Abstract
Background
Diabetic nephropathy (DN) is a main cause of end-stage renal disease with high mortality. Circular RNAs (circRNAs) are associated with the pathogenesis of DN. This study aimed to explore the role of circLARP1B in DN.
Methods
The levels of circLARP1B, miR-578, TLR4 in DN and high glucose (HG)-treated cells using quantitative real-time PCR. Their relationship was analyzed using dual-luciferase reporter assay. The biological behaviors were assessed by MTT assay, EDU assay, flow cytometry, ELISA, and western blot.
Results
The results indicated that circLARP1B and TLR4 were highly expressed, and miR-578 was low expressed in patients with DN and HG-induced cells. Knockdown of circLARP1B promoted the proliferation and cell cycle, and inhibited pyroptosis and inflammation of HG-induced cells. CircLARP1B is a sponge of miR-578, which targets TLR4. Rescue experiments showed that inhibition of miR-578 reversed the effects of circLARP1B knockdown, while TLR4 reversed the effects of miR-578.
Conclusion
CircLARP1B/miR-578/TLR4 axis suppressed the proliferation, blocked cell cycle at the G0-G1 phase, promoted pyroptosis, and inflammatory factor release of renal mesangial cells induced by HG. The findings suggested that circLARP1B may be a target for the treatment of DN.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diabetic nephropathy (DN) is an important complication in diabetic patients, leading to end-stage renal disease (ESRD). Hyperglycemia is a main cause of DN and a driving factor for DN to develop into ESRD [1]. It is characterized by glomerular hypertrophy, proteinuria, decreased glomerular filtration, and even renal fibrosis [2]. The incidence and mortality of DN are increasing [3]. The management of DN is to control blood glucose, blood pressure, and cardiovascular event [4]. However, due to the complex pathogenesis of DN, the current treatment strategy still cannot effectively reduce DN-caused mortality.
Non-coding RNAs are RNAs that cannot be translated to proteins that modulate inflammation, proliferation, and cell death in DN [5]. Circular RNAs (circRNAs) are a kind of non-coding RNAs that has cyclic structure without cup and tail. They commonly serve their functions by acting as miRNA sponges. CircRNAs play a crucial role in several renal diseases, associated with DN, hypertension, kidney injury, and renal cell cancer [6]. Accumulating evidence has reported that circRNAs regulate cellular functions in DN. For instance, circ_010383 knockdown promotes extracellular matrix (ECM) accumulation and kidney fibrosis [7]. Circ_0000285 is increased in high-glucose (HG) induced podocytes and promotes podocyte injury [8]. Overexpression of circ-LARP4 inhibited proliferation and induced apoptosis of mouse mesangial cells [9]. However, the role of circRNA La ribonucleoprotein 1B (circ-LARP1B) in DN remains unclear.
Pyroptosis is a newly discovered way of pro-inflammatory programmed cell death. It is mediated by the Gasdermins (GSDMs) protein family. Pyroptosis-induced inflammation is associated with DN. Inflammatory factors are abnormally expressed and act as biomarkers to identify DN [10]. Activation of inflammasome NLRP3 induces pyroptosis, and aberrant pyroptosis promotes the pathogenesis of DN [11]. Non-coding RNAs participate in inflammasome activation, regulating the pyroptosis mechanism in DN [12]. However, the effects of circLARP1B on pyroptosis and related inflammation are unknown.
Herein, we investigated the role of circLARP1B in DN. We established an HG-induced cell model and identified the effects of circLARP1B on cell proliferation, cell cycle, pyroptosis, and inflammation. We found circLARP1B inhibited the proliferation, blocked cell cycle at the G0-G1 phase, promoted pyroptosis and inflammation of HG-induced cells by the miR-578/TLR4 axis. The findings suggested that low expressed circLARP1B contributes to attenuating the progression of DN.
Materials and methods
Clinical specimens
Patients with DN (n = 24) and healthy subjects (n = 24) were enrolled in this study. Blood samples were collected from all subjects. Patients with obesity, other metabolic diseases, inflammation, blood diseases, and cancers were excluded. The study was approved by the Ethics Committee of The First Affiliated Hospital of Xi’an Medical University. Written informed consent was obtained from all subjects.
