted by Targetscan (http://www.targetscan.org/vert_72/), miRanda (http://www.microrna.org/), and miRDB (http://mirdb.org/) databases. The commonly targeted mRNAs of miR-873-3p were performed by using a Venn diagram overlap analysis (Bardou et al. 2014).
GC tissue samples
Primary GC samples, matched adjacent tumor samples, and normal tissue samples were obtained from 11 patients with GC undergoing radical gastrectomy at the Shaoxing People’s Hospital between August 2018 and August 2019. All the tissues were diagnosed by pathological examination by two independent reviewers. The present study was approved by the Ethical Review Board of the Shaoxing People’s Hospital and written informed consent was obtained from all patients.
Cell culture
AGS and SGC7901 cells were purchased from ATCC (American Type Culture Collection, Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone, Logan, UT, USA) including 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA) and with 1% penicillin (Gibco, Carlsbad, CA, USA) at 37 °C, 5% CO2 incubator.
Cell proliferation assay
Cells were seeded out in 96-well plates at 2.5 × 104/ml concentration (2500 cells/well) 48 h post transfection. Then, 10 μl of CCK8 reagent (Solarbio, Beijing, China) was added, and the absorbance of each well was determined by endpoint method at 450 nm after 2 h. The cell viability was normalized to control group.
qRT-PCR
Total RNA was extracted using the Bacterial RNA kit (Omega, USA). Reverse transcription was conducted using the PrimeScript RT reagent kit (TaKaRa, Dalian, China). qRT-PCR was performed using the SYBR Green PCR kit (Takara Biotechnology, Takara, Dalian, China). Primers for circRNA_0043691 synthesis were 5′-CTGGGAGACTCTATCGGGGA-3′(F) and 5′-TTCTCACACGCAGAGCTGTT-3′(R), respectively. For GAPDH it was 5′-CGCTCTCTGCTCCTCCTGTTC-3′ (F), 5′-ATCCGTTGACTCCGACCTT CAC-3′(R), respectively. For miR-873-3p, it was 5′-TTTGTGTGCATTTGCAGGAACT-3′ (F), 5′-GAAGATTTGTGGGTGTTCCCG-3′ (R).
For U6 it was 5′-GCTTCGGCAGCACATATACTAAAAT-3′ (F) and 5′-CGCTTCACGAATTTGCGTGTCAT-3′ (R), respectively. Real-time PCR was performed to determine relative gene expression using the 2−ΔΔCt method, and experiments were performed in triplicate.
RNAi and transfection
The siRNA of circRNA_0043691 corresponding negative control was purchased from GenePharma (Shanghai, China), and siRNA transfection was carried out using Lipofectamine 2000 (Thermo Fishier, Carlsbad, CA, USA) according to the manufacturer's protocols. The siRNA sequences for si-circRNA_0043691 were indicated as 5′- GACAAGGGTTTTCTCCAGGGA-3′.
Transwell invasion and migration assay
For cell invasion assay, 80 μl serum-free DMEM-diluted Matrigel (BD, San Jose, CA, USA) was added to the Transwell filter and incubated at 37 °C for 2 h to form a matrix gel. The opposite side of the upper chambers was coated with 0.2% gelatin. Then, 200 μl cell suspension (containing 6 × 105 cells) with serum-free medium was seeded in the upper chamber while the lower chamber was covered with 500 ul medium supplemented with 20% fetal calf serum. After 24 h the cells were investigated and photographed.
Dual-luciferase reporter assay
Dual-luciferase reporter assays were performed by co-transfection in the indicated cell lines with pmir-GLO-NT5C3L or pmir-GLO-NT5C3L-Mut vectors, and miR-873 mimics or miR-NC. Renilla relative luciferase activity was measured 48 h post transfection using a dual-luciferase reporter assay system according to the manufacturer’s protocol (Promega Corporation, Madison, WI, USA). Relative luciferase activity was normalized to the Renilla luciferase internal control.
