Abstract
Background
Breast cancer is an aggressive tumor, which poses a heavy burden to human health. Circular RNAs have been involved in the pathogenesis of breast cancer. This study aims to investigate whether circ_0008673 mediates breast cancer malignant progression by microRNA-153-3p (miR-153-3p)/cofilin 2 (CFL2) pathway.
Methods
The RNA levels of circ_0008673, miR-153-3p and CFL2 were detected by quantitative real-time polymerase chain reaction (qRT-PCR). The protein expression of CFL2, E-cadherin and N-cadherin was determined by western blot analysis. Cell proliferation was demonstrated through cell counting kit-8 and cell colony-formation assays. Cell apoptosis was detected by flow cytometry analysis. Cell migratory and invasive capacities were determined by transwell assay. The associated relationship between miR-153-3p and circ_0008673 or CFL2 was predicted by online databases, and testified by dual-luciferase reporter and RNA immunoprecipitation assays. In vivo assay was employed to demonstrate the effects of circ_0008673 silencing on tumor formation in vivo.
Results
Circ_0008673 and CFL2 expressions were upregulated, while miR-153-3p expression was downregulated in breast cancer tissues and cells compared with adjacent normal breast tissues and cells, respectively. Circ_0008673 overexpression promoted cell proliferation, migration and invasion, and repressed cell apoptosis, while circ_0008673 silencing had opposite effects. Additionally, circ_0008673 served as a sponge of miR-153-3p. And circ_0008673 was proved to regulate breast cancer cell malignancy by sponging miR-153-3p. MiR-153-3p was found to modulate breast cancer cell carcinogenesis via targeting CFL2. Furthermore, circ_0008673 silencing repressed tumor growth in vivo.
Conclusion
Circ_0008673 promoted breast cancer progression by upregulating CFL2 expression through sponging miR-153-3p. This study provides a theoretical basis for researching circRNA-directed treatment of breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer, a common malignant cancer, accounts for about 15% of cancer-caused deaths [1]. According to the data reported in 2018, more than 2 million people were diagnosed with breast cancer and approximately 30% of the patients died, with an age-standardized morbidity and mortality rates of 39.2 and 8.6 per one hundred thousand, respectively, in females [2]. Despite numerous diagnostic and therapeutic improvements, a number of sufferers still progress to metastatic stage after treatment. Thus, it is urgent to seek new molecules participating in the pathogenesis of breast cancer to improve unfavorable prognosis.
Circular RNA (circRNA) is a circular RNA without protein-coding capacity, characterized by stability, high expression and conservatism [3]. CircRNA usually acts as a competing endogenous RNA (ceRNA) to inhibit the activity of microRNA (miRNA) to further modulate the transcription or translation of target gene [4]. With the rapid development of next-generation sequencing technologies, multiple circRNAs have been unveiled to participate in modulating cancer evolution [5]. In breast cancer, lots of circRNAs also were identified to be involved. For example, circ_001783 was found to be closely related to heavier tumor burden, and its silencing greatly repressed cell proliferation and invasion [6]. Zhang and his colleagues also explained circ_0003645 absence hindered cell proliferation, and contributed to cell apoptosis [7]. Additionally, circ_0072309 [8] and circ_0000442 [9] were disclosed to act as cancer suppressors in breast cancer. Circ_0008673, a novel circRNA, has been indicated to be augmented in breast cancer tissues, and is related to larger tumor size and distant metastasis [10]. However, the underlying mechanism by which circ_0008673 mediated breast cancer process remains largely unknown. According to the ceRNA theory, we predicted the miRNA associated with circ_0008673, and the miRNA-interacted mRNA by online databases. Our data showed that circ_0008673 contained the binding sites of the seed sequence of miR-153-3p, and miR-153-3p carried the complementary sites of Cofilin 2 (CFL2).
MicroRNA (miRNA) is a small RNA with about 20 nucleotides and acts function via targeting the noncoding sequence of interest mRNA [11]. An increasing number of researchers reported that miRNAs were involved in the advancement of various cancers [12]. As reported, miR-153-3p played a tumor-repressing role in breast cancer development [13, 14]. CFL2 belongs to ADF/cofilins family, with encoding cofilin 2 protein. Yehl and his colleagues explained that CFL2 interacted with F-actin to mediate actin treadmilling [15]. In particular, in the study of Schwickert et al., CFL2 was found to participate in modulating breast cancer invasiveness [16], suggesting CFL2 participated in breast cancer progression. The above evidence suggests miR-153-3p/CFL2 may be responsible for circ_0008673-mediated breast cancer progression; however, the novel mechanism has not been reported.
Thus, the expression of circ_0008673 in breast cancer tissues and cells was quantified. In function, we studied the role of circ_0008673 on breast cancer cell processes, including cell proliferation, apoptosis, migration and invasion. Additionally, circ_0008673/miR-153-3p/CFL2 pathway was utilized to explain the molecular mechanism behind circ_0008673-mediated breast cancer progression.
Materials and methods
Specimen collection and storage
42 pairs of breast cancer and adjacent breast tissues were collected from breast cancer patients from Renmin Hospital, Hubei University of Medicine. Collected tissues were confirmed by two pathologists, and stored at – 80 °C in a freezer. Breast cancer suffers signed the written informed consent. The Ethics Committee of Renmin Hospital, Hubei University of Medicine approved this research.
