Abstract
Patients suffering from terminal-stage diabetic nephropathy (DN) are commonly diagnosed with kidney failure. The condition of DN patients gets generally improved by long-chain noncoding RNA (LncRNA) since it regulates microRNA (miR). The current study analyzes the role played by NEAT2/miR-206 upon cell death of renal tubular epithelial cells (RTECs), high glucose (HG)-induced inflammation and oxidative stress in diabetic nephropathy (DN). The researcher used high glucose (HG) to treat HK-2 cells in in vitro conditions to establish the DN cell model. qRT-PCR was used to confirm the transfection effect whereas the researcher also tested NEAT2, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain containing (NLRP3), caspase-1, interleukin IL-1β, gasdermin D (GSMDD)-N, and miR-206. To analyze the proteins in caspase-1, IL-1β, GSMDD-N, and NLRP3, Western blot technique was performed. The technique is also used to observe the pyroptosis. To identify TNF-α, IL-6, MCP-9, NEAT2, miR‐206, and NLRP3, dual‐luciferase reporter assay was conducted through ELISA kit to emphasize the correlation that exists among the above-mentioned factors. NEAT2 has been confirmed to have bound with miR-206 through double luciferase report experiments as well as RNA immunoprecipitation (RIP). NEAT2, present in HK-2 cells, was induced by HG. So, if NEAT2 is knocked down, it would mitigate TNF-α, IL-6, and MCP-9 as well. Among the HK-2 cells intervened with HG, the overexpressed miR-206 that was transfected into cells was in alignment with the modifications introduced in inflammatory factors and cytokines after NEAT2 is knocked down. The current study concludes that if NEAT2 is upregulated, it has the potential to retreat the inhibition of miR-206 on inflammatory response as well pyroptosis. Further, by targeting miR-206, NEAT2 has the potential to enhance HG-induced HK-2 pyroptosis. This miR-206 is predicted to be a latent target in the clinical treatment of DN.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The most common condition that leads to end-stage renal disease (ESRD) is diabetic nephropathy (DN) (Magee et al. 2017; Kanwar et al. 2011; Khajeh et al. 2021). DN is one of the most cost-incurring and life-threatening clinical burdens despite the availability of different treatment methods and diagnostic procedures. Studies have also identified medicinal plants for blood lipids and sugar control in diabetic and nondiabetic peoples (Kor et al. 2013; Zarei and Ghafaryan 2020; Ioannou 2017). Reports show that (Khajeh et al. 2021; Kor et al. 2013) about 30–40% of diabetic patients will deteriorate to DN, and about 50% of DN patients will eventually develop to ESRD. Patients can only undergo dialysis and kidney transplantation (Zarei and Ghafaryan 2020; Ioannou 2017; Kitada and Koya 2017) which undoubtedly increases the burden of patient environmental and genetic factors that are mutually influential in the incidence and exacerbation of this disease (Khajeh et al. 2021; Azizi et al. 2020).
In the progression of DN, as per recent study findings, noncoding RNAs (ncRNAs) play a crucial role owing to which it can potentially be utilized as biomarkers and therapeutic targets. ncRNA is a collective term that encompasses circular RNAs (circRNAs) (Azizi et al. 2020; St Laurent et al. 2015; Bhan et al. 2017), microRNAs (miRNAs), long noncoding RNAs (LncRNAs), and so on while these RNA species remain not to encode for proteins (St Laurent et al. 2015; Bhan et al. 2017). In both animal and human models of DN investigated earlier, the dysregulation of such ncRNA groups has been reported (St Laurent et al. 2015; Bhan et al. 2017; Zhang et al. 2019; Li et al. 2016). To be specific, few ncRNAs’ interaction attracts significant attention since it regulates the important steps in the progression of DN (St Laurent et al. 2015; Bhan et al. 2017). So, the current study discusses about the ncRNAs reported in DN earlier and how it interacts with critical genes in the progression of DN. The molecular mechanisms in DN can be understood precisely if the regulatory network of these ncRNAs is decoded. Further, this study may also help in leveraging the factors that regulate ncRNA network as novel biomarkers for DN and potential therapeutic targets. As per the literature, NEAT2-intervened high glucose (HG) tends to damage HK-2 cells and also induces epithelial–mesenchymal transition (Zhang et al. 2019; Li et al. 2016).