Cell culture and treatment
Human renal mesangial cells were purchased from Procell (Wuhan). The cells were cultured in the complete culture medium for human mesangial cells (Procell) in a 37 °C incubator with 5% CO2.
To establish high glucose (HG)-induced cell model, mesangial cells were treated with 30 mM d-glucose (Sigma-Aldrich). The cells in the control group were treated with 5 mM d-glucose.
Cell transfection
Vector, circLARP1B overexpression vector, TLR4 overexpression vector, sh-NC, sh-circLARP1B, miR-578 (miR-578 mimic), miR-NC (mimic NC), miR-578 inhibitor, inhibitor NC were acquired from Genepharma. Cell transfection was performed using Lipofectamine 3000 (Invitrogen) for 48 h.
RNase R treatment
Total RNA was isolated using TRIzol reagent (Invitrogen). Total RNA (2 μg) was incubated with 2 μL RNase R (20 U/μL). In the mock group, total RNA (2 μg) was incubated with 2 μL DEPC water. After incubation at 37 °C for 30 min, the expression of circLARP1B and linear LARP1B was measured using quantitative real-time PCR (qPCR).
Subcellular localization of circLARP1B
The nucleus and cytoplasm of renal mesangial cells were separated using the nuclear/cytosolic fractionation kit (Cell Biolabs) following the manufacturer’s protocol. The expression of circLARP1B was detected in the nucleus and cytoplasm using qPCR, respectively. U6 was the internal control of the nucleus, and GAPDH was the internal control of the cytoplasm.
qPCR
Total RNA was extracted from cells using the TRIzol reagent. Total RNA was reverse transcribed to cDNA using the Quant cDNA First Strand Synthesis Kit (TIANGEN). Quantitative PCR was performed using the SuperReal Premix Plus (SYBR Green) (TIANGEN) on ABI PRISM 7300 real-time PCR system. U6 was the internal control of miR-578, and GAPDH was the internal control of circLARP1B and TLR4. RNA levels were calculated using the 2−ΔΔCt method.
MTT assay
MTT assay was performed using the MTT Cell Proliferation and Cytotoxicity Assay Kit (Beyotime). Briefly, cells were seeded into 96-well plates and incubated at 37 °C with 5% CO2 for 24 h. MTT solution (5 mg/mL; 10 μL) was added to incubate for 2 h. Formazan solution (100 μL) was added until formazan dissolved. The absorbance was detected at 570 nm using a microplate reader (Bio-Tek).
EDU assay
EDU assay was performed using the BeyoClick™ EDU-488 cell proliferation detection kit (Beyotime). Briefly, cells were incubated with 10 μM EDU working solution for 2 h. Then, the cells were fixed with 4% paraformaldehyde and treated with 0.3% Triton X-100. After incubating with click additive solution, the cell nucleus was stained with DAPI for 10 min. The fluorescence signal was detected using a fluorescence microscope.
Cell cycle analysis
Cell cycle was conducted using a cell cycle detection kit (JianCheng). Cells were washed and resuspended using PBS. Then the cells were incubated with cold ethanol at 4 °C overnight. After centrifuging at 2000 rpm for 5 min, the cells were incubated with 100 μL RNase A at 37 °C for 30 min, followed by incubation with 400 μL PI at 4 °C for 30 min. The cell cycle was detected using a flow cytometer (BD science).
Pyroptosis determination
The cells were collected after centrifugation and incubated with 5 μL FLICA labeled caspase1 for 1 h, followed by incubation with 5 μL PI for 15 min. Pyroptosis was measured using a flow cytometer.
ELISA
According to the manufacturer’s protocols, the levels of IL-6, IL-10, TNF-α, IL-1β, and IL-18 in collected cell supernatants using specific ELISA kits.