Western blotting
After the cells were harvested, total cell protein was extracted with RIPA lysate containing PMSF. The protein concentration was quantified with BCA kit (Solarbio, Beijing, China). Samples were boiled in SDS–polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, and proteins were separated by 10% SDS-PAGE. Gels were transferred onto PVDF membranes (Millipore, Billerica, MA, USA). After blocking with 5% non-fat milk at room temperature for 2 h, the membranes were immunoblotted with primary antibody (anti-Keap1, 1:1000, Cell Signaling Technology, 4678S; anti-STIM1, 1:1000, Cell Signaling Technology, 5668S). Membranes were subsequently incubated with the horseradish peroxidase-conjugated goat anti-rabbit second antibody (Invitrogen) at a dilution of 1:5000 for 2 h at room temperature.
Statistical analysis
All data from the present study are shown as the mean ± SD. The difference between two groups was calculated using the student’s t test, and the differences among three groups were analyzed by one-way ANOVA analysis followed by Tukey’ analyses. P < 0.05 indicated statistically significant difference.
Results
Bioinformatic analysis of GSE141977
After normalization, the medians became consistent and were at an identical level, suggesting that the normalization process is valid, and the normalized data may be used for additional analysis (Fig. 1).
A total of 211 circRNAs were significantly differentially expressed, including 143 remarkably downregulated circRNAs and 68 significantly upregulated circRNAs visualized by volcano plots (Fig. 2) and a cluster heatmap (Fig. 3). We selected circRNA_0043691, the first upregulated circRNA, for further study. Then, we explored the structure of circRNA_0043691. CircRNA_0043691 was a circRNA with 201 nt and consisted from exon 7 (Fig. 4).
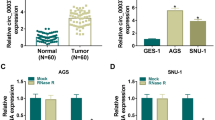
CircRNA_0043691 was upregulated in GC
We firstly detected circRNA_0043691 expression in 50 paired GC samples and adjacent normal tissues by RT-qPCR. The qRT-PCR analysis using the 50 paired GC samples and confirmed that circRNA_0043691 mRNA expression levels were significantly upregulated in GC tissues compared with adjacent normal tissues (P < 0.05, Fig. 5A).
CircRNA_0043691 was upregulated in GC tissues. A CircRNA_0043691 was upregulated in GC tissues; B Relative expression of circRNA_0043691 after knockdown of circRNA_0043691 in AGS cells; C Relative cell proliferation in AGS cells between si-NC and si-circRNA_0043691 at 0, 24, 48, 72, and 96 h. D Relative expression of circRNA_0043691 after knockdown of circRNA_0043691 in SGC7901 cells; E Relative cell proliferation in SGC7901 cells between si-NC and si-circRNA_0043691 at 0, 24, 48, 72, and 96 h. Transwell migration (F) and invasion (G) assay results between si-NC and si-circRNA_0043691 in AGS cells. Transwell migration (H) and invasion (I) assay results between si-NC and si-circRNA_0043691 in SGC7901 cells. *P < 0.05
Knockdown of circRNA_0043691 reduced the AGS and SGC7901 cells proliferation, migration, and invasion
To illustrate the function of circRNA_0043691 in GC, RNA silencing was performed to knockdown the expression of circRNA_0043691 in GC cells by using siRNA (Fig. 5B, D). In vitro experiment further confirmed that knockdown of circRNA_0043691 inhibited GC cell proliferation at 96 h (Fig. 5C, E). Consistently, the Transwell migration indicated that circRNA_0043691 knockdown significantly suppressed the migration and invasion abilities of AGS and SGC7901 cells (Fig. 5F–I). The results of the cell invasion assay were consistent with the cell migration assay (Fig. 5J–M).
CircRNA_0043691 directly binds to miR-873-3p and acts as a sponge
Venn diagram analysis revealed one miRNAs (miR-873-3p) overlapped with Circular RNA Interactome, TargetScan, and miRanda (Fig. 6).