Inflammatory cell necrosis, or otherwise known as pyroptosis, is a newly found (Wang et al. 2020; Vande Walle and Lamkanfi 2016; Livak and Schmittgen 2001) programmed cell death method. According to the literature, nuclear paraspeckle assembly transcript (NEAT2) or otherwise called LncRNA metastasis‐associated lung adenocarcinoma transcript‐1 (MALTA1) tends to increment with DM in HK-2 cells treated with HG. So, cell pyroptosis (Zhang et al. 2019; Wang et al. 2020; Vande Walle and Lamkanfi 2016) gets inhibited, when NEAT2 gets downregulated. In recent years, a paradigm shift occurred toward the investigation of noncoding RNA (ncRNAs) especially long-chain noncoding RNA (LncRNA) has gained much attention (Wang et al. 2020; Vande Walle and Lamkanfi 2016). LncRNA-miR axis has been proved to increase oxidative stress as well as pyroptosis of renal tubular cells (Zhang et al. 2019; Vande Walle and Lamkanfi 2016). But, it remains opaque whether NEAT2 exhibits the same impact of inhibiting pyroptosis in renal tubular epithelial cells (RTECs), caused by HG. So, the current study investigates the role played by NEAT2 upon the HG-induced pyroptosis of RETCs and proposes novel therapy regimens for clinical applications.
Methods and Materials
Collection of Clinical Samples
For this study, the authors selected a total of 85 patients including 47 males and 38 females with average age of 46 ± 3.5 years, diagnosed with DN based on criteria defined before (Zhang et al. 2019) between January 2016 and December 2019 and grouped them under ‘patient group.’ On the other hand, healthy control group had a total of 45 healthy individuals including 27 males and 18 females with average age of 45 ± 5.5 years. Peripheral blood sample was collected from the patients and centrifuged to obtain serum samples for the purpose of detection. Informed consent was obtained from all study participants and the approval for conducting the study was obtained from Medical Ethics Committee. Patients who met this inclusion criteria were included: > 30 mg/g urinary protein/creatinine ratio with positive urine protein (the amount of urine protein was higher than 0.5 g/24 h) or urine albumin excretion rate was continuously greater than 200 μg/min and < 60 mL/min/1.73 m2 glomerular filtration rate (GFR) (Song et al. 2020; López-Campos et al. 2012 Mar). Exclusion criteria for the study are as follows: patients with malignant tumors and those who cannot cooperate for the investigation and statistics. Table 1 shows the baseline data of DN patients who are matched with healthy controls in terms of age and sex.
Cell Culture and Model Construction
The authors procured HK-2 from ATCC while the purchased cells were then cultured in RPMI 1640 (Cat. No. 11879020, Thermo fisher) with 10% fetal bovine serum, 100 μg/mL streptomycin, and 100 U/mL penicillin at 5% CO2 and 37 °C temperature. After the culture was made to starve for 24 h in glucose and serum-free media, control group (CG) and HG groups were established to incorporate HK-2 cells, induced by HG. Between the groups, CG received 5.5 mM glucose for 12 h, 24 h, and 48 h whereas HG received 30 mM glucose for 12 h, 24 h, and 48 h (Livak and Schmittgen 2001).