Western blot
Cells were lysed in RIPA lysis buffer (KeyGEN). Following measuring protein concentration using a BCA kit (Beyotime). Equal proteins were run on SDS–polyacrylamide gel and transferred to the PVDF membrane (Millipore). After blocking with 5% skim milk, the membranes were incubated with primary antibodies at 4 °C overnight, followed by incubated with secondary antibody at 37 °C for 1 h. The bands were observed using the ECL reagent (Beyotime).
Dual-luciferase reporter analysis
The 3′-UTR of circLARP1B or TLR4 containing the potential binding sites were cloned into pGL3 vectors (Promega) to obtain wild-type plasmids (WT-circLARP1B or WT-ESR1), Similarly, the mutant sequences of circLARP1B or TLR4 were synthesized and inserted into pGL3 vectors (MUT-circLARP1B or MUT-ESR1). Renal mesangial cells were seeded into 24-well plates and co-transfected with WT or MUT plasmids and mimic or mimic NC using Lipofectamine 3000. Luciferase activity was measured 48 h post-transfection using the Dual-Luciferase reporter assay system (Promega).
Data analysis
All experiments were repeated three times. Data analyses were performed using GraphPad Prism 7.0 software and the results are shown as mean ± SD. Comparisons were analyzed using the unpaired Student's t-test (two groups) or one-way ANOVA (multiple groups). P < 0.05 means a statistically significant difference.
Results
CircLARP1B is highly expressed in DN and HG-induced cells
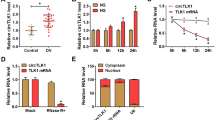
We first detected the levels of circLARP1B in clinical samples. The data showed that circLARP1B expression was increased in the blood of patients with DN, compared with normal individuals (Fig. 1A). Similarly, circLARP1B expression was higher in HG-treated cells than the control cells (Fig. 1B). CircLARP1B cannot be degraded by RNase R, whereas liner LARP1B was significantly degraded by RNase R (Fig. 1C). CircLARP1B was mostly present in the cytoplasm rather than in the nucleus (Fig. 1D).
CircLARP1B is highly expressed in DN and HG-induced cells. A The blood samples were collected from patients with DN and healthy control, and circLARP1B expression was detected using qPCR. B Renal mesangial cells were treated with 5 mM or 30 mM d-glucose (control or HG), and circLARP1B expression was examined using qPCR. C The levels of circLARP1B and linear LARP1B were measured by qPCR after treating with RNase R treatment. D The levels of circLARP1B were detected in the nucleus and cytoplasm using qPCR. U6 and GAPDH were the internal control in the nucleus and cytoplasm. ***P < 0.001
CircLARP1B inhibited HG-induced proliferation and cell cycle, and promoted pyroptosis and inflammation
To explore the functional role of circLARP1B in HG-induced cells, we overexpressed or knockdown circLARP1B. The levels of circLARP1B were upregulated after transfection with circLARP1B overexpression vector, whereas circLARP1B was downregulated after transfection with sh-circLARP1B (Fig. 2A). HG inhibited cell viability, and circLARP1B inhibited HG-induced cell viability, whereas knockdown of circLARP1B promoted HG-induced cell viability (Fig. 2B). Cell proliferation was inhibited by HG and further inhibited by circLARP1B and promoted by knockdown of circLARP1B (Fig. 2C and D). Overexpression of circLARP1B increased HG-induced cells in the G0-G1 phase and decreased HG-induced cells in the S phase, whereas knockdown of circLARP1B had the opposite results (Fig. 2E). Inversely, cell pyroptosis was induced by HG, which was facilitated by circLARP1B and was suppressed by sh-circLARP1B (Fig. 2F and G). The levels of IL-6, TNF-α, IL-1β, and IL-18 were elevated and the levels of IL-10 were reduced by circLARP1B overexpression in HG-induced cells, whereas knockdown of circLARP1B has the opposite results on inflammatory factors levels (Fig. 2H–L). The levels of IL-18, IL-1β, caspase 1, and NLRP3 were upregulated by overexpression of circLARP1B and were downregulated by knockdown of circLARP1B in HG-treated cells (Fig. 2M).
