Abstract
The involvement of circular RNAs (circRNAs) in the progression of diabetic nephropathy (DN) has been reported. However, the functions of circ_0060077 in DN remain unclear. HK-2 cells were treated with high glucose (HG) to establish DN cell model. Quantitative real-time polymerase chain reaction (qRT-PCR) was proceeded to determine the levels of circ_0060077, microRNA-145-5p (miR-145-5p) and vasorin (VASN). Cell counting kit-8 (CCK-8) assay, 5-ethynyl-2’-deoxyuridine (EdU) assay and colony formation assay were conducted to assess cell proliferation ability. Flow cytometry analysis was employed for cell apoptosis. The oxidative stress level was evaluated by commercial kits. Enzyme-linked immunosorbent assay (ELISA) was adopted to examine the concentrations of inflammatory factors. Western blot assay was utilized for protein levels. Dual-luciferase reporter assay and RNA pull-down assay were manipulated to analyze the relationships among circ_0060077, miR-145-5p and VASN. Circ_0060077 level was increased in DN patients and HG-stimulated HK-2 cells. Circ_0060077 knockdown ameliorated the inhibitory effect of HG on HK-2 cell proliferation and the promotional effects on cell apoptosis, oxidative stress, inflammation and fibrosis. MiR-145-5p was identified as the target for circ_0060077 and miR-145-5p inhibition ameliorated the effect of circ_0060077 silencing on HG-induced HK-2 cell injury. Moreover, miR-145-5p directly bound to VASN. Overexpression of miR-145-5p facilitated cell proliferation and repressed apoptosis, oxidative injury, inflammation and fibrosis in HG-induced HK-2 cells by targeting VASN. Circ_0060077 silencing protected HK-2 cells from HG-induced damage by regulating miR-145-5p/VASN axis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diabetic nephropathy (DN) is a serious complication that commonly existed in diabetic patients [1, 2]. A typical feature of DN is the accumulation of extracellular matrix (ECM) proteins, such as various collagen, laminin, fibronectin (FN) and the thickening of the glomerulus and tubular basement membrane, ultimately leading to tubulointerstitial fibrosis and glomerular sclerosis [3]. Although some progress has been made in recent years, the pathogenesis of DN has not been entirely understood [4, 5]. Therefore, investigating the potential molecular mechanisms of DN and discovering effective molecular therapeutic targets for DN are crucial.
Circular RNAs (circRNAs) are non-coding RNAs (ncRNAs) that are formed by back-splicing and distinguished by covalently-closed loops [6]. CircRNAs take part in the governing of gene expression via functioning as microRNA (miRNA) sponges [7]. Up to date, circRNAs have been gradually identified to play crucial regulatory functions in human diseases, including DN [8]. For instance, circ_0000712 knockdown alleviated high-glucose (HG)-stimulated SV40-MES13 cell damage in DN depending on the regulation of miR-879-5p and SOX6 [9]. Circ_0003928 deficiency relieved HG-caused HK-2 cell apoptosis and inflammatory injury by decoying miR-151-3p and modulating Anxa2 [10]. Circ_0060077 derived from the gene of centrosomal protein 250 (CEP250) and was upregulated in type 1 diabetes (T1DM) patients [11]. However, it is still unclear whether circ_0060077 participates in the development of DN.
In this work, the level of circ_0060077 in DN patients and DN cell model was determined. Furthermore, the function and potential mechanism of circ_0060077 in the progression of DN cell model were investigated.
Materials and Methods
Clinical Sample Acquisition
The peripheral blood samples were harvested from 25 DN patients and 25 healthy volunteers at The First Affiliated Hospital of Henan University of Science and Technology. The samples were stored at -80 °C until use. The study was approved by the Ethics Committee of The First Affiliated Hospital of Henan University of Science and Technology. The participants offered the written informed consents.
Cell Culture
Human kidney-2 (HK-2) cells were offered by Procell (CL-0109; Wuhan, China) and maintained at MEM (PM150410; Procell) plus 10% FBS (164,210; Procell) and 1% penicillin/streptomycin (PB180120; Procell) in a humid incubator containing 5% CO2.
Glucose was purchased from Sigma-Aldrich (49,163; St. Louis, MO, USA). For cell treatment, HK-2 cells were treated with high glucose (HG) (30 mM) for 12 h, 24 h and 48 h. The cells exposed to 30 mM glucose for 24 h were used to establish the cell model of DN. The cells exposed to normal glucose (NG) (5.5 Mm) were used as controls.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The RNA was obtained from cells and peripheral blood samples by using TRIzol (15596018; Invitrogen, Carlsbad, CA, USA), and then DNAs were eliminated by adding DNase (9003–98-9; Solarbio, Beijing, China). Thereafter, the RNAs were reversely transcribed into cDNAs via PrimeScript™ Reverse Transcriptase Kit (RR014A; Takara, Dalian, China) or miRNA 1st Strand cDNA Synthesis Kit (MR101-01; Vazyme, Nanjing, China) and either Random hexamer primers or Oligo (dT)18 primers. Next, SYBR green reagent (MQ101-01; Vazyme) was utilized for qRT-PCR and the 2−ΔΔCt strategy was utilized to assess the data with β-actin or U6 as the internal control. The primers are presented in Table 1. To analyze the ring feature of circ_0060077, RNase R assay was conducted on total RNA through the usage of RNase R (Epicenter Biotechnologies, Madison, WI, USA) and then the enrichment of circ_0060077 and CEP250 was detected.