Cell Transfection
The authors used Lipofectamine 2000 kit for transfection purpose. Further, miR-206-mimics, -NEAT2#1, PcDNA3.1-NC, -NEAT2, si-NC, miR-NC, and -NEAT2#2 were procured from Sangon Biotech. The control group remained untransfected whereas HG group cells were transfected with these vectors. qRT-PCR was utilized to verify the transfection effect. Table 2 shows the primer sequences used in this study. The authors followed Song et al. (2020) for transfection procedures and rescue experiment (Song et al. 2020).
qRT-PCR Detection
As per the established protocol, Trizol reagent was used to extract serum and cells from the collected samples, in which RNA integrity was detected (Wang et al. 2019a). iScript™ Reverse Transcription Supermix kit was used to perform reverse transcription of total RNA (Bio-Rad, USA) to obtain cDNA. In this step, TransScript Green miRNA Two-Step qRT-PCR SuperMix was amplified with the help of TB Green™ Fast qPCR Mix (Takara, Japan) and ABI 7500 PCR (ABI, USA). At last, the results for qPCR were determined by 2−ΔΔCt. In this study, U6 was used as an internal control for miR whereas GAPDH was used for mRNA and LncRNA, respectively. Table 2 lists the primer sequences used in this study.
ELISA for Inflammatory Markers
After 48 h of transfection, the cells were collected and centrifuged and the supernatant was considered further to identify TNF-α, IL-6, and MCP-9. As per the studies conducted earlier (López-Campos et al. 2012), ELISA procedure was conducted following the recommended protocol. Serum samples were diluted to a final dilution of 1:50. 100 μL of each sample was added to a microtiter plate that was pre-coated with a monoclonal antibody specific for targets and incubated for 1 h. After five washes, a horseradish peroxidase-conjugated polyclonal antibody specific for targets was added for 1 h. The plate was washed five times, and the substrate solution was added for 15 min. Finally, a stop solution was added, and the optical density was read at 450 nm using a microplate reader (Tecan, Männedorf, Switzerland). The standard curve provided in the kit was used to determine the final concentration of targets in serum, and the concentration read from the standard curve was multiplied by the dilution factor.
Western Blot Test
After collecting the transfected cells, it was lysed using protein extraction reagent. Bovine serum albumin (BCA) kit was used in this study to test the protein. For this investigation, 35 μg protein sample was considered. The protein sample was made to undergo 10% SDS-PAGE gel electrophoresis. Afterward, the protein was shifted to PVDF, rinsed, and sealed with 5% defatted milk for an hour at 25 °C. Then, the membrane was put with primary antibodies and incubated. To counter horse radish peroxidase (HRP 1:5000)-coupled secondary antibodies, the membrane was cultured for an hour at 4 °C. Enhanced chemiluminescent reagent (procured from Thermo Fisher Science, Inc.) was used to observe the band. As per the study conducted earlier (Liu et al. 2019a), Western blotting was conducted.
Double Fluorescein Report
The researcher treated the complementary DNA fragments, which are inclusive of NEAT2-WT or NEAT2-mut fragments, with the downstream of luciferase gene in psi-CHECK2. The reagent procured from Invitogen, USA, was used to co-transfect the miR-206-mimicsNEAT2-WT or NEAT2-mut reporter vector. After 48 h of transfection, the authors conducted experiments for fireflies and renin luciferase activities (Wang et al. 2019a) 25.
RNA Immunoprecipitation (RIP)
The authors used EZMagna RIP kit and applied RIP to find the role played by NEAT2 and miR-206 upon latent binding protein Ago2. In this step, lysis of HK-2 was performed followed by its cultivation with protein A magnetic beads that were conjugated at 4 ℃. To wipe-away the proteins, after 6 h, the beads were rinsed, placed with 0.1% SDS/ 0.5 mg/mL protease K at 55 ℃ for 30 min. Finally, qRT-PCR was utilized to analyze the immunoprecipitated RNA so as to establish the presence of NEAT2 and miR-206 (Wang et al. 2019a, b).