Transfection of si_circRNA_0043691 obviously decreased the miR-873-3p levels in AGS and SGC7901 cells (Fig. 7A). AGS and SGC7901 cells were transfected with miR-NC, miR-873-3p, or anti-miR-873-3p, and the infection efficiency was confirmed by RT-PCR (Fig. 7B). Potential miRNAs targeting circRNA_0043691 sequences were predicted using the circInteractome database (https://circinteractome.nia.nih.gov/index.html) and found that circRNA_0043691 has binding sites with miR-873-3p (Fig. 7C). A dual-luciferase reporter assay showed that miR-873-3p significantly decreased the luciferase activity of the reporter with wild-type NT5C3L1-3ʹ UTR, but unaffected the activity of the mutant-type vector in AGS cell (Fig. 7D) and SGC7901 cells (Fig. 7E).
miR-873-3p is a target of circRNA_0043691. A Relative expression of miR-873-3p between si-NC and si-circ_0043691 in AGS and SGC7901 cells. B Relative miR-873 expression between miR-NC, miR-873, and anti-miR-873 groups. C The binding sites between circ_0043691 and miR-873-3p. D Luciferase activity was detected in AGS cells that were co-transfected with both a luciferase reporter system and either miR-873 or miR-NC. E Luciferase activity was detected in SGC7901 cells that were co-transfected with both a luciferase reporter system and either miR-873 or miR-NC
CircRNA_0043691 regulates GART via sponging miR-873-3p in GC cells
Venn diagram analysis revealed 16 mRNAs overlapped with Targetscan, miRanda, and miRDB (Fig. 8). The miR-873-3p and GART had highest binding energy and were thus selected for further analysis.
Putative miR-873-3p binding sequence in the 3′-UTR of GART mRNA was predicted using the TargetScan database (Fig. 9A). After transfection for 48 h, a dual-luciferase reporter assay was performed, revealing that in GART 3'UTR WT-transfected cells, the luciferase activity of miR-873-3p-overexpressing cells was significantly decreased when compared to that of miR-NC-transfected cells, which was abolished when the GART 3'UTR MUT luciferase reporter vector was used (Fig. 9B), suggesting that miR-873-3p directly binds to the 3'UTR of GART mRNA.
GART is a target of miR-873-3p. A The binding sites between miR-873-3p and GART. B Luciferase activity of GART-WT and GART-MUT in the miR-873-3p and miR-NC groups in AGS and SGC7901 cells. C Relative GART expression in miR-NC, miR-873-3p, and anti-miR-873-3p groups in AGS and SGC7901 cells. D Relative GART protein expression between miR-NC, miR-873-3p, and anti-miR-873-3p groups in AGS and SGC7901 cells. E Relative GART Mrna expression between si-NC, si-circ_0043691, and si-circ_0043691 + anti-miR-873-3p groups in AGS and SGC7901 cells. F Relative GART protein expression between si-NC, si-circ_0043691, and si-circ_0043691 + anti-miR-873-3p groups in AGS and SGC7901 cells
As shown in Fig. 9C, D, transfection of AGS and SGC7901 with miR-873-3p led to a decrease, whereas transfection with the anti-miR resulted in increased GART gene and protein expression, respectively, when compared with miR-NC. Compared with si-NC, si-circRNA_0043691 significantly decreased the GART expression, while co-transfection with miR-873-3p could partially reverse the inhibitory effect of si-circRNA_0043691 on GART expression (Fig. 9E, F).
Knockdown GART inhibited GC cell proliferation and invasion
Additionally, we conducted loss-of-function assays and confirmed the oncogenic roles of GART in GC. Our results from the RT-PCR analysis showed that, compared with the si-NC group, the GART mRNA expression levels were significantly reduced in both the AGS and SGC7901 cells knockdown of GART (P < 0.05) (Fig. 10A).