Cell Transfection
To silence circ_0060077 expression, short interfering RNA against circ_0060077 (si-circ_0060077) was transiently transfected into HK-2 cells and related scramble control (si-NC) was used as control. MiR-145-5p mimic or miR-145-5p inhibitor was transiently transfected into HK-2 cells to increase or reduce miR-145-5p level with miR-NC mimic and miR-NC inhibitor used as controls. VASN overexpression vector (VASN) was transiently transfected into HK-2 cells to enhance VASN expression with vector used as a control. Cell transfection was done via lipofectamine 2000 (11668019; Invitrogen) according to the manufacturers’ instructions.
Cell Counting Kit-8 (CCK-8) Assay
To test cell viability, CCK-8 assay reagent (C0037; Beyotime, Shanghai, China) was utilized. In brief, the transfected HK-2 cells were added into 96-well plates and treated with HG for 48 h. Afterward, 10 μL CCK-8 was supplemented into the well for 2 h. Thereafter, the absorption at 450 nm was recorded.
5-Ethynyl-2’-deoxyuridine (EdU) Assay
To assess cell proliferation, the EdU kit (C10310; RIBOBIO, Guangzhou, China) was adopted. Shortly, after relevant treatment, HK-2 cells were plated into 24-well plates. Following EdU was added, the cells were fixed using paraformaldehyde (158127; Sigma-Aldrich) and maintained with 0.5% Triton-X-100 (93,443; Sigma-Aldrich). After that, EdU and DAPI (D9542; Sigma-Aldrich) were utilized to stain the nuclei. By using the fluorescence microscope (Olympus, Tokyo, Japan), the images were obtained and EdU-positive cells was quantified.
Colony Formation Assay
After HK-2 cells were inoculated into 12-well plates for 12 days, the formed colonies were stained with 0.1% crystal violet (V5265; Sigma-Aldrich). At last, the colony numbers were manually counted.
Flow Cytometry Analysis
By utilizing Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) Apoptosis Kit (A211-01; Vazyme), the apoptosis of HK-2 cells was estimated. In short, the collected HK-2 cells were resuspended in binding buffer and dyed with Annexin V-FITC and PI in the dark. After 15 min, the apoptotic cells were analyzed with flow cytometry.
Western Blot Assay
After isolation using RIPA buffer (P0013C; Beyotime), the proteins were subjected to SDS-PAGE electrophoresis for separation. Then the proteins were electroblotted onto PVDF membranes. After that, the membranes were blocked with 5% non-fat milk for 2 h, kept with primary antibodies overnight and secondary antibody (ab6721; 1:10,000; Abcam, Cambridge, MA, USA) for 2 h. The bands were exposed using ECL kit (E411-03; Vazyme). The primary antibodies included Bax (ab104156; 1:1000; Abcam), Bcl-2 (ab32124; 1:1000; Abcam), cleaved-caspase-3 (ab2302; 1:500; Abcam), collagen I (Col I; ab34710; 1:1000; Abcam), Col IV (ab236640; 1:1000; Abcam), fibronectin (FN; ab2413; 1:1000; Abcam),VASN (ab156868; 1:1000; Abcam) and β-actin (ab5694; 1:5000; Abcam).
Measurement of Malondialdehyde (MDA) Content and Superoxide Dismutase (SOD) Activity
The MDA content and SOD activity were examined utilizing MDA and SOD assay kits (MAK085; 19160; Sigma-Aldrich) according to the protocols.
Enzyme-Linked Immunosorbent Assay (ELISA)
The levels of IL-6, TNF-α, and IL-1β were examined with ELISA kits (ab178013; ab181421; ab214025; Abcam) in line with the manufacturers’ protocols.
Dual-Luciferase Reporter Assay
The sequences of circ_0060077 or VASN 3’UTR consisting of the wild-type or mutant miR-145-5p binding sites were introduced into pmirGLO (E1330; Promega, Fitchburg, WI, USA). Then, the generated vectors were introduced into HK-2 cells in combination with miR-NC mimic/miR-145-5p mimic. The luciferase signal was examined with the Dual-Luciferase Reporter Assay Reagent (E1910; Promega).