Statistical Analysis
The study made use of GraphPad Prism software package 7.02 to illustrate the figures and to perform analysis. All the measurements are written as means ± SD. For inter-group comparison and multi-group comparison, the authors used independent sample t test and one-way ANOVA (denoted via F), respectively. For multi-time analysis, repeated measurement ANOVA was used while LSD-t test was utilized for post-event pairwise comparison. Further, Bonferroni was used for back testing while Pearson test was conducted to correlate the genes. If P < 0.05, the difference is said to be statistically significant.
Results
Elevated NEAT2 Expression in DN Patients and HG-intervened HK-2 Cells
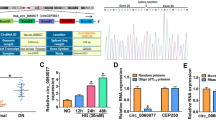
The findings from the qRT-PCR detection reveal NEAT2 in patients with DN is higher when compared with healthy individuals (Fig. 1a). Furthermore, the researchers identified HK-2 cultured with HG levels at different time period, wherein NEAT2 increased in a time-dependent manner in HK-2. Note that the increases in NEAT2 in HK-2 increased based on concentration and time dependency (Fig. 1b).
Knocking Down NEAT2 Obstructed HG-Intervened Inflammatory Factors in HK-2
The researchers further attempted to construct a si-NEAT2 sequence as a basis to observe NEAT2 on HK-2 which has HG intervention. Since si-NEAT2 has a silencing role, si-NEAT2#2 is considered by the researchers, which has considerable transfection effect as shown in Fig. 2a. We further examined the inflammatory factors and their concentration, which revealed that TNF-α, IL-6, and MCP-9 in HK-2, which were intervened by HG, tend to have increased; however, NF-α, IL-6, and MCP-9 in HG were obstructed post-si-NEAT2#2 transfection (Fig. 2b–d). The findings reveal the fact that NEAT2 inhibition could be considered as a key mechanism that reduces HK-2-based inflammatory reaction.
Effect of NEAT2 knock-down on HG-induced inflammatory response of HK-2 cells. a qRT-PCR detection of NEAT2 relative expression level after construction of si-NEAT2. b qRT-PCR detection of NEAT2 relative expression level in cells transfected with si-NEAT2#2. c-d Elsia detection of the relative expression levels of TNF-α, IL-6 and MCP-1 in cells transfected with si-NEAT2#2. ** indicates P < 0.01, **** indicates P < 0.001
Knocking Down NEAT2 Inhibited HG-induced Pyroptosis of HK-2 Cells
The researchers also carried out experiments that attempted to determine NEAT2 on HK-2 pyroptosis which has undergone HG intervention. Based on tests that examined cell pyroptosis, the researchers found significant increases in the levels of caspase-1, IL-1β, NLRP3, mRNA, and GSDMD-N protein. However, an important note is that the elevation in the expression levels of the components reversed because of NEAT2 knockdown. Furthermore, the results of the tests reveal that HG-induced protein expression level upregulation of the caspase-1, IL-1β, and NLRP3 in HK cells reverse due to si-NEAT2 (Fig. 3a, b, Supplementary Fig. 1a). All these findings suggest that NEAT2 could be a viable component that can enhance HG-induced HK-2 pyroptosis.
Effect of NEAT2 knock-down on HG-induced pyroptosis of HK-2 cells. a WB detection of the effect of NEAT2 knock-down on HG-induced pyroptosis protein in HK-2 cells. b qRT-PCR detection of the effect of NEAT2 knock-down on HG-induced pyroptosis-related factors in HK-2 cells. ** indicates P < 0.01, *** indicates P < 0.001
NEAT2 Targeted miR-206
The specific binding site of miR-206 and NEAT2 is revealed by bioinformatics study (data not shown) and expression of miR-206 in patients with DN displayed a downward trend, (Fig. 4a). The researchers identified the miR differences in HK-2 with HG intervention which revealed that miR-206 is the most significant when compared to others (Fig. 4b). Furthermore, the researchers performed double luciferase and RIP, which revealed that both NEAT2 and miR-206 are precipitated by Ago2 antibody as shown in Fig. 4c. Double luciferase reports revealed the obstruction of NEAT2-WT fluorescence activity by miR-206, which is revealed in Fig. 4. All these findings clearly reveal that miR-206 could be targeted by NEAT2.