CircLARP1B inhibited HG-induced proliferation and cell cycle, promoted pyroptosis and inflammation. A The transfection efficiency was detected using qPCR after transfection with circLARP1B overexpression vector and sh-circLARP1B. B Cell viability was analyzed using MTT assay. C, D EDU assay was carried out to evaluate cellular proliferation. (E) Cell cycle was detected using flow cytometry. F, G Cell pyroptosis was detected using flow cytometry by double staining with caspase1 and PI. ELISA was used to measure the levels of H IL-6, I IL-10, J TNF-α, K IL-1β, and L IL-18. M The protein expression of IL-18, IL-1β, caspase 1, and NLRP3 was examined using western blot. **P < 0.01 vs. vector group in (A). **P < 0.01 and ***P < 0.001 vs. control group in (B–M). ###P < 0.001 vs. sh-NC group in (A). #P < 0.05 and ##P < 0.01 vs. HG + vector group in (B–M). &&P < 0.01 and &&&P < 0.001 vs. HG + sh-NC group
CircLARP1B directly targets miR-578
The data of bioinformatics showed that miR-578 was found to have binding sites of circLARP1B (Fig. 3A). MiR-578 reduced the luciferase activity of WT-circLARP1B rather than MUT-circLARP1B (Fig. 3B). MiR-578 was downregulated in patients with DN (Fig. 3C). The expression of miR-578 was negatively related to circLARP1B expression (Fig. 3D). MiR-578 was lower expressed in HG-treated cells than the control cells (Fig. 3E). Overexpression of circLARP1B decreased miR-578 expression, whereas knockdown of circLARP1B increased miR-578 expression (Fig. 3F).
CircLARP1B directly targets miR-578. A Bioinformatic analysis predicted the binding sites between circLARP1B and miR-578. B The relationship between circLARP1B and miR-578 was affirmed by dual-luciferase reporter assay. C MiR-578 expression was detected in the blood of patients with DN and healthy subjects using qPCR. D The relationship between circLARP1B and miR-578 in the blood of patients with DN was evaluated by the Pearson correlation coefficient. E MiR-578 expression was detected in mesangial cells treated with 5 mM or 30 mM d-glucose (control or HG) using qPCR. F MiR-578 expression was detected using qPCR after transfection with circLARP1B overexpression vector and sh-circLARP1B. ***P < 0.001. ###P < 0.001
MiR-578 inhibitor reversed the effects of circLARP1B silencing on cell proliferation, cycle, pyroptosis, and inflammation
To investigate the role of miR-578. miR-578 mimic and miR-NC mimic were transfected. The expression of miR-578 was upregulated after transfection with miR-578 (Fig. 4A). Silencing of circLARP1B promoted cell viability and proliferation, whereas inhibition of miR-578 rescued the promotion (Fig. 4B–D). The cells in the G0-G1 phase were reduced and in the S phase were increased by knockdown of circLARP1B, which were abrogated by inhibition of miR-578 (Fig. 4E). Knockdown of circLARP1B suppressed pyroptosis of HG-treated cells, and miR-578 downregulation abolished the suppression (Fig. 4F and G). Additionally, depletion of circLARP1B decreased the levels of IL-6, TNF-α, IL-1β, and IL-18, and increased the levels of IL-10, while downregulation of miR-578 abrogated the effects on inflammatory factors levels induced by sh-circLARP1B (Fig. 4H–L). As shown in Fig. 4M, knockdown of circLARP1B reduced IL-18, IL-1β, caspase 1, and NLRP3 levels, and reduced miR-578 abrogated the reduction.