RNA Pull-Down Assay
The biotin-labeled probe of miR-145-5p, mutant miR-145-5p and control (Biotin-miR-145-5p, Biotin-miR-145-5p MUT and Biotin-NC) was synthesized by GenePharma (Shanghai, China) and then cultivated with streptavidin-coated magnetic beads (88,816; Invitrogen) for the generation of probe-covered beads. Next, the biotin-coupled RNA complexes were pulled down and bound RNAs were extracted. The enrichment of circ_0060077 and VASN in the bound fractions was quantified.
Statistical Analysis
All data were acquired from three independent experiments and analyzed using GraphPad Prism 7. The results were presented as mean ± SD. The difference between 2 groups was analyzed by Student’s t-test, while that among 3 groups was analyzed by one-way analysis of variance. P < 0.05 was thought as significant.
Results
Circ_0060077 Was Upregulated in DN Patients and HG-Treated HK-2 cells
Circ_0060077 derived from the exons 20–25 of CEP250 gene and possessed a mature length of 939 nt (Fig. 1A). The expression of circ_0060077 in DN patients was determined. The results showed that circ_0060077 was upregulated in the peripheral blood samples of DN patients compared to normal controls (Fig. 1B). Moreover, circ_0060077 level was increased in HG-treated HK-2 cells in a time-dependent manner (Fig. 1C). CircRNAs do not have 3’ polyadenylated tail; thus, we detected the existence of circ_0060077 in the reverse transcription products using random primers or oligo (dT)18 primers. The results showed that Oligo (dt)18 primers reversely transcribed cDNA could barely amplify circ_0060077 compared with Random primers reversely transcribed cDNA (Fig. 1D). RNase R assay indicated that circ_0060077 was resistant to RNase R digestion, while CEP250 was markedly digested by RNase R (Fig. 1E). These results suggested that circ_0060077 might play a role in DN development.
Circ_0060077 was highly expressed in DN patients and HG-stimulated HK-2 cells. (A) Circ_0060077 originated from the exons 20–25 of CEP250. (B) Circ_0060077 expression in DN patients and healthy volunteers was detected by qRT-PCR assay. (C) The expression of circ_0060077 in HG-treated HK-2 cells was determined by qRT-PCR assay. (D) The RNA in HK-2 cells was reversely transcribed into cDNA using random primers or Oligo (dt)18 primers, and then circ_0060077 and CEP250 levels were detected by qRT-PCR. (E) The RNA in HK-2 cells was treated with or without RNase R, and then circ_0060077 and CEP250 levels were determined by qRT-PCR. *P < 0.05.
Circ_0060077 Knockdown Reversed the Effects of Hg on Cell Proliferation, Apoptosis, Oxidative Stress, Inflammation and Fibrosis in HK-2 Cells
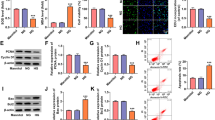
To explore the exact functions of circ_0060077, si-circ_0060077 was transfected into HK-2 cells to silence circ_0060077 expression. As indicated by qRT-PCR assay, the introduction of si-circ_0060077 markedly reduced circ_0060077 expression in HK-2 cells (Fig. 2A). CCK-8 assay showed that HG treatment led to a suppression in HK-2 cell viability, while circ_0060077 silencing reversed the effect (Fig. 2B). EdU assay showed that HG exposure repressed the proliferation of HK-2 cells, with circ_0060077 knockdown abrogated the effect (Fig. 2C). The results of colony formation assay exhibited the colony formation of HK-2 cells was inhibited by HG treatment, while the effect was weakened by reducing circ_0060077 (Fig. 2D). HG treatment led to a promotion in HK-2 cell apoptosis, whereas circ_0060077 deficiency reversed this effect (Fig. 2E). Then, we detected the levels of apoptosis-related proteins via western blot assay. It was found that HG treatment increased Bax and cleaved-caspase-3 levels and decreased Bcl-2 level in HK-2 cells, with circ_0060077 knockdown ameliorated the effects (Fig. 2F). We also found that HG exposure elevated MDA level and inhibited SOD activity in HK-2 cells, but circ_0060077 knockdown relieved the effects (Fig. 2G and H). The concentrations of IL-6, TNF-α and IL-1β were increased in HG-treated HK-2 cells, whereas si-circ_0060077 transfection abated the effects (Fig. 2I). Additionally, HG treatment increased the protein levels of fibrosis-related markers (FN, Col I and Col IV) in HK-2 cells, but circ_0060077 downregulation rescued the impacts (Fig. 2J). Taken together, silencing of circ_0060077 relieved HG-induced HK-2 cell damage.