Targeted binding of NEAT2 and miR-206. a qRT-PCR detection of miR-206 relative expression in DN patients and control group. b qRT-PCR detection of miR relative expression level in DN patients and control group. c Pearson test for the correlation between NEAT2 and miR-206 in serum of DN patients. d. The targeted binding of NEAT2 and miR-206 was confirmed by dual luciferase report. * indicates P < 0.05, *** indicates P < 0.001
miR-206 Elevation Could Obstruct Inflammatory Response to Cells After NEAT2 Overexpression and Enhance Cell Apoptosis
The researchers tested post-transfection efficiency and then performed construction of co-transfected cells to determine whether NEAT2 regulates miR-206 to influence the inflammatory responses based on HG intervention and pyroptosis which is caused due to HK-2 cells. The cells with mimic 206a had decreased in expression compared to negative control and PcDNA. The maximum expression was seen in PcDNA indicating that decreases in TNF-α, IL-6, and MCP-9 post-transfection of miR-206-mimics in HK-2 cells (Fig. 5a); however, WB and qRT-PCR findings revealed that NLRP3, caspase-1, IL-1β, and GSDMD-N protein and mRNA showed a significant increase in expression post-transfection of miR-206-mimics. It is important to note that the expression was brought down post-co-transfection of PcDNA. Note that when PcDNA-NEAT2 was compared with miR-NC and PcDNA, there were no significant differences (Fig. 5b, c and supplementary Fig. 1b.); however, miR-206a-mimic had high expression which was significant. The results hence reveal that through the regulation of miR-206, NEAT2 could considerably involve in RTEC pyroptosis.
NEAT2 participated in pyroptosis of renal tubular epithelial cells through miR-206. a Elisa was used to detect the relative expression of HG-induced inflammatory factors in HK-2 cells after co-transfection. b qRT-PCR detection of HG-induced pyroptosis related factor expression in HK-2 cells after co-transfection. * indicates P < 0.05, *** indicates P < 0.001
Discussion
Literature studies revealed the role played by LncRNAs and miRNA in the development of several conditions and diseases, such as inflammation, epithelial dysfunction, hypertrophy, and inflammation in the cellular level, as in diabetic neuropathy. Among all these conditions and diseases, it is important to note that LncRNA is examined with great attention and analyzed crucially, owing to its regulatory role that it plays in several diseases, especially in diseases dealing with metabolism. Findings from Liu et al. (2019) revealed that LncRNA NEAT2/ microRNA-382-3p/Resistin axis considerably reduces insulin resistance in patients with type-II diabetes (Liu et al. 2019a). However, other research claim that NEAT2 tends to have increased expression in several tumors and has elevations especially in patients with diabetes (Liu et al. 2019a; Zhang et al. 2020). NEAT2 mechanism in DN patients was not clear and had open venues for research. In this study, the researchers attempted to detect serum in HG-induced HK-2 and patients with DN, which revealed that through the knockdown of NEAT2, pyroptosis and inflammation reduced, and NEAT2 was found to be higher when compared with CG. Based on the findings, the researchers suggest that NEAT2 could be the possible target for the treatment of DN.