MiR-578 inhibitor reversed the effects of circLARP1B silencing on cell proliferation, pyroptosis, and inflammation. A The levels of miR-578 were detected using qPCR following mimic transfection. B Cell viability was analyzed using MTT assay. C, D EDU assay was carried out to evaluate cellular proliferation. E Cell cycle was detected using flow cytometry. F, G Cell pyroptosis was detected using flow cytometry by double staining with caspase1 and PI. ELISA was used to measure the levels of H IL-6, I IL-10, J TNF-α, K IL-1β, and L IL-18. M The protein expression of IL-18, IL-1β, caspase 1, and NLRP3 was examined using western blot. ***P < 0.001 vs. miR-NC group in (A) and control group in (B-M). ##P < 0.01 and ###P < 0.001 vs. HG group. &P < 0.05 and &&P < 0.01 vs. HG + sh-circLARP1B + miR-NC group
TLR4 is a miR-578 target
Then, we predicted the targets of miR-578, and found TLR4 is one of the targets of miR-578 (Fig. 5A). MiR-578 reduced the luciferase activity of WT-TLR4 instead of MUT-TLR4 (Fig. 5B). The levels of TLR4 were higher in patients with DN than normal subjects (Fig. 5C). TLR4 expression was negatively related to circLARP1B expression (Fig. 5D). In addition, TLR4 was upregulated in HG-treated cells (Fig. 5E). Overexpression of miR-578 decreased TLR4 expression (Fig. 5F).
TLR4 is a miR-578 target. A Bioinformatic analysis predicted the binding sites between TLR4 and miR-578. B The relationship between TLR4 and miR-578 was confirmed by dual-luciferase reporter assay. C TLR4 expression was detected in the blood of patients with DN and healthy subjects using qPCR. D The correlation between TLR4 and miR-578 in the blood of patients with DN was evaluated by the Pearson correlation coefficient. E TLR4 expression was detected in cells treated with 5 mM or 30 mM d-glucose (control or HG) using qPCR. F TLR4 expression was detected using qPCR after transfection with miR-578 mimic. ***P < 0.001
TLR4 reversed the effects of miR-578 on cell proliferation, pyroptosis, and inflammation
To investigate the role of TLR4. TLR4 overexpression vector and empty vector were transfected. The expression of TLR4 was upregulated after transfection with TLR4 overexpression vector (Fig. 6A). MiR-578 promoted cell viability and proliferation, whereas TLR4 rescued the promotion induced by miR-578 (Fig. 6B–D). MiR-578 decreased the cells in the G0-G1 phase and increased S phase cells, while TLR4 abrogated the effects induced miR-578 (Fig. 6E). MiR-578 inhibited pyroptosis of HG-treated cells, and TLR4 reversed the inhibition (Fig. 6F and G). Overexpression of miR-578 decreased the levels of IL-6, TNF-α, IL-1β, and IL-18, and elevated the levels of IL-10, and overexpression of TLR4 abrogated the effects induced by miR-578 (Fig. 6H–L). Additionally, miR-578 decreased IL-18, IL-1β, caspase 1, and NLRP3 levels, while TLR4 abrogated the decrease (Fig. 6M).
TLR4 reversed the effects of miR-578 on cell proliferation, pyroptosis, and inflammation. A The levels of TLR4 were detected using qPCR after transfection with TLR4 overexpression vector and empty vector. B Cell viability was analyzed using MTT assay. C, D EDU assay was used to evaluate cellular proliferation. E Cell cycle was detected using flow cytometry. F, G Cell pyroptosis was detected using flow cytometry by double staining with caspase1 and PI. ELISA was used to measure the levels of H IL-6, I IL-10, J TNF-α, K IL-1β, and L IL-18. M The protein expression of IL-18, IL-1β, caspase 1, and NLRP3 was examined using western blot. ***P < 0.001 vs. vector group in (A). **P < 0.01 and ***P < 0.001 vs. control group in (B–M). ##P < 0.01 and ###P < 0.001 vs. HG group. &&P < 0.01 vs. HG + miR-578 + vector group
Discussion
In recent years, research on DN has developed, and its pathogenesis is more and more understood. The researchers found that the molecular mechanism of DN is very complex. Almost all kidney cells, including endothelial cells, tubulointerstitial cells, podocytes, and mesangial cells, have abnormal functions involved in the pathogenesis and progression of DN [13]. Hypertrophy and death of mesangial cells are key to the fate of renal cells in patients with DN [14]. High glucose induces inflammation and fibrosis, leading to glomerulosclerosis [15]. CircRNAs regulate cellular processes such as proliferation, programmed cell death, and metastasis, acting as biomarkers and potential therapeutic targets in renal diseases including DN [16]. CircLARP1B expression is upregulated in hepatocellular carcinoma. Knockdown of circLARP1B inhibits tumor cell growth and metastasis, and enhances radiosensitivity [17]. Silencing of circLARP1B suppresses cell proliferation, invasion, and glycolysis, and facilitates apoptosis of cutaneous squamous cell carcinoma [18]. However, the role of circLARP1B in DN is little known. In this study, circLARP1B was highly expressed in patients with DN. HG treatment increased circLARP1B in mesangial cells. Then, we explored the role of circLARP1B in DN. The results showed that overexpression of circLARP1B inhibited the proliferation, blocked cell cycle, and promoted pyroptosis and inflammation of HG-induced mesangial cells, whereas knockdown of circLARP1B had the opposite results. The findings suggested that silencing of circLARP1B attenuated the progression of DN.