Cell purchase and culture
Human breast cancer cell lines (MDA-MB-231, MDA-MB-468, MCF-7) and human normal breast cell line (MCF-10A) were purchased from American Type Culture Collection (Manassas, VA, USA). Human breast cancer cell line (MCF-10AT) was provided by Karmanos Cancer Institute (Detroit, Michigan, USA). Purchased cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Procell, Wuhan, China) with 10% fetal bovine serum (FBS; Thermo Fisher, Waltham, MA, USA) and 1% penicillin/streptomycin (Procell) at 37 °C in a humid incubator with 5% CO2.
Plasmid construction, oligonucleotide synthesis and cell transfection
Small interfering RNAs against circ_0008673 (si-circ_0008673#1 and si-circ_0008673#2) and CFL2 (si-CFL2), small hairpin RNA targeting circ_0008673 (sh-circ_0008673), miR-153-3p mimics (miR-153-3p), miR-153-3p inhibitors (in-miR-153-3p) and control groups (si-con, si-NC, sh-con, miR-NC and in-miR-NC) were synthesized by GenePharma (Shanghai, China). The over-expression plasmids of circ_0008673 (circ_0008673) and CFL2 (CFL2) as well as control groups (Vector, pcDNA) were provided by Geneseed Co., Ltd. (Guangzhou, China). The wild-type (WT) plasmids of circ_0008673 (circ_0008673 WT) and the 3ʹ-untranslated region of CFL2 (CFL2 3ʹUTR WT) as well as the mutant (MUT) vectors of circ_0008673 (circ_0008673 MUT) and CFL2 (CFL2 3ʹUTR MUT) were built by Geneseed Co., Ltd. Plasmids or oligonucleotides were transfected into MCF-10AT and MCF-7 cells with TurboFect reagent (Thermo Fisher) according to the instruction of manufacturer. The sequences of synthesized oligonucleotides were displayed as below. Si-circ_0008673#1 5ʹ-AAAGGTTCATTGGAACAGAAA-3ʹ, si-circ_0008673#2 5ʹ-AAAAGGTTCATTGGAACAGAA-3ʹ, miR-153-3p 5ʹ-UUGCAUAGUCACAAAAGUGAUC-3ʹ, in-miR-153-3p 5ʹ-GAUCACUUUUGUGACUAUGCAA-3ʹ, si-CFL2 5ʹ-GCAGTTCTCTTCTGTTTAA-3ʹ, si-con 5ʹ-CCTCTACCTGTCGCTGAGCTGTAAT-3ʹ, miR-NC 5ʹ-UUUGUACUACACAAAAGUACUG-3ʹ, in-miR-NC 5ʹ-CAGUACUUUUGUGUAGUACAAA-3ʹ and si-NC 5ʹ-GCATCTTTCGTCTTGTTAA-3ʹ.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Tissues and cells were first lysed using TransZol (TransGen, Beijing, China). RNA was isolated using RNAsimple kit (Tiangen, Beijing, China). Then, cDNA was amplified with FastKing RT Kit (Tiangen) and TQiagen reverse transcription kit (Hilden, Germany), respectively. For quantification analysis, SuperReal PreMix Color (Tiangen) was employed with a Mx3000P system (Stratagene, Santa Clara, CA, USA). Data were assessed with the 2−∆∆Ct method. The sense and antisense sequences of primers were listed as below. Circ_0008673 5ʹ-AGGAACCAGGGATGAAAT-3ʹ and 5ʹ-CCAGACAGATGGGACACT-3ʹ; BRCA1 DNA repair associated (BRCA1) 5ʹ-AAAAGACATGACAGCGATAC-3ʹ and 5ʹ-CTTGGAAGGCTAGGATTG-3ʹ; miR-153-3p 5ʹ-ACACTCCAGCTGGGTTGCATAGTCACAAAA-3ʹ and 5ʹ-TGGTGTCGTGGAGTCG-3ʹ; CFL2 5ʹ-CGAGGGCACTATGGCTTCT-3ʹ and 5ʹ-ACCAAGATCTGCTTTGCTTCC-3ʹ; U6 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ; glyceraldehyde 3-phosphate dehydrogenase (GAPDH) 5ʹ-GGTCACCAGGGCTGCTTT-3ʹ and 5ʹ-GGAAGATGGTGATGGGATT-3ʹ. U6 and GAPDH were employed as controls.
Cytoplasmic and nuclear circ_0008673 analysis
Cell nucleus was separated away from cytoplasm using PARIS™ Kit (Thermo Fisher). Briefly, cultured MCF-10AT and MCF-7 cells were harvested and instantly placed on ice. After that, the cells were suspended in 300 μL Cell Fractionation Buffer (Thermo Fisher) by pipetting. Cell supernatant was collected up by centrifugation at 500 g for 6 min. The fractions containing cell nucleus and cytoplasm were severally separated by carefully aspirating. Obtained cell nucleus and cytoplasm fractions were lysed with Lysis solution (Thermo Fisher) and RNA was extracted. Circ_0008673 content was assessed by qRT-PCR. GAPDH and U6 served as references.
RNase R resistance analysis assay
Cultured MCF-10AT and MCF-7 cells were harvested and lysed. RNA was isolated and incubated with RNase R (3 U/μg RNA, Geneseed) at 37 °C. After 30 min, RNA was purified with Qiagen RNeasy Cleaning Kit (Valencia, CA, USA). And circ_0008673 content was determined by qRT-PCR. Linear BRCA1 was employed as a reference.