Circ_0060077 knockdown promoted cell proliferation and inhibited apoptosis, oxidative stress, inflammation and fibrosis in HG-treated HK-2 cells. (A) The expression of circ_0060077 in HK-2 cells transfected with si-NC or si-circ_0060077 was detected by qRT-PCR assay. (B-J) HK-2 cells were divided into 4 groups: NG, HG, HG + si-NC and HG + si-circ_0060077. (B-E) The viability, proliferation, colony formation and apoptosis of HK-2 cells were assessed by CCK-8 assay, EdU assay, colony formation assay and flow cytometry analysis, respectively. (F) The protein levels of Bax, Bcl-2 and cleaved-caspase-3 in HK-2 cells were measured via western blot assay. (G and H) MDA content and SOD activity were examined by related commercial kits. (I) The concentrations of IL-6, TNF-α and IL-1β were examined with ELISA kits. (J) The protein levels of FN, Col I and Col IV in HK-2 cells were measured through western blot assay. *P < 0.05.
Circ_0060077 Directly Targeted miR-145-5p
To explore the potential mechanism of circ_0060077 in regulating HG-induced HK-2 cell damage, we searched circinteractome (https://circinteractome.irp.nia.nih.gov/) and found miR-145-5p was a target of circ_0060077 (Fig. 3A). As we observed in Fig. 3B, miR-145-5p mimic transfection increased miR-145-5p expression in HK-2 cells. Dual-luciferase reporter assay showed that miR-145-5p mimic transfection repressed the luciferase activity of circ_0060077 WT in HK-2 cells, but did not change the luciferase activity of circ_0060077 MUT (Fig. 3C). RNA pull-down assay showed that Biotin-miR-145-5p markedly pulled down circ_0060077 compared to Biotin-NC and Biotin-miR-145-5p MUT control groups (Fig. 3D). These results demonstrated the interaction between circ_0060077 and miR-145-5p. Furthermore, circ_0060077 knockdown remarkably increased miR-145-5p expression in HK-2 cells (Fig. 3E). Indeed, miR-145-5p was lowly expressed in DN patients’ peripheral blood samples and HG-treated HK-2 cells (Fig. 3F and G). Taken together, circ_0060077 directly targeted miR-145-5p to regulate miR-145-5p expression.
MiR-145-5p was targeted by circ_0060077. (A) Circ_0060077 contained miR-145-5p binding sites. (B) The expression of miR-145-5p in HK-2 cells transfected with miR-NC mimic or miR-145-5p mimic was determined by qRT-PCR. (C and D) Dual-luciferase reporter assay and RNA pull-down assay were performed to analyze the relationship between miR-145-5p and circ_0060077. (E) The expression of miR-145-5p in HK-2 cells transfected with si-NC or si-circ_0060077 was determined by qRT-PCR. (F) The expression of miR-145-5p in the peripheral blood samples of DN patients and healthy volunteers was detected by qRT-PCR. (G) The expression of miR-145-5p in HG-treated HK-2 cells was detected by qRT-PCR. *P < 0.05.
Circ_0060077 Knockdown Remitted HG-Induced HK-2 Cell Injury by Targeting miR-145-5p
As presented in Fig. 4A, miR-145-5p expression was repressed in HK-2 cells after miR-145-5p inhibitor transfection compared to miR-NC inhibitor transfection groups. As illustrated by CCK-8 assay, EdU assay and colony formation assay, circ_0060077 knockdown promoted cell viability, proliferation and colony formation in HG-exposed HK-2 cells, whereas these effects were overturned by downregulating miR-145-5p (Fig. 4B-D). Flow cytometry analysis showed that the suppressive role of circ_0060077 knockdown on HG-treated HK-2 cell apoptosis was reversed by miR-145-5p inhibition (Fig. 4E). Circ_0060077 knockdown decreased the protein levels of Bax and cleaved-caspase-3 and increased the protein level of Bcl-2 in HG-mediated HK-2 cells, whereas the addition of miR-145-5p inhibitor rescued the effect (Fig. 4F). Knockdown of circ_0060077 led to a reduction in MDA content and an enhancement of SOD activity in HG-triggered HK-2 cells, but these effects were rescued by miR-145-5p downregulation (Fig. 4G and H). Circ_0060077 deficiency-mediated suppressive effects on IL-6, TNF-α and IL-1β levels were also recovered by the inhibition of miR-145-5p in HG-treated HK-2 cells (Fig. 4I). Besides, the levels of FN, Col I and Col IV were decreased by circ_0060077 knockdown in HG-treated HK-2 cells, while miR-145-5p inhibition reversed the impacts (Fig. 4J). Collectively, circ_0060077 silencing relieved HG-induced HK-2 cell damage through regulating miR-145-5p expression.