Knockdown of GART inhibited GC cells proliferation, migration, and invasion. A Relative GART expression between si-NC and si-GART groups in AGS and SGC7901 cells. B Relative cell proliferation between si-NC and si-GART groups in AGS and SGC7901 cells. C Relative invasion capacity between si-NC and si-GART groups in AGS and SGC7901 cells
The results of the CCK8 assay revealed that proliferation rates in the si-GART-transfected group were significantly lower compared with the si-NC group in AGS and SGC7901 cells at 48, 74, and 96 h (Fig. 10B). Transwell assays showed that the invasive activities of GC cells (AGS and SGC7901) were suppressed by the knockdown of GART than that of si-NC group (Fig. 10C).
GART could be a potential biomarker for GC
Furthermore, public datasets were analyzed to evaluate the prognostic value of GART in GC. GEPIA database profiling showed that GART was significantly overexpressed in GC samples relative to the normal sample (P < 0.05, Fig. 11A), there was no significant difference between the GC tissues at different clinical stage (P > 0.05) (Fig. 11B). Kaplan–Meier analyses indicated that higher GART expression level correlated highly with shorter overall survival; however, the difference has no statistical significance (Fig. 11C).
Bioinformatic analysis of GART in GC progression. A GART expression in GC and normal tissue by using the GEPIA database. B GART expression in GC and normal tissue at different clinical stage by using the GEPIA database. C In the GEPIA database, the expression levels of GART were found to be significantly correlated with shorter overall survival of GC patients
Discussion
CircRNAs have been reported to exert a crucial role in many types of tumors, serving as oncogenes or tumor suppressors. For example, Cao et al. (2021) reported that Circular RNA circLMO7 acts as a microRNA-30a-3p sponge to promote gastric cancer progression via the WNT2/β-catenin pathway. Lin et al. (2020) revealed that CircRNA_0043691 is upregulated in GC and promotes the GC cells’ growth, migration, and invasion via miR-665/YAP1 signaling pathway. In this study, we identified that circRNA_0043691 was upregulated in GC tissues. Moreover, circRNA_0043691 sponges miR-873-3p to promote proliferation and metastasis of GC by upregulating GART expression.
Accumulating studies have demonstrated that circRNAs serve as crucial mediators in regulating the progression of diverse human cancers, including GC. Firstly, bioinformatics analysis was performed to identify differentially expressed circRNAs from GEO database (GSE78092). We found that circRNA_0043691 was upregulated in GC tissues than normal tissues. We then construct the circRNA_0043691 knockdown GC cells and find that knockdown of circRNA_0043691significantly decreased GC cells proliferation, migration, and invasion. CircRNAs can function as miRNA sponge via their miRNA binding sites. A total of 212 miRNAs was thereafter obtained by taking the intersection of the two generated databases.
Previous studies demonstrated that miR-873-3p plays critical role in tumor progression. Deng et al. (2021) revealed that circTP63 enhances estrogen receptor-positive breast cancer progression and malignant behaviors through the miR-873-3p/FOXM1 axis. Malavika et al. (2020) found that miR-873-3p targets HDAC4 to stimulate matrix metalloproteinase-13 expression upon parathyroid hormone exposure in rat osteoblasts. Gao et al. (2019) suggested that miR-873/PDL1 axis regulates the stemness of breast cancer cells. Liang and Li (2020) found that miR-873, as a suppressor in cervical cancer, inhibits cells proliferation, invasion, and migration via negatively regulating ULBP2. Zhang et al. (2017b) revealed that miR-873 suppresses H9C2 cardiomyocyte proliferation by targeting GLI1. In this study, we firstly identified that miR-873-3p was a target of circRNA_0043691 and suppresses the migration and invasion of GC cells in vitro. Moreover, knockdown of miR-873-3p could partially reverse the effects of si-circRNA_0043691 on GC cells proliferation, migration, and invasion.