Cell counting kit-8 (CCK-8) assay
The viability of MCF-10AT and MCF-7 cells was determined by CCK-8 assay. In brief, cells were grown in 96-well plates and transfected with circ_0008673, si-circ_0008673#1, si-circ_0008673#2, in-miR-153-3p, miR-153-3p, si-CFL2, CFL2, Vector, si-con, in-miR-NC, miR-NC, si-NC or pcDNA based on different purposes. Cells were grown for 1, 2, 3, 4, 5 and 6 days, respectively. Then, CCK-8 reagent (Beyotime, Shanghai, China) was used to incubate the cells for 3 h. The samples were analyzed with a microplate reader (Thermo Fisher). Cell viability was determined by assessing the output of wavelength at 450 nm.
Cell colony-formation assay
MCF-10AT and MCF-7 cells were seeded in 6-well plates, and transfected with various treatments as shown in CCK-8 assay. And cells were continued to be grown for about 2 weeks. Media were renewed every 3 days. After supernatant was removed, the cells were fixed with paraformaldehyde (Beyotime) and stained with crystal violet (Beyotime). Finally, cell colony-forming ability was determined via figuring up the number of colonies. A colony was deemed when cell numbers over 50.
Flow cytometry analysis
The apoptotic rate of MCF-10AT and MCF-7 cells was revealed by Annexin V-fluorescein isothiocyanate (Annexin V-FITC)/propidium iodide (PI) double staining kit (Solarbio, Beijing, China). In brief, grown cells were collected, and suspended in Binding buffer (Solarbio). After that, Annexin V-FITC (Solarbio) and PI (Solarbio) were singly employed to incubate the cells in dark. The samples were assessed with flow cytometry (Thermo Fisher).
Transwell assay
Transwell chambers (8.0 µm, Millipore, Bradford, MA, USA) were employed to assess cell migratory and invasive capacities. In brief, MCF-10AT and MCF-7 cells were seeded in the upper chambers supplemented with FBS-free DMEM (Procell), and DMEM containing 15% FBS (Thermo Fisher) was pipetted into the lower chambers. At 24 h after culture, cells were fixed and stained with methanol (Corning, Madison, New York, USA) and crystal violet (Beyotime), respectively. Finally, results were analyzed by counting cell numbers in the lower chambers under microscope (Olympus, Tokyo, Japan) with 100 × magnification. For cell invasion assay, the chambers were pre-coated with Matrigel (Corning).
Western blot analysis
Tissues and cells were first lysed by incubation with RIPA buffer (Beyotime). The lysates were mixed with loading buffer (Thermo Fisher), which were subsequently boiled in the boiling water to denature proteins. After that, protein samples were loaded by 12% bis–tris-acrylamide gel (Thermo Fisher). And the separated protein bands were transferred onto nitrocellulose membranes (GE Healthcare, Westborough, MA, USA), and blocked with nonfat dry milk (Solarbio). The membranes were incubated with primary antibodies and secondary antibody (1:8000; Affinity, Nanjing, China), respectively. Finally, protein bands were emerged by RapidStep ECL Reagent (Millipore). The primary antibodies were anti-E-cadherin (1:2000; Affinity), anti-N-cadherin (1:800; Affinity), anti-CFL2 (1:1500; Affinity) and anti-GAPDH (1:15,000; Affinity). GAPDH served as a control.
Dual-luciferase reporter assay
The binding sites of miR-153-3p and circ_0008673 or CFL2 were severally predicted by starBase online database (http://starbase.sysu.edu.cn/agoClipRNA.php?source=circRNA&flag=target&clade=mammal&genome=human&assembly=hg19&miRNA=all&clipNum=1&deNum=0&target=hsa_circ_0008673) and Targetscan online database (http://www.targetscan.org/cgi-bin/targetscan/vert_71/view_gene.cgi?rs=ENST00000354662.1&taxid=9606&members=miR-153-3p&showcnc=1&shownc=1&shownc_nc=1&showncf1=1&subset=1). The putative binding sites were identified by dual-luciferase reporter assay kit (Promega, Madison, WI, USA). In brief, constructed plasmids were mixed with miR-153-3p or miR-NC, and transfected into MCF-10AT and MCF-7 cells with TurboFect reagent (Thermo Fisher). 48 h later, the cells were collected up and lysed using Passive lysis buffer (Promega). Luciferase activities were evaluated after cells were incubated with Luciferase assay regent II (Promega) or Stop&Glo Reagent (Promega). Results were expressed as the value of firefly luciferase activity/Renilla luciferase activity.
RNA immunoprecipitation (RIP) assay
Magna RIP kit (Millipore) was utilized to demonstrate whether miR-153-3p was directly associated with circ_0008673 or CFL2. In brief, fresh MCF-10AT and MCF-7 cells were collected and lysed with RIP lysis buffer (Millipore) containing protease and RNase inhibitors (Millipore). And antibodies against argonaute2 (Anti-Ago2; 1:50; Abcam) and immunoglobulin G (Anti-IgG; 1:100; Abcam) were incubated with the lysates for 4 h before pre-treated with magnetic beads (Millipore). Twenty-four hours later, magnetic beads were washed, and the contents of circ_0008673, miR-153-3p and CFL2 were determined by qRT-PCR.
In vivo assay
Female BALB/c nude mice (6-week-old) were purchased from Charles River (Beijing, China), and fed in pathogen-free environment. These mice were divided into two groups (sh-circ_0008673 group and sh-con group, N = 6, respectively). Constructed sh-circ_0008673 and sh-con were severally packaged as lentivirus to infect MCF-7 cells. Then, 5 × 106 MCF-7 cells were subcutaneously injected into the right back of mice. Tumor volume was measured every 4 days until the 24th day after injection. The forming tumors were photographed before excised, and weight of every tumor was weighted. Additionally, circ_0008673 and miR-153-3p expressions were detected by qRT-PCR, and the protein levels of CFL2, E-cadherin and N-cadherin were determined by western blot analysis. The Animal Care and Use Committee of Renmin Hospital, Hubei University of Medicine approved this research.