Circ_0060077 regulated HG-mediated HK-2 cell proliferation, apoptosis, oxidative stress, inflammation and fibrosis by interacting with miR-145-5p. (A) The expression of miR-145-5p in HK-2 cells with miR-NC inhibitor or miR-145-5p inhibitor transfection was examined via qRT-PCR. (B-J) HK-2 cells were transfected with si-NC, si-circ_0060077, si-circ_0060077 + miR-NC inhibitor or si-circ_0060077 + miR-145-5p inhibitor and then treated with HG for 48 h. (B-E) HK-2 cell viability, proliferation, colony formation and apoptosis were evaluated by CCK-8 assay, EdU assay, colony formation assay and flow cytometry analysis, respectively. (F) The protein levels of Bax, Bcl-2 and cleaved-caspase-3 in HK-2 cells were measured via western blot assay. (G and H) MDA content and SOD activity in HK-2 cells were examined with kits. (I) The levels of IL-6, TNF-α and IL-1β in HK-2 cells were examined with ELISA kits. (J) The protein levels of FN, Col I and Col IV in HK-2 cells were measured through western blot assay. *P < 0.05.
VASN Served as the Target Gene of miR-145-5p
Subsequently, VASN was predicted to be the target gene of miR-145-5p by analyzing targetscan (http://www.targetscan.org/vert_71/), and the complementary sequences are exhibited in Fig. 5A. As indicated by dual-luciferase reporter assay, the luciferase activity of VASN 3’UTR WT but not VASN 3’UTR MUT was repressed by miR-145-5p mimic in HK-2 cells (Fig. 5B). RNA pull-down assay showed that VASN was enriched by Biotin-miR-145-5p compared to Biotin-NC and Biotin-miR-145-5p MUT groups (Fig. 5C). MiR-145-5p overexpression led to a reduction in VASN expression in HK-2 cells at mRNA and protein levels (Fig. 5D and E). Moreover, VASN mRNA and protein levels were increased in the peripheral blood samples of DN patients relative to normal controls (Fig. 5F and G). HG treatment also increased VASN mRNA and protein levels in HK-2 cells in a time-dependent way (Fig. 5H and I). To summarize, miR-145-5p regulated VASN expression in HK-2 cells by direct interaction.
VASN was the target of miR-145-5p. (A) The binding sites between VASN and miR-145-5p were exhibited. (B and C) The combination between VASN and miR-145-5p was demonstrated by dual-luciferase reporter assay and RNA pull-down assay. (D and E) The mRNA and protein levels of VASN in HK-2 cells transfected with miR-NC mimic or miR-145-5p mimic were measured by qRT-PCR or western blot assay. (F and G) The mRNA and protein levels of VASN in the peripheral blood samples of DN patients and normal controls were measured by qRT-PCR or western blot assay. (H and I) The mRNA and protein levels of VASN in HG-treated HK-2 cells were measured by qRT-PCR or western blot assay. *P < 0.05.
Overexpression of miR-145-5p Promoted Cell Proliferation and Suppressed Cell Apoptosis, Oxidative Stress, Inflammation and Fibrosis in HG-Treated HK-2 Cells
The overexpression vector of VASN transfection markedly increased VASN expression in HK-2 cells compared to vector control groups (Fig. 6A). Then the relationship between miR-145-5p and VASN in regulating HG-induced HK-2 cell progression was explored. The results of CCK-8 assay, EdU assay and colony formation assay exhibited that miR-145-5p overexpression facilitated cell viability, proliferation and colony formation in HG-stimulated HK-2 cells, while these effects were overturned by increasing VASN (Fig. 6B-D). As demonstrated by flow cytometry analysis, miR-145-5p overexpression led to an inhibition in HG-treated HK-2 cell apoptosis, while VASN upregulation weakened the effect (Fig. 6E). The protein levels of Bax and cleaved-caspase-3 were decreased, and the protein level of Bcl-2 was increased in HG-mediated HK-2 cells transfected with miR-145-5p mimic, with VASN overexpression abrogated the effects (Fig. 6F). The enhancement of miR-145-5p reduced MDA content and facilitated SOD activity in HG-treated HK-2 cells, whereas the effects were rescued by increasing VASN (Fig. 6G and H). The suppressive roles of miR-145-5p overexpression in IL-6, TNF-α and IL-1β levels in HG-mediated HK-2 cells were reversed by VASN overexpression vector transfection (Fig. 6I). Additionally, miR-145-5p overexpression decreased the protein levels of FN, Col I and Col IV in HG-triggered HK-2 cells, while VASN upregulation rescued the effects (Fig. 6J). Taken together, miR-145-5p overexpression relieved HG-mediated HK-2 cell injury by targeting VASN.
Overexpression of miR-145-5p suppressed HG-induced HK-2 cell damage by regulating VASN. (A) The protein level of VASN in HK-2 cells transfected with vector or VASN was measured via western blot assay. (B-J) HK-2 cells were transfected with miR-NC mimic, miR-145-5p mimic, miR-145-5p mimic + vector or miR-145-5p mimic + VASN and then treated with HG. (B-E) HK-2 cell viability, proliferation, colony formation and apoptosis were assessed by CCK-8 assay, EdU assay, colony formation assay and flow cytometry analysis, respectively. (F) The protein levels of Bax, Bcl-2 and cleaved-caspase-3 in HK-2 cells were measured through western blot assay. (G and H) The content of MDA and the activity of SOD were examined with kits. (I) The concentrations of IL-6, TNF-α and IL-1β were examined with ELISA kits. (J) The protein levels of FN, Col I and Col IV were measured by western blot assay. *P < 0.05.