One of the important pathways for LncRNA for its participation in bodily functions in ceRNA (Salmena et al. 2011; Li et al. 2014). Findings of the several research studies in the past revealed that LncRNA could involve in the treatment of diabetes through the regulation of downstream binding miR (St Laurent et al. 2015; Bhan et al. 2017; Song et al. 2020). For instance, LncRNA TUG1 is deemed to enhance diabetic nephropathy through the obstruction of miR-21 (Wang et al. 2019a), which elevates TIMP3. Previous studies have also ascertained the fact that NEAT2 could enhance HK-2 cell inflammation and pyroptosis through HG intervention (Zhang et al. 2019; Song et al. 2020; Wang et al. 2019a); however, such an underlying mechanism is still not clear and requires research. Based on these findings, the researchers in the present paper postulated the fact that miR is combined with NEAT2 downstream. Five potential miRNAs were considered through the analysis of online websites (St Laurent et al. 2015; Zhang et al. 2019; Song et al. 2020). Furthermore, the researchers detected HK-2 with HG intervention and revealed that the difference showed by miR-206 is found to be significant when compared to miRNAs (Samaeekia et al. 2017; Yan et al. 2018a). Furthermore, miR-206 was found in regulated activity associated with glucokinase and in islets (Yan et al. 2018a; He et al. 2016). The findings of the study revealed the reduction in miR-206 in the serum of patients with DN, wherein the analysis of correlation revealed that NEAT2 and miR-206 have negative correlation, thereby suggesting a regulatory relationship between the two components (Samaeekia et al. 2017; Yan et al. 2018a). In addition, the researchers revealed the existence of a targeting relationship between miR-206 and NEAT2, as per the RIP and fluorescence activity reports. A newly identified focal cell death apoptosis pattern report is based on the release of several factors associated with inflammation post-rupture, which revealed that organs are subject to inflammation which aggravates the disease (He et al. 2016).
Studies also revealed that LncRNA NEAT2 activates the kidney tissue samples in HK-2 cells that are treated with HG, thereby promoting HK-injury that is induced through HG intervention through several other signaling pathways (Liu et al. 2019; Yan et al. 2018b). Also reported is that fact that LncRNA NEAT2 tends to facilitate HK-cell fibrosis by miR0145/ZEB2 axis that is HG-induced (Liu et al. 2019). LncRNA NEAT2 is also known for its renal tubular epithelial pyroptosis modulation in DN (Azizi et al. 2020; Song et al. 2020), which is also in line with other studies in the same arena, and per the findings of the present study. Several studies in the past have also revealed the inhibition of HK-2 cell apoptosis through LncRNA UCA1 in DN through the targeting of miR-206 (Yan et al. 2018b). However, it is important to note that no considerable research is available as a basis to prove the relationship that exists between pyroptosis and miR-206. To further explore the role of miR-206 on anti-pyroptosis, the researchers identified the presence of NLRP3, caspase-1, IL-1β, and GSDMD-N, which are pyroptosis markers, which indicate pyroptosis. Identification of biomarkers in an early stage of diabetic nephropathy (DN) can effectively inhibit its progression as study reports two novel renal tubular proteins (cyclophilin A and periostin) as potential markers for early prediction of DN relative to albuminuria with a higher diagnostic accuracy than urinary cyclophilin A specifically in overt DN stage (Abdel Ghafar et al. 2020).
Several previous research revealed that NLRP3 could act as a viable component for the activation of pro-caspase-1 for further cracking down into caspase-1, thereby activating pro-IL-1β for the acceleration of IL-1 β secretion; however, it is important to note that pro-IL-1β secretions in large amounts tend to induce inflammatory cascading reactions and tissue damages (Hughes and O'Neill 2018). The findings of the study also revealed that miR-206 upregulation can obstruct pyroptosis and could mitigate the appearance of inflammation-based reactions (He et al. 2016; Abdel Ghafar et al. 2020; Hughes and O'Neill 2018). However, it is important to note that NEAT2 in an overexpressed manner and elevations in miR-206 co-transfected cells tend to reverse pyroptosis. All these findings suggest the mediating effect of NEAT2 on miR-206 for the regulation of HG-induced pyroptosis of HK-2 cells (Wu et al. 2019; Yunqiao et al. 2014).