There are many binding sites between circRNAs and miRNAs, which combination enhanced the effects of miRNAs on target mRNAs [19]. We predicted that miR-578 has potential binding sites with circLARP1B. Then, miR-578 was confirmed as a target of circLARP1B, consistent with a previous study [17]. The role of miR-578 in tumors is very complex, and it can promote or inhibit cancer development and progression in different types of tumors [20, 21]. Additionally, miR-578 is also involved in intervertebral disc degeneration, acute kidney injury, and macular degeneration [22,23,24]. In this study, we investigated the role of miR-578 in DN. MiR-578 was low expressed in patients with DN. Downregulation of miR-578 rescued the effects of circLARP1B on cell proliferation, cell cycle, pyroptosis, and inflammation. The data indicated that silencing of circLARP1B decelerated the progression of DN by sponging miR-578.
MiRNAs regulate targeted mRNAs expression in human diseases, and the miRNAs-mRNAs regulatory axis is related to DN [25]. TLRs are pattern recognition receptors that exist in the innate immune system rather than in the host [26]. TLR4 can induce a large increase in pro-inflammatory factors, promoting the inflammatory process, including infection-induced inflammation and sterile inflammation [27, 28]. Due to DN is associated with inflammation, TLR4 has been identified to be involved in DN [29, 30]. Herein, TLR4 was found to be upregulated in DN. Overexpression of TLR4 rescued the effects of miR-578 on cell proliferation, cell cycle, pyroptosis, and inflammation. The data suggested that miR-578 attenuated the progression of DN by targeting TLR4.
In conclusion, we elaborated on the effects of circLARP1B on the biological behaviors of renal mesangial cells in DN and the molecular mechanism. We found that silencing of circLARP1B promoted the proliferation, blocked cell cycle, and suppressed pyroptosis and inflammation of HG-induced mesangial cells via the miR-578/TLR4 axis, thereby may attenuate the progression of DN. The findings suggested that circLARP1B may be a target for DN therapy.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Wada J, Makino H (2013) Inflammation and the pathogenesis of diabetic nephropathy. Clin Sci (Lond) 124(3):139–152. https://doi.org/10.1042/CS20120198
Lu Z, Liu N, Wang F (2017) Epigenetic regulations in diabetic nephropathy. J Diabetes Res 2017:7805058. https://doi.org/10.1155/2017/7805058
Valencia WM, Florez H (2017) How to prevent the microvascular complications of type 2 diabetes beyond glucose control. BMJ 356:i6505. https://doi.org/10.1136/bmj.i6505
Selby NM, Taal MW (2020) An updated overview of diabetic nephropathy: diagnosis, prognosis, treatment goals and latest guidelines. Diabetes Obes Metab 22(Suppl 1):3–15. https://doi.org/10.1111/dom.14007
Lv J, Wu Y, Mai Y, Bu S (2020) Noncoding RNAs in diabetic nephropathy: pathogenesis, biomarkers, and therapy. J Diabetes Res 2020:3960857. https://doi.org/10.1155/2020/3960857
van Zonneveld AJ, Kölling M, Bijkerk R, Lorenzen JM (2021) Circular RNAs in kidney disease and cancer. Nat Rev Nephrol 17(12):814–826. https://doi.org/10.1038/s41581-021-00465-9