Statistical analysis
Data were analyzed by SPSS 21.0 software (IBM, Somers, NY, USA) based on the results from three independent duplicate tests. Results were shown as means ± standard deviations (SD). The significant differences were compared with Spearman’s correlation test in Spearman correlation analysis, with two-tailed Student’s t tests or Wilcoxon rank-sum test between the two groups, and with one-way analysis of variance (ANOVA) among three or more groups. Statistical significance was deemed when P value < 0.05.
Results
Circ_0008673 promoted breast cancer cell malignancy
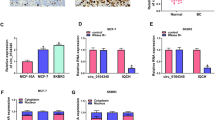
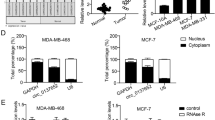
Circ_0008673 expression was first determined in breast cancer tissues. Data displayed that circ_0008673 expression was dramatically upregulated in breast cancer tissues compared with paracancerous normal breast tissues (Fig. 1A). Subsequently, qRT-PCR data showed circ_0008673 expression was also obviously increased in MCF-10AT, MDA-MB-231, MDA-MB-468 and MCF-7 cells as compared to human normal breast cell line (MCF-10A cells), respectively (Fig. 1B). These data implicated circ_0008673 might be involved in the progression of breast cancer. MCF-10AT and MCF-7 cells were employed in subsequent study based on the lowest or highest circ_0008673 expression in the two cell types. Subsequent analysis presented circ_0008673 was mainly located in cytoplasm (Fig. S1A, B). Furthermore, result displayed that circ_0008673 expression had little change after RNase R treatment, but linear BRCA1 expression was apparently downregulated (Fig. S1C, D), suggesting the high stability of circ_0008673.
Circ_0008673 promoted breast cancer cell malignancy. A and B Circ_0008673 expression was detected by qRT-PCR in 42 pairs of breast cancer and matched normal breast tissues as well as MCF-10A, MCF-10AT, MDA-MB-231, MDA-MB-468 and MCF-7 cells. C and D The effects of circ_0008673 overexpression on cell viability and colony-forming ability were revealed by CCK-8 assay and cell colony-formation assay, respectively, in MCF-10AT cells. E Flow cytometry analysis was employed to analyze the effects of circ_0008673 overexpression on the apoptosis of MCF-10AT cells. F The effects of circ_0008673 overexpression on cell migration and invasion were presented by transwell assay in MCF-10AT cells. G The effects of circ_0008673 overexpression on the protein expression of E-cadherin and N-cadherin were presented by western blot analysis in MCF-10AT cells. H–J The effects of circ_0008673 silencing on tumor volume and weight were revealed. **P < 0.01 and ***P < 0.001
To demonstrate whether circ_0008673 was involved in the development of breast cancer, the effects of circ_0008673 overexpression and silencing on cell processes of breast cancer were revealed. Results first presented that constructed circ_0008673, si-circ_0008673#1 and si-circ_0008673#2 were effective in increasing or decreasing circ_0008673 expression (Fig. S2A, B). Subsequently, data showed circ_0008673 overexpression promoted cell viability and colony-forming ability (Fig. 1C, D), but inhibited cell apoptosis (Fig. 1E). Transwell assay presented cell migratory and invasive ability were enhanced after circ_0008673 overexpression (Fig. 1F). Meanwhile, western blot analysis showed that E-cadherin protein expression was downregulated and N-cadherin protein expression was upregulated after circ_0008673 overexpression in MCF-10AT cells (Fig. 1G). The study then investigated the impacts of circ_0008673 depletion on breast cancer cell processes. To validate this, MCF-7 cells were chosen as the highest expression of the circRNA in the cell line. As displayed, circ_0008673 knockdown repressed cell viability and cell colony-forming ability (Fig. S3A, B), induced cell apoptosis (Fig. S3C), and repressed cell metastasis (Fig. S3D–F). In support, circ_0008673 knockdown inhibited tumor volume and weight in vivo (Fig. 1H–J), decreased circ_0008673 and N-cadherin expression, and increased E-cadherin expression when compared with sh-con group (data not shown). Collectively, the above data demonstrated circ_0008673 served as an oncogene in breast cancer progression. Si-circ_0008673#2 was employed for subsequent study because of its significantly inhibitory effects on MCF-7 cell malignancy.
Circ_0008673 regulated breast cancer cell malignancy by binding to miR-153-3p
The study continued to explore the mechanism of circ_0008673 in regulating breast cancer cell malignancy. The miRNAs containing the binding sites of circ_0008673 were predicted using online databases. Based on the prediction of circBank, circInteractome and starBase online databases, we found that only miR-153-3p contained the binding sites of circ_0008673 (data not shown). Subsequently, dual-luciferase reporter assay exhibited that miR-153-3p apparently repressed the relative luciferase activity of circ_0008673 WT, but not that of circ_0008673 MUT (Fig. 2A). RIP assay also presented both circ_0008673 and miR-153-3p were dramatically enriched by Anti-Ago2 compared with Anti-IgG in MCF-10AT and MCF-7 cells (data not shown). Additionally, we found circ_0008673 overexpression obviously decreased miR-153-3p expression, and circ_0008673 knockdown apparently upregulated miR-153-3p expression in vitro and in vivo (data not shown). An obviously low miR-153-3p expression was also found in breast cancer tissues and MCF-10AT and MCF-7 cells as compared to adjacent normal breast tissues and MCF-10A cells, respectively (Fig. 2B, C). Furthermore, it was revealed that miR-153-3p expression was negatively related to circ_0008673 expression in breast cancer tissues (data not shown). All in all, these evidences demonstrated circ_0008673 directly bound to miR-153-3p.