Circ_0060077 Regulated VASN Expression by Targeting miR-145-5p
Thereafter, the relationships among circ_0060077, miR-145-5p and VASN were estimated. Our results exhibited that circ_0060077 silencing reduced the mRNA and protein levels of VASN in HG-treated HK-2 cells, while miR-145-5p inhibitor transfection ameliorated the effects (Fig. 7A and B). These results indicated that circ_0060077 positively regulated VASN expression by sponging miR-145-5p.
Circ_0060077 targeted miR-145-5p to regulate VASN expression. (A and B) HK-2 cells were transfected with si-NC, si-circ_0060077, si-circ_0060077 + miR-NC inhibitor or si-circ_0060077 + miR-145-5p inhibitor and stimulated with HG, and then VASN mRNA and protein levels were measured by qRT-PCR and western blot assay, respectively. *P < 0.05.
Discussion
Currently, abundant circRNAs have been discovered and demonstrated to serve as vital regulators in the development of renal diseases [20, 21]. In this work, we mainly explored the impacts of circ_0060077 on DN development. HK-2 cells were stimulated with HG to mimic DN in vitro, as previously reported [22, 23]. In this study, HG treatment resulted in a loss of cell proliferation and a stimulation of apoptosis, oxidative stress, inflammation and fibrosis in HK-2 cells, indicating DN cell model was constructed.
Several circRNAs were reported to be abnormally expressed in DN and involved in DN progression. For instance, circ_0080425 was elevated in DN in vivo and in vitro model and facilitated cell fibrosis in MCs by targeting FGF11 [24]. Circ-AKT3 was weakly expressed DN mice model and HG-treated MCs, and inhibited the accumulation of extracellular matrix in MCs depending on miR-296-3p/E-cadherin pathway [25]. The heatmap showed that circ_0060077 was upregulated in T1DM patients [11]. However, the exact roles of circ_0060077 in DN are not clear. Herein, circ_0060077 level was enhanced in DN patients and HG-triggered HK-2 cells. Moreover, circ_0060077 deficiency provoked HG-mediated HK-2 cell proliferation and suppressed cell apoptosis, oxidative damage, inflammatory response and fibrosis. Taken together, circ_0060077 knockdown alleviated HG-induced HK-2 cell impairments, illustrating that circ_0060077 might play a promotional role in DN.
CircRNAs can sponge miRNAs to alter gene expression and then regulate diverse biological progresses in human diseases [7]. Moreover, miRNAs play a crucial role in DN progression by targeting related mRNAs [12]. For example, miR-483-5p mitigated the renal tubular impairment in DN by interacting with HDCA4 [13]. MiR-17-5p interference ameliorated HG-caused cell apoptosis and fibrosis in human mesangial cells (HMCs) by binding to KIF23 and inactivating Wnt/β-catenin pathway [14]. Thus, we further explored the underlying mechanism of circ_0060077 in DN. As a result, circ_0060077 was found to target miR-145-5p. Wei et al. reported that miR-145-5p relieved HG-stimulated podocyte apoptosis in DN via Notch pathway [15]. Moreover, miR-145 participated in HG-mediated MC proliferation, fibrosis and apoptosis via lncRNA MEG3/miR-145 pathway [26] or lncRNA MALAT1/miR-145/ZEB2 pathway [27]. In this research, miR-145-5p deletion was able to rescue the impact of circ_0060077 silencing on HG-induced HK-2 damage. Additionally, overexpression of miR-145-5p aggravated cell growth and restrained apoptosis, oxidative stress, inflammation and fibrosis in HG-triggered HK-2 cells.
Vasorin (VASN) is a type I transmembrane glycoprotein and plays vital regulatory roles in human diseases, such as glioma [16], thyroid cancer [17] and prostate cancer [18]. Moreover, VASN level was found to be abnormally increased in DN [19]. In the present study, VASN was demonstrated to be the target gene of miR-145-5p. Furthermore, VASN overexpression ameliorated miR-145-5p-mediated cell damage in HG-treated HK-2 cells.
However, there were some limitations of this study, For example, the number of samples was limited and there was the lack of the animal experiments.
In summary, circ_0060077 sponged miR-145-5p to promote VASN level, thereby suppressing cell viability and promoting apoptosis, oxidative stress, inflammation and fibrosis in HG-stimulated HK-2 cells (Fig. 8). These findings might provide a preclinical basis for DN therapy.
Availability of Data and Materials
All data generated or analyzed during this study are included in the article.