Peng F, Gong W, Li S, Yin B, Zhao C, Liu W, Chen X, Luo C, Huang Q, Chen T, Sun L, Fang S, Zhou W, Li Z, Long H (2021) circRNA_010383 acts as a sponge for miR-135a, and its downregulated expression contributes to renal fibrosis in diabetic nephropathy. Diabetes 70(2):603–615. https://doi.org/10.2337/db20-0203
Yao T, Zha D, Hu C, Wu X (2020) Circ_0000285 promotes podocyte injury through sponging miR-654–3p and activating MAPK6 in diabetic nephropathy. Gene 747:144661. https://doi.org/10.1016/j.gene.2020.144661
Wang Y, Qi Y, Ji T, Tang B, Li X, Zheng P, Bai S (2021) Circ_LARP4 regulates high glucose-induced cell proliferation, apoptosis, and fibrosis in mouse mesangial cells. Gene 765:145114. https://doi.org/10.1016/j.gene.2020.145114
Rayego-Mateos S, Morgado-Pascual JL, Opazo-Ríos L, Guerrero-Hue M, García-Caballero C, Vázquez-Carballo C, Mas S, Sanz AB, Herencia C, Mezzano S, Gómez-Guerrero C, Moreno JA, Egido J (2020) Pathogenic pathways and therapeutic approaches targeting inflammation in diabetic nephropathy. Int J Mol Sci 21(11):3798. https://doi.org/10.3390/ijms21113798
Al Mamun A, Ara Mimi A, Wu Y, Zaeem M, Abdul Aziz M, Aktar Suchi S, Alyafeai E, Munir F, Xiao J (2021) Pyroptosis in diabetic nephropathy. Clin Chim Acta 523:131–143. https://doi.org/10.1016/j.cca.2021.09.003
Wen S, Li S, Li L, Fan Q (2020) circACTR2: a novel mechanism regulating high glucose-induced fibrosis in renal tubular cells via pyroptosis. Biol Pharm Bull 43(3):558–564. https://doi.org/10.1248/bpb.b19-00901
Maezawa Y, Takemoto M, Yokote K (2015) Cell biology of diabetic nephropathy: roles of endothelial cells, tubulointerstitial cells and podocytes. J Diabetes Investig 6(1):3–15. https://doi.org/10.1111/jdi.12255
Manda G, Checherita AI, Comanescu MV, Hinescu ME (2015) Redox signaling in diabetic nephropathy: hypertrophy versus death choices in mesangial cells and podocytes. Mediators Inflamm 2015:604208. https://doi.org/10.1155/2015/604208
Tung CW, Hsu YC, Shih YH, Chang PJ, Lin CL (2018) Glomerular mesangial cell and podocyte injuries in diabetic nephropathy. Nephrology (Carlton) 23(Suppl 4):32–37. https://doi.org/10.1111/nep.13451
Jin J, Sun H, Shi C, Yang H, Wu Y, Li W, Dong YH, Cai L, Meng XM (2020) Circular RNA in renal diseases. J Cell Mol Med 24(12):6523–6533. https://doi.org/10.1111/jcmm.15295
Zhu S, Chen Y, Ye H, Wang B, Lan X, Wang H, Ding S, He X (2022) Circ-LARP1B knockdown restrains the tumorigenicity and enhances radiosensitivity by regulating miR-578/IGF1R axis in hepatocellular carcinoma. Ann Hepatol 27(2):100678. https://doi.org/10.1016/j.aohep.2022.100678
Wang L, Hou S, Li J, Tian T, Hu R, Yu N (2022) Circular RNA circ-LARP1B contributes to cutaneous squamous cell carcinoma progression by targeting microRNA-515-5p/TPX2 microtubule nucleation factor axis. Bioengineered 13(1):1209–1223. https://doi.org/10.1080/21655979.2021.2019172
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388. https://doi.org/10.1038/nature11993
Chen Z, Wang F, Xiong Y, Wang N, Gu Y, Qiu X (2020) CircZFR functions as a sponge of miR-578 to promote breast cancer progression by regulating HIF1A expression. Cancer Cell Int 20:400. https://doi.org/10.1186/s12935-020-01492-5