Through the present study, the researchers confirm the mediating effect of NEAT2 on miR-206 for the regulation of HG-induced HK-cell pyroptosis (Fig. 6). However, the study has several limitations. Firstly, the study has not conducted tests on the target genes that are miR-206 downstream. Secondly, the research samples considered for the study are few and hence the role of NEAT2 on DN is not explored. As a venue for future studies, the researchers open path for future researchers to further improve the findings of the study. To summarize, the researchers once again ascertain that NEAT2 can improve H-induced HK-2 pyroptosis through miR-206 targeting and reveals NEAT2 to be a latent DN target.
References
Abdel Ghafar MT, Shalaby KH, Okda HI et al (2020) Assessment of two novel renal tubular proteins in type 2 diabetic patients with nephropathy. J Investig Med 68:748–755
Azizi S, Andohjerdi RB, Mohajerani H (2020) Evaluation of two types of vitamin D receptor gene morphism in patients with type 2 diabetes and obesity. Int J Adv Biol Biomed Res 8(1):86–91
Bhan A, Soleimani M, Mandal SS (2017) Long noncoding RNA and cancer: a new paradigm. Cancer Res 77:3965–3981
He Y, Hara H, Nunez G (2016) Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci 41:1012–1021
Hughes MM, O’Neill LAJ (2018) Metabolic regulation of NLRP3. Immunol Rev 281:88–98
Ioannou K (2017) Diabetic nephropathy: is it always there? Assumptions, weaknesses and pitfalls in the diagnosis. Hormones (athens) 16:351–361
Kanwar YS, Sun L, Xie P, Liu FY, Chen S (2011) A glimpse of various pathogenetic mechanisms of diabetic nephropathy. Annu Rev Pathol 6:395–423
Khajeh H, Bahari A, Rashki A (2021) TCF7L2 polymorphisms in type 2 diabetes, insight from HRM and ARMS techniques. Int J Adv Biol Biomed Res 9(3):204–214
Kitada M, Koya D (2017) Diabetic nephropathy. Nihon Jinzo Gakkai Shi 59:38–42
Kor NM, Didarshetaban MB, Pour HRS (2013) Fenugreek (Trigonella foenum-graecum L.) as a valuable medicinal plant. Int J Adv Biol Biomed Res 1(8):922–931
Li JH, Liu S, Zhou H, Qu LH, Yang JH (2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42:D92-97
Li S, Geng J, Xu X, Huang X, Leng D, Jiang D, Liang J, Wang C, Jiang D, Dai H (2016) miR-130b-3p modulates epithelial-mesenchymal crosstalk in lung fibrosis by targeting IGF-1. PLoS ONE 11:e0150418
Liu SX, Zheng F, Xie KL, Xie MR, Jiang LJ, Cai Y (2019) Exercise reduces insulin resistance in type 2 diabetes mellitus via mediating the LncRNA MALAT1/MicroRNA-382-3p/Resistin axis. Mol Ther Nucleic Acids 18:34–44
Liu B, Qiang L, Wang GD, Duan Q, Liu J (2019) LncRNA MALAT1 facilities high glucose induced endothelial to mesenchymal transition and fibrosis via targeting miR-145/ZEB2 axis. Eur Rev Med Pharmacol Sci 23(8):3478–3486
Liu SX, Zheng F, Xie KL, Xie MR, Jiang LJ, Cai Y (2019a) Exercise reduces insulin resistance in type 2 diabetes mellitus via mediating the LncRNA MALAT1/MicroRNA-382-3p/resistin axis. Mol Ther Nucleic Acids 18:34–44
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
López-Campos JL, Arellano E, Calero C, Delgado A, Márquez E, Cejudo P, Ortega F, Rodríguez-Panadero F, Montes-Worboys A (2012) Determination of inflammatory biomarkers in patients with COPD: a comparison of different assays. BMC Med Res Methodol 31(12):40. https://doi.org/10.1186/1471-2288-12-40