Change history
16 September 2022
A Correction to this paper has been published: https://doi.org/10.1007/s10753-022-01740-y
References
Magee, C., D.J. Grieve, C.J. Watson, and D.P. Brazil. 2017. Diabetic Nephropathy: A Tangled Web to Unweave. Cardiovascular Drugs and Therapy 31 (5–6): 579–592. https://doi.org/10.1007/s10557-017-6755-9.
Rossing P. 2006. Prediction, progression and prevention of diabetic nephropathy. The Minkowski Lecture 2005. Diabetologia 49 (1): 11–19. https://doi.org/10.1007/s00125-005-0077-3.
Loeffler, I., and G. Wolf. 2015. Epithelial-to-Mesenchymal Transition in Diabetic Nephropathy: Fact or Fiction? Cells 4 (4): 631–652. https://doi.org/10.3390/cells4040631.
Ni, W.J., L.Q. Tang, and W. Wei. 2015. Research progress in signalling pathway in diabetic nephropathy. Diabetes/Metabolism Research and Reviews 31 (3): 221–233. https://doi.org/10.1002/dmrr.2568.
Alicic, R.Z., M.T. Rooney, and K.R. Tuttle. 2017. Diabetic Kidney Disease: Challenges, Progress, and Possibilities. Clinical Journal of the American Society of Nephrology 12 (12): 2032–2045. https://doi.org/10.2215/CJN.11491116.
Ebbesen, K.K., T.B. Hansen, and J. Kjems. 2017. Insights into circular RNA biology. RNA Biology 14 (8): 1035–1045. https://doi.org/10.1080/15476286.2016.1271524.
Hansen, T.B., T.I. Jensen, B.H. Clausen, J.B. Bramsen, B. Finsen, C.K. Damgaard, and J. Kjems. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495 (7441): 384–388. https://doi.org/10.1038/nature11993.
Han, B., J. Chao, and H. Yao. 2018. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacology & Therapeutics 187: 31–44. https://doi.org/10.1016/j.pharmthera.2018.01.010.
Zhao, L., H. Chen, Y. Zeng, K. Yang, R. Zhang, Z. Li, T. Yang, and H. Ruan. 2021. Circular RNA circ_0000712 regulates high glucose-induced apoptosis, inflammation, oxidative stress, and fibrosis in (DN) by targeting the miR-879-5p/SOX6 axis. Endocrine Journal. https://doi.org/10.1507/endocrj.EJ20-0739.
An, L., D. Ji, W. Hu, J. Wang, X. Jin, Y. Qu, and N. Zhang. 2020. Interference of Hsa_circ_0003928 Alleviates High Glucose-Induced Cell Apoptosis and Inflammation in HK-2 Cells via miR-151-3p/Anxa2. Diabetes, Metabolic Syndrome and Obesity 13: 3157–3168. https://doi.org/10.2147/DMSO.S265543.
Yang, L., X. Han, C. Zhang, C. Sun, S. Huang, W. Xiao, Y. Gao, Q. Liang, F. Luo, W. Lu, J. Fu, and Y. Zhou. 2020. Hsa_circ_0060450 Negatively Regulates Type I Interferon-Induced Inflammation by Serving as miR-199a-5p Sponge in Type 1 Diabetes Mellitus. Frontiers in Immunology 11: 576903. https://doi.org/10.3389/fimmu.2020.576903.
Tang, J., D. Yao, H. Yan, X. Chen, L. Wang, and H. Zhan. 2019. The Role of MicroRNAs in the Pathogenesis of Diabetic Nephropathy. International Journal of Endocrinology 2019: 8719060. https://doi.org/10.1155/2019/8719060.
Liu, L., H. Chen, J. Yun, L. Song, X. Ma, S. Luo, and Y. Song. 2021. miRNA-483-5p Targets HDCA4 to Regulate Renal Tubular Damage in Diabetic Nephropathy. Hormone and Metabolic Research. https://doi.org/10.1055/a-1480-7519.
Chen, X., L. Gu, X. Cheng, J. Xing, and M. Zhang. 2021. MiR-17-5p downregulation alleviates apoptosis and fibrosis in high glucose-induced human mesangial cells through inactivation of Wnt/beta-catenin signaling by targeting KIF23. Environmental Toxicology. https://doi.org/10.1002/tox.23280.
Wei, B., Y.S. Liu, and H.X. Guan. 2020. MicroRNA-145-5p attenuates high glucose-induced apoptosis by targeting the Notch signaling pathway in podocytes. Experimental and Therapeutic Medicine 19 (3): 1915–1924. https://doi.org/10.3892/etm.2020.8427.
Liang, W., B. Guo, J. Ye, H. Liu, W. Deng, C. Lin, X. Zhong, and L. Wang. 2019. Vasorin stimulates malignant progression and angiogenesis in glioma. Cancer Science 110 (8): 2558–2572. https://doi.org/10.1111/cas.14103.