Circ_0008673 promoted cell proliferation, migration and invasion, whereas repressed cell apoptosis by binding to miR-153-3p. A Dual-luciferase reporter assay was conducted to demonstrate circ_0008673 was associated with miR-153-3p in MCF-10AT and MCF-7 cells. B and C MiR-153-3p expression was detected by qRT-PCR in 42 pairs of breast cancer and adjacent normal breast tissues as well as MCF-10A, MCF-10AT and MCF-7 cells. D–I MCF-10AT cells were transfected with Vector, circ_0008673, circ_0008673 + miR-NC and circ_0008673 + miR-153-3p, respectively. D Cell viability was detected by CCK-8 assay. E Cell colony-forming ability was determined by cell colony-formation assay. F Cell apoptosis was demonstrated by flow cytometry analysis. G and H The migration and invasion of MCF-10AT cells were detected by transwell assay. I The protein expression of E-cadherin and N-cadherin was detected by western blot analysis. **P < 0.01 and ***P < 0.001
Given the associated relationship between circ_0008673 and miR-153-3p, whether circ_0008673 repressed breast cancer cell malignancy via interacting with miR-153-3p was revealed in this part. Results first showed the synthesized miR-153-3p mimics and inhibitors were effective in upregulating or downregulating miR-153-3p expression (Fig. S2C). After that, results displayed that miR-153-3p mimics reversed circ_0008673 overexpression-mediated promotion of cell viability and cell colony-forming ability (Fig. 2D, E). Flow cytometry analysis presented circ_0008673 overexpression repressed cell apoptosis, whereas this effect was hindered after transfection of miR-153-3p mimics (Fig. 2F). Additionally, the promotion of circ_0008673 overexpression on cell migration and invasion were attenuated by miR-153-3p mimics (Fig. 2G, H). Furthermore, western blot analysis showed circ_0008673 overexpression decreased E-cadherin protein expression and increased N-cadherin protein expression, while these effects were reversed after transfection of miR-153-3p mimics (Fig. 2I). In support, miR-153-3p inhibitors also overturned the inhibitory effects of circ_0008673 knockdown on cell viability, colony-forming ability and metastasis, and the stimulatory impact of that on cell apoptosis (Fig. S4). Taken together, the above results demonstrated circ_0008673 could regulate cell malignancy of breast cancer via interacting with miR-153-3p.
MiR-153-3p repressed cell malignancy of breast cancer by targeting CFL2
Targetscan online database showed CFL2 3’UTR possessed the binding sequence of miR-153-3p (data not shown). Subsequent results presented the relative luciferase activity was apparently inhibited after co-transfection of CFL2 3ʹUTR WT and miR-153-3p, whereas that had little change in CFL2 3ʹUTR MUT and miR-153-3p group (Fig. 3A). RIP assay exhibited miR-153-3p and CFL2 contents were higher in Anti-Ago2 group than in Anti-IgG group (data not shown), implicated miR-153-3p could directly bind to CFL2. Afterwards, we found CFL2 protein expression was dramatically downregulated by miR-153-3p mimics, but upregulated by miR-153-3p inhibitors in MCF-10AT or MCF-7 cells (data not shown). Furthermore, our data exhibited the protein expression of CFL2 was significant upregulated in breast cancer tissues and MCF-10AT and MCF-7 cells when compared with paracancerous normal breast tissues and MCF-10A cells, respectively (Fig. 3B, C). Also, western blot analysis presented CFL2 expression was upregulated after circ_0008673 overexpression, which was attenuated by miR-153-3p mimics (Fig. S6A). Meanwhile, CFL2 expression was downregulated by circ_0008673 silencing; however, this effect was restored by miR-153-3p inhibitors (Fig. S6B). In support, in vivo assay further showed circ_0008673 knockdown downregulated CFL2 expression in vivo (data not shown). These data presented circ_0008673 modulated CFL2 expression by binding to miR-153-3p.
MiR-153-3p regulated breast cancer cell processes by binding to CFL2. A Dual-luciferase reporter assay was performed to demonstrate that miR-153-3p was directly associated with CFL2 in MCF-10AT and MCF-7 cells. B and C The protein level of CFL2 was detected by western blot in 42 pairs of breast cancer and paracancerous normal breast tissues as well as MCF-10A, MCF-10AT and MCF-7 cells. D–I MCF-10AT cells were transfected with in-miR-NC, in-miR-153-3p, in-miR-153-3p + si-NC and in-miR-153-3p + si-CFL2, respectively. D Cell viability was determined by CCK-8 assay. E Cell colony-forming ability was detected by cell colony-formation assay. F Cell apoptosis was analyzed by flow cytometry analysis. G and H Cell migratory and invasive capacities were determined by transwell assay. I The protein levels of E-cadherin and N-cadherin were determined by western blot analysis. A: adjacent normal breast tissues. T: breast cancer tissues. **P < 0.01 and ***P < 0.001
We then explored whether miR-153-3p regulated the malignancy of MCF-10AT and MCF-7 cells by binding to CFL2. Results initially presented that the high efficiency of in-miR-153-3p and miR-153-3p in decreasing or increasing miR-153-3p expression (Fig. S2D). The data from western blot analysis also showed that si-CFL2 and CFL2 were effective in downregulating or upregulating CFL2 expression (Fig. S2E). Subsequently, miR-153-3p inhibitors promoted cell viability and colony-forming ability, whereas these effects were restrained after CFL2 silencing (Fig. 3D, E). Flow cytometry analysis showed miR-153-3p inhibitors repressed cell apoptosis, but CFL2 absence attenuated this effect (Fig. 3F). Additionally, it was found that MiR-153-3p inhibitors contributed to cell migration and invasion; however, these effects were reversed by CFL2 silencing (Fig. 3G, H). Results presented miR-153-3p inhibitors downregulated E-cadherin protein expression, and upregulated N-cadherin protein level, which was reversed after CFL2 silencing (Fig. 3I). Also, CFL2 overexpression also overturned the inhibitory effects of miR-153-3p mimics on cell viability, colony-forming ability and metastasis, and the stimulatory impact of that on cell apoptosis (Fig. S5). Collectively, the above data demonstrated miR-153-3p regulated the malignancy of MCF-7 and MCF-10AT cells by targeting CFL2.