Magee C, Grieve DJ, Watson CJ, Brazil DP (2017) Diabetic nephropathy: a tangled web to unweave. Cardiovasc Drugs Ther 31:579–592
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–358
Samaeekia R, Adorno-Cruz V, Bockhorn J, Chang YF, Huang S, Prat A, Ha N, Kibria G, Huo D, Zheng H, Dalton R, Wang Y, Moskalenko GY, Liu H (2017) miR-206 inhibits stemness and metastasis of breast cancer by targeting MKL1/IL11 pathway. Clin Cancer Res 23:1091–1103
Song J, Su ZZ, Shen QM (2020) Long non-coding RNA MALAT1 regulates proliferation, apoptosis, migration and invasion via miR-374b-5p/SRSF7 axis in non-small cell lung cancer. Eur Rev Med Pharmacol Sci 24:1853–1862
St Laurent G, Wahlestedt C, Kapranov P (2015) The Landscape of long noncoding RNA classification. Trends Genet 31:239–251
Vande Walle L, Lamkanfi M (2016) Pyroptosis. Curr Biol 26:R568–R572
Wang F, Gao X, Zhang R, Zhao P, Sun Y, Li C (2019a) LncRNA TUG1 ameliorates diabetic nephropathy by inhibiting miR-21 to promote TIMP3-expression. Int J Clin Exp Pathol 12:717–729
Wang T, Zhang Q, Liu M, Lu H, Lu H, Zhu J, Yuan Z, Li J (2019b) suPAR as a marker of diabetic nephropathy in patients with type 2 diabetes. Int J Clin Exp Med 12:4218–4225
Wang W, Jia Y-J, Yang Y-L, Xue M, Zheng Z-J, Wang L, Xue Y-M (2020) LncRNA GAS5 exacerbates renal tubular epithelial fibrosis by acting as a competing endogenous RNA of miR-96–5p. Biomed Pharmacotherapy. 121:109411
Wu K, Li J, Qi Y, Zhang C, Zhu D, Liu D, Zhao S (2019) SNHG14 confers gefitinib resistance in non-small cell lung cancer by up-regulating ABCB1 via sponging miR-206–3p. Biomed Pharmacother 116:108995
Yan Q, Tian Y, Hao F (2018a) Downregulation of LncRNA UCA1 inhibits proliferation and invasion of cervical cancer cells through miR-206 expression. Oncol Res. https://doi.org/10.3727/096504018X15185714083446
Yan Q, Tian Y, Hao F (2018b) Downregulation of LncRNA UCA1 inhibits proliferation and invasion of cervical cancer cells through miR-206 expression. Oncol Res 9:1
Yunqiao L, Vanke H, Jun X, Tangmeng G (2014) MicroRNA-206, down-regulated in hepatocellular carcinoma, suppresses cell proliferation and promotes apoptosis. Hepatogastroenterology 61:1302–1307
Zarei M, Ghafaryan H (2020) Alfa-Glucosidase inhibitory and antioxidant activity of hexane extract of flowers, leave and stems of Haplophyllum acutifolium DC. and Ferula haussknechtii Wolff ex Rech. Int J Adv Biol Biomed Res 8(2):153–164
Zhang J, Jiang T, Liang X, Shu S, Xiang X, Zhang W, Guo T, Xie W, Deng W, Tang X (2019) LncRNA MALAT1 mediated high glucose-induced HK-2 cell epithelial-to-mesenchymal transition and injury. J Physiol Biochem 75:443–452
Zhang Z, Li M, Zhang Z (2020) LncRNA MALAT1 modulates oxaliplatin resistance of gastric cancer via sponging miR-22-3p. Onco Targets Ther 13:1343–1354
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
El-Lateef, A.E.A., El-Shemi, A.G.A., Alhammady, M.S. et al. LncRNA NEAT2 Modulates Pyroptosis of Renal Tubular Cells Induced by High Glucose in Diabetic Nephropathy (DN) by via miR-206 Regulation. Biochem Genet 60, 1733–1747 (2022). https://doi.org/10.1007/s10528-021-10164-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-021-10164-6