A previous study suggested that circRNA might act as ceRNAs to interact with miRNAs and influence the expression of miRNA target genes (Chen and Yang 2015). In this study, the target gene of miR-873-3p was GART. Luciferase activity was performed and identified that miR-873-3p has potential target sites with GART. A recent study showed that knockdown of the predicted targets such as TYMS, DHFR, and GART resulted in decreased cytotoxicity of the drug combination in the cancer cell (Lee et al. 2017). In this study, we revealed that knockdown of GART significantly decreased the cell proliferation and invasion in GC cells. And this reduction was further confirmed by GEPIA database. However, from bioinformatic results, GART expression has no difference in clinical stage. Though higher expression of GART was associated with a reduction of overall survival in GC patients, the difference has no statistical significance. These findings need for more studies to verify.
However, a limitation of the present study was the lack of animal experiments, and thus, these results require further validation in vivo. Another limitation of this study is that downstream signaling pathway in GC cells was not studied.
Conclusion
In conclusion, this is the first study that revealed that circRNA_0043691 was found to act as a sponge of miR-873-3p, which directly bound to GART and regulated the GART expression. Knockdown of circRNA_0043691 could be a potential therapeutic target for GC. Our findings might provide new insight into GC development and provided a novel potential strategy for GC treatment.
Data availability
We state that the data will not be shared since all the raw data are present in the figures included in the article.
Change history
31 May 2023
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s00335-023-10000-9
References
Ang TL, Fock KM (2014) Clinical epidemiology of gastric cancer. Singapore Med J 55:621–628. https://doi.org/10.11622/smedj.2014174
Bardou P, Mariette J, Escudié F et al (2014) jvenn: an interactive Venn diagram viewer. BMC Bioinform 15:293. https://doi.org/10.1186/1471-2105-15-293
Cao J, Zhang X, Xu P et al (2021) Circular RNA circLMO7 acts as a microRNA-30a-3p sponge to promote gastric cancer progression via the WNT2/β-catenin pathway. J Exp Clin Cancer Res 40:6. https://doi.org/10.1186/s13046-020-01791-9
Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12:381–388. https://doi.org/10.1080/15476286.2015.1020271
Correa P (2013) Gastric cancer: overview. Gastroenterol Clin N Am 42:211–217. https://doi.org/10.1016/j.gtc.2013.01.002
De Re V (2018) Molecular features distinguish gastric cancer subtypes. Int J Mol Sci. https://doi.org/10.3390/ijms19103121
Deng Y, Xia J, Xu YE (2021) Circular RNA circTP63 enhances estrogen receptor-positive breast cancer progression and malignant behaviors through the miR-873-3p/FOXM1 axis. Anticancer Drugs 32:44–52. https://doi.org/10.1097/cad.0000000000001010
Feng W, Ding Y, Zong W et al (2019) Non-coding RNAs in regulating gastric cancer metastasis. Clin Chim Acta 496:125–133. https://doi.org/10.1016/j.cca.2019.07.003
Gao L, Guo Q, Li X et al (2019) MiR-873/PD-L1 axis regulates the stemness of breast cancer cells. EBioMedicine 41:395–407. https://doi.org/10.1016/j.ebiom.2019.02.034
Lee YH, Choi H, Park S et al (2017) Drug repositioning for enzyme modulator based on human metabolite-likeness. BMC Bioinform 18:226. https://doi.org/10.1186/s12859-017-1637-5
Liang HX, Li YH (2020) MiR-873, as a suppressor in cervical cancer, inhibits cells proliferation, invasion and migration via negatively regulating ULBP2. Genes Genomics 42:371–382. https://doi.org/10.1007/s13258-019-00905-8
Lin X, Huang C, Chen Z et al (2020) CircRNA_100876 is upregulated in gastric cancer (GC) and promotes the GC cells’ growth, migration and invasion via miR-665/YAP1 signaling. Front Genet 11:546275. https://doi.org/10.3389/fgene.2020.546275