Discussion
Breast cancer is an aggressive malignant tumor and is the second most common cancer in females, posing a heavy burden to human health [17]. However, the nosogenesis of breast cancer has not been fully investigated. Previous researches have shown that circRNA plays a vital part in breast cancer progression [18,19,20], and can be employed as a predictive biomarker or therapeutic target of various cancers [21]. Therefore, this research was designed to reveal the role of circRNA in breast cancer evolution. And we found circ_0008673 contributed to cell malignancy via modulating miR-153-3p/CFL2 axis in breast cancer.
A growing body of evidences have explained the importance of circRNAs in breast cancer progression. For instance, circ_103809 was proved to facilitate cell proliferation and repress cell apoptosis via controlling phosphoinositide 3-kinase (PI3K)/protein kinase B pathway [22]. Circ_0136666 could enhance cell proliferative and metastatic abilities via augmenting cyclin-dependent kinase 6 (CDK6) through binding to miR-1299 [23]. In contrast, circ-cyclin B1 (CCNB1) overexpression repressed tumor growth and extended mouse viability [24]. In this paper, we found that circ_0008673 was increased in specimen and cell lines, and circ_0008673 overexpression enhanced cell proliferation and migration capacities. In contrast, circ_0008673 silencing restrained cell proliferation and migration. These results were consistent with the findings of Hu and his colleagues [10]. Beyond that, our data also demonstrated that circ_0008673 repressed cell apoptosis and contributed to cell invasion, and that circ_0008673 promoted tumor formation in nude mice. Additionally, cytoplasmic and nuclear circ_0008673 analysis assay presented circ_0008673 was chiefly located in cytoplasm, implicating that circ_0008673 chiefly participated in post-transcriptional regulation. These data suggested that circ_0008673 could promote breast cancer progression.
Based on the features of miRNAs in cell biological behaviors that they acted function at the post-transcriptional level [25], we screened the miRNAs associated with circ_0008673 by circBank, circInteractome and starBase online databases. As a result, we found only miR-153-3p contained the complementary sites of circ_0008673, and the miRNA was thereby employed in subsequent study. The binding relationship was proved by mechanism assays. MiR-153-3p, a miRNA, also played a vital part in cancer development [26, 27]. In terms of mechanism underneath breast cancer development, previous research indicated cancer susceptibility candidate 15 (CASC15) contributed to cell proliferation and invasion by sponging miR-153-3p [14]. Sun et al. also presented miR-153-3p repressed cell proliferation, migration and invasion by binding to Rho-associated coiled-coil containing protein kinase 1 (ROCK1) [13]. These evidences demonstrated miR-153-3p repressed cell proliferation and metastasis of breast cancer. In agreement with these findings, we also observed the repressive impacts of miR-153-3p on cell proliferation and metastasis. Besides, qRT-PCR data displayed the low expression of miR-153-3p in tissue samples and cell lines, and miR-153-3p led to upregulation of cell apoptotic rate. Moreover, circ_0008673 silencing-mediated repressive impacts on cell malignancy could be reversed after transfection of miR-153-3p inhibitors, which meant circ_0008673 regulated breast cancer cell carcinogenesis by sponging miR-153-3p.
MiRNA commonly regulates cancer progression by targeting mRNA [28]. Based on this mechanism, we further sought the mRNA interacted with miR-153-3p. And we found CFL2 was a target gene of miR-153-3p. CFL2 was classified as actin-binding protein, and contributed to cell proliferation and migration of prostate cancer [29] and gastric cancer [30]. Additionally, Yu and his colleagues presented CFL2 had the potential as a therapeutic target in nasopharyngeal cancer owing to its promoting effect on cell tumor or cell growth [31]. In this study, we also found CFL2 was overexpressed in breast cancer tissues and cell lines, which was in line with the results of Luo et al. [32]. In addition, rescue experiments showed CFL2 overexpression attenuated the effects of miR-153-3p mimics on cell proliferation, metastasis and apoptosis. These data not only suggested that CFL2 promoted cell malignancy of breast cancer, but also demonstrated that miR-153-3p regulated cell carcinogenesis by targeting CFL2. In vivo assay further indicated that circ_0008673 upregulated CFL2 expression. Importantly, circ_0008673 controlled CFL2 via interacting with miR-153-3p.
All in all, circ_0008673 was highly expressed in breast cancer tissues and cells. Circ_0008673 overexpression enhanced cell proliferative and metastatic capacities, but downregulated cell apoptotic rate in breast cancer cells. Circ_0008673 could upregulate CFL2 expression by binding to miR-153-3p. Additionally, circ_0008673 knockdown repressed tumor formation in vivo. Collectively, circ_0008673 facilitated breast cancer progression by miR-153-3p/CFL2 axis. Our findings demonstrate that circ_0008673 has the potential as a predictive biomarker or therapeutic target for breast cancer.