Wang M, Ma M, Yang Y, Li C, Wang Y, Sun X, Wang M, Sun Y, Jiao W (2021) Overexpression of hsa_circ_0008274 inhibited the progression of lung adenocarcinoma by regulating HMGA2 via sponging miR-578. Thorac Cancer 12(16):2258–2264. https://doi.org/10.1111/1759-7714.14059
Yan P, Sun C, Luan L, Han J, Qu Y, Zhou C, Xu D (2022) Hsa_circ_0134111 promotes intervertebral disc degeneration via sponging miR-578. Cell Death Discov 8(1):55. https://doi.org/10.1038/s41420-022-00856-2
Gao Q, Zheng Y, Wang H, Hou L, Hu X (2022) circSTRN3 aggravates sepsis-induced acute kidney injury by regulating miR-578/ toll like receptor 4 axis. Bioengineered 13(5):11388–11401. https://doi.org/10.1080/21655979.2022.2061293
Zhou RM, Shi LJ, Shan K, Sun YN, Wang SS, Zhang SJ, Li XM, Jiang Q, Yan B, Zhao C (2020) Circular RNAZBTB44 regulates the development of choroidal neovascularization. Theranostics 10(7):3293–3307. https://doi.org/10.7150/thno.39488
Regulatory Network in Diabetic Nephropathy. J Healthc Eng. 2021; 2021:8161701. doi: https://doi.org/10.1155/2021/8161701
Linde A, Mosier D, Blecha F, Melgarejo T (2007) Innate immunity and inflammation–new frontiers in comparative cardiovascular pathology. Cardiovasc Res 73(1):26–36. https://doi.org/10.1016/j.cardiores.2006.08.009
Sheu JJ, Chang LT, Chiang CH, Youssef AA, Wu CJ, Lee FY, Yip HK (2008) Prognostic value of activated toll-like receptor-4 in monocytes following acute myocardial infarction. Int Heart J 49(1):1–11. https://doi.org/10.1536/ihj.49.1
Rocha DM, Caldas AP, Oliveira LL, Bressan J, Hermsdorff HH (2016) Saturated fatty acids trigger TLR4-mediated inflammatory response. Atherosclerosis 244:211–215. https://doi.org/10.1016/j.atherosclerosis.2015.11.015
Wu Y, Zhao Y, Yang HZ, Wang YJ, Chen Y (2021) HMGB1 regulates ferroptosis through Nrf2 pathway in mesangial cells in response to high glucose. Biosci Rep 41(2):BSR20202924. https://doi.org/10.1042/BSR20202924
Zhu L, Han J, Yuan R, Xue L, Pang W (2018) Berberine ameliorates diabetic nephropathy by inhibiting TLR4/NF-κB pathway. Biol Res 51(1):9. https://doi.org/10.1186/s40659-018-0157-8
Funding
This work was supported by Xi’an Science and Technology Planning Project Medical Research Project: General Research under grant number 2023JH-YXYB-0302.
Author information
Authors and Affiliations
Contributions
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript. YD drafted the work and revised it critically for important intellectual content; YF was responsible for the acquisition, analysis and interpretation of data for the work; YC and CT made substantial contributions to the conception or design of the work.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
The study was approved by the Ethics Committee of The First Affiliated Hospital of Xi’an Medical University.
Consent to participate
Written informed consent was obtained from all subjects.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Du, Y., Feng, Y., Cai, Y. et al. CircLARP1B promotes pyroptosis of high glucose-induced renal mesangial cells by regulating the miR-578/TLR4 axis. Int Urol Nephrol 56, 283–293 (2024). https://doi.org/10.1007/s11255-023-03672-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11255-023-03672-4