Malavika D, Shreya S, Raj Priya V et al (2020) miR-873-3p targets HDAC4 to stimulate matrix metalloproteinase-13 expression upon parathyroid hormone exposure in rat osteoblasts. J Cell Physiol 235:7996–8009. https://doi.org/10.1002/jcp.29454
Orman S, Cayci HM (2019) Gastric cancer: factors affecting survival. Acta Chir Belg 119:24–30. https://doi.org/10.1080/00015458.2018.1453437
Qi R, Wang DT, Xing LF et al (2018) miRNA-21 promotes gastric cancer growth by adjusting prostaglandin E2. Eur Rev Med Pharmacol Sci 22:1929–1936. https://doi.org/10.26355/eurrev_201804_14717
Tan Z (2019) Recent advances in the surgical treatment of advanced gastric cancer: a review. Med Sci Monit 25:3537–3541. https://doi.org/10.12659/msm.916475
Toro-Domínguez D, Martorell-Marugán J, López-Domínguez R et al (2019) ImaGEO: integrative gene expression meta-analysis from GEO database. Bioinformatics 35:880–882. https://doi.org/10.1093/bioinformatics/bty721
Venerito M, Link A, Rokkas T et al (2016) Gastric cancer—clinical and epidemiological aspects. Helicobacter 21(Suppl 1):39–44. https://doi.org/10.1111/hel.12339
Vohlonen I, Härkönen M, Malila N et al (2017) Challenges in evaluation of screening for gastric cancer among men based on nonrandomized design. Acta Oncol 56:917–922. https://doi.org/10.1080/0284186x.2016.1278304
Wu JQ, Mao LB, Liu LF et al (2021) Identification of key genes and pathways of BMP-9-induced osteogenic differentiation of mesenchymal stem cells by integrated bioinformatics analysis. J Orthop Surg Res 16:273. https://doi.org/10.1186/s13018-021-02390-w
Yang F, Hu A, Li D et al (2019) Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer 18:158. https://doi.org/10.1186/s12943-019-1094-z
Yu X, Xiao W, Song H et al (2020) CircRNA_100876 sponges miR-136 to promote proliferation and metastasis of gastric cancer by upregulating MIEN1 expression. Gene 748:144678. https://doi.org/10.1016/j.gene.2020.144678
Zhang Y, Liu H, Li W et al (2017a) CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging 9:1585–1594. https://doi.org/10.18632/aging.101254
Zhang JS, Zhao Y, Lv Y et al (2017b) miR-873 suppresses H9C2 cardiomyocyte proliferation by targeting GLI1. Gene 626:426–432. https://doi.org/10.1016/j.gene.2017.05.062
Zhang Z, Dong Y, Hua J et al (2019a) A five-miRNA signature predicts survival in gastric cancer using bioinformatics analysis. Gene 699:125–134. https://doi.org/10.1016/j.gene.2019.02.058
Zhang X, Wang S, Wang H et al (2019b) Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer 18:20. https://doi.org/10.1186/s12943-018-0935-5
Zhu Z, Rong Z, Luo Z et al (2019a) Circular RNA circNHSL1 promotes gastric cancer progression through the miR-1306-3p/SIX1/vimentin axis. Mol Cancer 18:126. https://doi.org/10.1186/s12943-019-1054-7
Zhu Y, Zhang X, Qi M et al (2019b) miR-873-5p inhibits the progression of colon cancer via repression of tumor suppressor candidate 3/AKT signaling. J Gastroenterol Hepatol 34:2126–2134. https://doi.org/10.1111/jgh.14697
Funding
There is no funding for this article.
Author information
Authors and Affiliations
Contributions
YZ, GYH, and ZXZ designed the study and conducted the experiments. YMJ and FT performed the statistical analyses. MFY, YZ, and GYH wrote the draft. GYH and ZXZ edited and confirmed the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
The present study was approved by the Ethical Review Board of the Shaoxing People’s Hospital.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1007/s00335-023-10000-9"
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Hu, G., Zhang, Z. et al. RETRACTED ARTICLE: CircRNA_0043691 sponges miR-873-3p to promote metastasis of gastric cancer. Mamm Genome 32, 476–487 (2021). https://doi.org/10.1007/s00335-021-09900-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-021-09900-5