References
Siegel RL, Miller KD, Jemal A (2017) Cancer statistics, 2017. CA Cancer J Clin 67(1):7–30
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Kolakofsky D (1976) Isolation and characterization of Sendai virus DI-RNAs. Cell 8(4):547–555
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–338
Wang F, Nazarali AJ, Ji S (2016) Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res 6(6):1167–1176
Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, Andrews R, Zhong W, Zhang X, Song E, Gong C (2019) Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis 10(2):55–55
Zhang J, Ke S, Zheng W, Zhu Z, Wu Y (2020) Hsa_circ_0003645 promotes breast cancer progression by regulating miR-139-3p/HMGB1 axis. Onco Targets Ther 13:10361–10372
Yan L, Zheng M, Wang H (2019) Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag Res 11:1033–1041
Zhang XY, Mao L (2021) Circular RNA Circ_0000442 acts as a sponge of MiR-148b-3p to suppress breast cancer via PTEN/PI3K/Akt signaling pathway. Gene 766:145113
Hu Y, Song Q, Zhao J, Ruan J, He F, Yang X, Yu X (2020) Identification of plasma hsa_circ_0008673 expression as a potential biomarker and tumor regulator of breast cancer. J Clin Lab Anal 34(9):e23393
Moya L, Meijer J, Schubert S, Matin F, Batra J (2019) Assessment of miR-98-5p, miR-152-3p, miR-326 and miR-4289 expression as biomarker for prostate cancer diagnosis. Int J Mol Sci 20(5):1154
Shin VY, Chu K-M (2014) MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol 20(30):10432–10439
Sun L, Wang H, Jiang J, Bi X (2020) miR-153-3p inhibits proliferation and migration of breast cancer cells via down-regulating ROCK1. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 36(2):138–144
Yu L, Xu Q, Yu W, Duan J, Dai G (2018) LncRNA cancer susceptibility candidate 15 accelerates the breast cancer cells progression via miR-153-3p/KLF5 positive feedback loop. Biochem Biophys Res Commun 506(4):819–825
Yehl J, Kudryashova E, Reisler E, Kudryashov D, Polenova T (2017) Structural analysis of human cofilin 2/filamentous actin assemblies: atomic-resolution insights from magic angle spinning NMR spectroscopy. Sci Rep 7:44506–44506
Schwickert A, Weghake E, Brüggemann K, Engbers A, Brinkmann BF, Kemper B, Seggewiß J, Stock C, Ebnet K, Kiesel L, Riethmüller C, Götte M (2015) microRNA miR-142-3p inhibits breast cancer cell invasiveness by synchronous targeting of WASL integrin alpha V, and additional cytoskeletal elements. PLoS One 10(12):e0143993–e0143993
Tang Q, Hann SS (2020) Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Ther 13:2067–2092
Chen D, Ma W, Ke Z, Xie F (2018) CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle 17(16):2080–2090
Chang P, Wang F, Li Y (2018) Hsa_circ_0000673 is down-regulated in gastric cancer and inhibits the proliferation and invasion of tumor cells by targetting miR-532-5p. Biosci Rep 38(5):BSR20180538
Geng Z, Wang W, Chen H, Mao J, Li Z, Zhou J (2019) Circ_0001667 promotes breast cancer cell proliferation and survival via Hippo signal pathway by regulating TAZ. Cell Biosci 9:104–104
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu Y-M, Dhanasekaran SM, Engelke CG, Cao X, Robinson DR, Nesvizhskii AI, Chinnaiyan AM (2019) The landscape of circular RNA in cancer. Cell 176(4):869-881.e13
Qiu X, Wang Q, Song H, Shao D, Xue J (2020) circ_103809 promotes breast cancer progression by regulating the PI3K/AKT signaling pathway. Oncol Lett 19(6):3725–3730
Liu LH, Tian QQ, Liu J, Zhou Y, Yong H (2019) Upregulation of hsa_circ_0136666 contributes to breast cancer progression by sponging miR-1299 and targeting CDK6. J Cell Biochem 120(8):12684–12693
Fang L, Du WW, Lyu J, Dong J, Zhang C, Yang W, He A, Kwok YSS, Ma J, Wu N, Li F, Awan FM, He C, Yang BL, Peng C, MacKay HJ, Yee AJ, Yang BB (2018) Enhanced breast cancer progression by mutant p53 is inhibited by the circular RNA circ-Ccnb1. Cell Death Differ 25(12):2195–2208
Acunzo M, Romano G, Wernicke D, Croce CM (2015) MicroRNA and cancer—a brief overview. Adv Biol Regul 57:1–9
Luan W, Shi Y, Zhou Z, Xia Y, Wang J (2018) circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun 502(1):22–29
Li C, Zhang Y, Zhao W, Cui S, Song Y (2019) miR-153-3p regulates progression of ovarian carcinoma in vitro and in vivo by targeting MCL1 gene. J Cell Biochem 120(11):19147–19158
Lee YS, Dutta A (2009) MicroRNAs in cancer. Annu Rev Pathol 4:199–227
Wo Q, Zhang D, Hu L, Lyu J, Xiang F, Zheng W, Shou J, Qi X (2019) Long noncoding RNA SOX2-OT facilitates prostate cancer cell proliferation and migration via miR-369-3p/CFL2 axis. Biochem Biophys Res Commun 520(3):586–593
Bian Y, Guo J, Qiao L, Sun X (2018) miR-3189–3p mimics enhance the effects of S100A4 siRNA on the inhibition of proliferation and migration of gastric cancer cells by targeting CFL2. Int J Mol Sci 19(1):236
Yu BB, Lin GX, Li L, Qu S, Liang ZG, Chen KH, Zhou L, Lu QT, Sun YC, Zhu XD (2018) Cofilin-2 acts as a marker for predicting radiotherapy response and is a potential therapeutic target in nasopharyngeal carcinoma. Med Sci Monit 24:2317–2329
Luo D, Wilson JM, Harvel N, Liu J, Pei L, Huang S, Hawthorn L, Shi H (2013) A systematic evaluation of miRNA:mRNA interactions involved in the migration and invasion of breast cancer cells. J Transl Med 11:57–57
Funding
None.