Bhandari, A., Y. Guan, E. Xia, Q. Huang, and Y. Chen. 2019. VASN promotes YAP/TAZ and EMT pathway in thyroid carcinogenesis in vitro. American Journal of Translational Research 11 (6): 3589–3599.
Cui F.L., A.N., Mahmud, Z.P. Xu, Z.Y. Wang, and J.P. Hu. 2020. VASN promotes proliferation of prostate cancer through the YAP/TAZ axis. European Review for Medical and Pharmacological Sciences 24 (12): 6589–6596. https://doi.org/10.26355/eurrev_202006_21644.
Ahn, J.M., B.G. Kim, M.H. Yu, I.K. Lee, and J.Y. Cho. 2010. Identification of diabetic nephropathy-selective proteins in human plasma by multi-lectin affinity chromatography and LC-MS/MS. Proteomics. Clinical Applications 4 (6–7): 644–653. https://doi.org/10.1002/prca.200900196.
Jin, J., H. Sun, C. Shi, H. Yang, Y. Wu, W. Li, Y.H. Dong, L. Cai, and X.M. Meng. 2020. Circular RNA in renal diseases. Journal of Cellular and Molecular Medicine 24 (12): 6523–6533. https://doi.org/10.1111/jcmm.15295.
Zhang, J.R., and H.J. Sun. 2020. Roles of circular RNAs in diabetic complications: From molecular mechanisms to therapeutic potential. Gene 763: 145066. https://doi.org/10.1016/j.gene.2020.145066.
Meng, Q., X. Zhai, Y. Yuan, Q. Ji, and P. Zhang. 2020. lncRNA ZEB1-AS1 inhibits high glucose-induced EMT and fibrogenesis by regulating the miR-216a-5p/BMP7 axis in diabetic nephropathy. Brazilian Journal of Medical and Biological Research 53 (4): e9288. https://doi.org/10.1590/1414-431X20209288.
Wang, J., and S.M. Zhao. 2021. LncRNA-antisense non-coding RNA in the INK4 locus promotes pyroptosis via miR-497/thioredoxin-interacting protein axis in diabetic nephropathy. Life Sciences 264: 118728. https://doi.org/10.1016/j.lfs.2020.118728.
Liu, H., X. Wang, Z.Y. Wang, and L. Li. 2020. Circ_0080425 inhibits cell proliferation and fibrosis in diabetic nephropathy via sponging miR-24-3p and targeting fibroblast growth factor 11. Journal of Cellular Physiology 235 (5): 4520–4529. https://doi.org/10.1002/jcp.29329.
Tang, B., W. Li, T.T. Ji, X.Y. Li, X. Qu, L. Feng, and S. Bai. 2020. Circ-AKT3 inhibits the accumulation of extracellular matrix of mesangial cells in diabetic nephropathy via modulating miR-296-3p/E-cadherin signals. Journal of Cellular and Molecular Medicine 24 (15): 8779–8788. https://doi.org/10.1111/jcmm.15513.
Li, J., X. Jiang, L. Duan, and W. Wang. 2019. Long non-coding RNA MEG3 impacts diabetic nephropathy progression through sponging miR-145. American Journal of Translational Research 11 (10): 6691–6698.
Liu B., L. Qiang, G.D. Wang, Q. Duan, and J. Liu. 2019. LncRNA MALAT1 facilities high glucose induced endothelial to mesenchymal transition and fibrosis via targeting miR-145/ZEB2 axis. European Review for Medical and Pharmacological Sciences 23 (8): 3478–3486. https://doi.org/10.26355/eurrev_201904_17713.
Acknowledgements
None.
Funding
No funding support.
Author information
Authors and Affiliations
Contributions
Jinjin Zhou participated in the design of the work, methodology, data interpretation, and analysis for the work, carried out the statistical analysis and drafted the manuscript. Xia Peng participated in the methodology, data interpretation and analysis for the work. Yanhai Ru and Jiayun Xu participated in data interpretation and methodology. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical Approval
The research related to human use has been complied with all the relevant national regulations, institutional policies and in accordance the tenets of the Helsinki Declaration and has been approved by the Ethics Committee of The First Affiliated Hospital of Henan University of Science and Technology.
Consent for Publication
All patients in this study provided their consent for publication.
Competing Interests
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The corresponding author of this article should be Jiayun Xu, jiayunxu23560@163.com.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, J., Peng, X., Ru, Y. et al. Circ_0060077 Knockdown Alleviates High-Glucose-Induced Cell Apoptosis, Oxidative Stress, Inflammation and Fibrosis in HK-2 Cells via miR-145-5p/VASN Pathway. Inflammation 45, 1911–1923 (2022). https://doi.org/10.1007/s10753-022-01649-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-022-01649-6