Author information
Authors and Affiliations
Contributions
LZ, WZ, XC conceived and designed the experiments; ZZ and JT performed the experiments, Funding acquisition; YS and FC contributed reagents/materials/analysis tools; XY wrote the paper, SL revised the paper All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no financial conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
404_2021_6149_MOESM1_ESM.tif
Supplementary file1 Fig. S1 Identification of location and stability of circ_0008673 in MCF-10AT and MCF-7 cells. A and B Cytoplasmic and nuclear circ_0008673 analysis assay was employed to demonstrate that circ_0008673 was chiefly located in cytoplasm of MCF-10AT and MCF-7 cells. C and D RNase R resistance analysis assay was used to assess the stability of circ_0008673. ***P < 0.001 (TIF 316 KB)
404_2021_6149_MOESM2_ESM.tif
Supplementary file2 Fig. S2 The expression of circ_0008673, miR-153-3p and CFL2 was detected by qRT-PCR or western blot in MCF-10AT or MCF-7. A and B The efficiency of circ_0008673, si-circ_0008673#1 and si-circ_0008673#2 in increasing or decreasing circ_0008673 expression was detected by qRT-PCR in MCF-10AT or MCF-7 cells. C MiR-153-3p expression was detected by qRT-PCR. D The effects of miR-153-3p inhibitors and mimics on miR-153-3p expression were detected by qRT-PCR in MCF-10AT or MCF-7 cells. E The efficiency of si-CFL2 and CFL2 in affecting CFL2 expression was determined by western blot assay in MCF-10AT or MCF-7 cells. ***P < 0.001 (TIF 438 KB)
404_2021_6149_MOESM3_ESM.tif
Supplementary file3 Fig. S3 Circ_0008673 silencing repressed breast cancer cell malignancy. A The effect of circ_0008673 silencing on cell viability was revealed by CCK-8 assay in MCF-7 cells. B Cell colony-formation assay was employed to determine the impact of circ_0008673 silencing on cell colony-forming ability in MCF-7 cells. C Flow cytometry analysis was employed to analyze the effect of circ_0008673 silencing on cell apoptosis in MCF-7 cells. D and E The effects of circ_0008673 silencing on cell migration and invasion were presented by transwell assay in MCF-7 cells. F The effects of circ_0008673 silencing on the protein expression of E-cadherin and N-cadherin were presented by western blot analysis in MCF-7 cells. **P < 0.01 and ***P < 0.001. (TIF 4681 KB)
404_2021_6149_MOESM4_ESM.tif
Supplementary file4 Fig. S4 Circ_0008673 depletion inhibited cell malignancy of breast cancer by interacting with miR-153-3p. A–F MCF-7 cells were severally transfected with si-con, si-circ_0008673#2, si-circ_0008673#2+in-miR-NC and si-circ_0008673#2+in-miR-153-3p. A Cell viability was detected by CCK-8 assay. B Cell colony-forming ability was determined by cell colony-formation assay. C Cell apoptosis was demonstrated by flow cytometry analysis. D and E The migration and invasion of MCF-7 cells were detected by transwell assay. F The protein expression of E-cadherin and N-cadherin was detected by western blot analysis. **P < 0.01 and ***P < 0.001 (TIF 585 KB)
404_2021_6149_MOESM5_ESM.tif
Supplementary file5 Fig. S5 MiR-153-3p mimics inhibited cell proliferation, migration and invasion, whereas promoted cell apoptosis by binding to CFL2. A–F MCF-7 cells were transfected with miR-NC, miR-153-3p, miR-153-3p+pcDNA and miR-153-3p+CFL2, respectively. A Cell viability was determined by CCK-8 assay. B Cell colony-forming ability was detected by cell colony-formation assay. C Cell apoptosis was analyzed by flow cytometry analysis. D and E Cell migratory and invasive ability were determined by transwell assay. F The protein levels of E-cadherin and N-cadherin were determined by western blot analysis. ***P < 0.001 (TIF 545 KB)
404_2021_6149_MOESM6_ESM.tif
Supplementary file6 Fig. S6 Circ_0008673 regulated CFL2 expression by associating with miR-153-3p. A The effects between circ_0008673 overexpression and miR-153-3p mimics on CFL2 protein expression were determined by western blot analysis in MCF-10AT cells. B The impacts between circ_0008673 silencing and miR-153-3p inhibitors on CFL2 protein expression were revealed by western blot analysis in MCF-7 cells. ***P < 0.001 (TIF 230 KB)
Rights and permissions
About this article
Cite this article
Zhang, L., Zhang, W., Zuo, Z. et al. Circ_0008673 regulates breast cancer malignancy by miR-153-3p/CFL2 axis. Arch Gynecol Obstet 305, 223–232 (2022). https://doi.org/10.1007/s00404-021-06149-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00404-021-06149-w