Abstract
This systematic review and meta-analysis were conducted to investigate the association between methylenetetrahydrofolate reductase (MTHFR) C677T and A1298C polymorphisms with breast cancer (BC) in Asians. Systematic searches were conducted in PubMed, EMBASE, Web of Science, and Scopus by May 2020. Inter-study heterogeneity was also assessed with a Q test, along with I2 statistics. Random-effects models were applied to pooled crude ORs with corresponding 95% CIs for the genetic models. A total of 1097 identified results, along with 36 qualified studies were included: for MTHFR C677T polymorphism, a total of 36 studies was comprised of 11,261 cases and 13,318 controls and for MTHFR A1298C polymorphism, a number of 19 studies contained 7424 cases and 8204 controls. Likewise, for C677T polymorphism, an increased risk of BC was seen for the allelic (OR 1.21, 95% CI 1.09–1.33, P < 0.01, I2 = 78.9%), dominant (OR 1.17, 95% CI 1.05–1.30, P < 0.01, I2 = 71.8%), recessive (OR 1.43, 95% CI 1.23–1.67, P < 0.01, I2 = 55.8%), and homozygous models (OR 1.48, 95% CI 1.25–1.75, P < 0.01, I2 59.9%) among BC patients compared to controls. Also, in terms of A1298C polymorphism, an association was found between the allelic (OR 1.15, 95% CI 1.04–1.28, P < 0.01, I2 70.4%) and homozygous models (OR 1.38, 95% CI 1.15–1.66, P < 0.01, I2 44.2%) with the risk of BC. In conclusion, findings revealed that MTHFR C677T variant might be a factor that predisposes BC in Asians. Furthermore, it was found that A1298C variant acts as a BC risk factor, particularly in a Western Asia population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Considered as a major cause of death all over the world, breast cancer (BC) is seen as one of the most common malignancies among women, accounting for 30% of all new cancer diagnoses. Moreover, although the malignancy is observed in both developed Western countries and developing Asian countries, BC is estimated to have a higher mortality percentage in Asian countries than in Western ones (Mubarik et al. 2019; Paydar et al. 2019). The reason for the higher rate of mortalities in these countries may be due to a variety of factors such as increased longevity, late diagnosis, insufficient access to care, higher exposure to risk factors, high-fat foods consumption, reduced pregnancy rates, and obesity (Ghoncheh et al. 2016; Yip 2009).
According to what was previously stated, the incidence of BC is intensified by hereditary and genetic factors. More than 170 genomic loci harboring common variants related to BC have been recognized by genome-wide association studies (GWAS). The most well known are mutations in the BRCAs genes (Ferreira et al. 2019; Momenimovahed and Salehiniya 2019). Also, the mutation in methylenetetrahydrofolate reductase (MTHFR) has been shown to have a positive correlation with BC (Meneses-Sanchez et al. 2019). Located on chromosome 1 (p36.22), the MTHFR gene with 20.373 base pair contains 13 exons (Antonaros et al. 2019). This enzyme has been indicated to catalyze the irreversible conversion of 5.10-methylenetetrahydrofolate (methyleneTHF) to 5-methyltetrahydrofolate (methylTHF). The latter is involved in the conversion of homocysteine to methionine, thereby the formation of S-adenosylmethionine (SAM). Therefore, the MTHFR is involved in the methylation of DNA, indicating an association with cancer development (Odin et al. 2006). It has been shown that among the MTHFR mutations, C677T (rs1801133) and A1298C (rs1801131) are the most reported ones (Wan et al. 2018). The C677T polymorphism (rs1801133) transforms alanine into valine, resulting in reduced enzyme activity (Kumar et al. 2015; Zhu et al. 2014). Numerous studies have revealed that the MTHFR C677T polymorphism is a risk factor for BC (Ergul et al. 2003; He and Shen 2017; Waseem et al. 2016). As enzyme activity decreases by this mutation, the plasma levels of homocysteine increase, as a result of which methylation of DNA reduces, hence the development of cancer (Waseem et al. 2016). However, other studies have shown contradictory results (Floris et al. 2020; Hekim et al. 2007; Kalyankumar and Jamil 2006). Another common SNP, A1298C (rs1801133), transforms glutamate to alanine at 429 residues and reduces enzyme activity (Wan et al. 2018). Similar to C677T, there is still controversy as to the association of this mutation with BC, particularly in Asian population (Castiglia et al. 2019; Kaya et al. 2016).
Given the higher rate of breast cancer among Asians and the contradictory results as to the role that MTHFR C677T and A1298C variants play in susceptibility to BC among Asians, the aim of this systematic review and meta-analysis study was to investigate the role of MTHFR C677T and A1298C gene polymorphisms in BC in Asian populations.
Methods
Literature Search
We ran a systematic search of electronic databases such as PubMed, EMBASE, Web of Science, Scopus, and Google Scholar in order to identify the related studies which investigated the association between MTHFR C677T and A1298C variants in an Asian population up to May 2020. The searches for the related literature were conducted using a combination of the following MeSH terms and relevant keywords: "methylenetetrahydrofolate reductase (MTHFR)," "polymorphism," "genetic," "mutation," "point mutation," "mutation," "missense," "breast cancer," and "breast neoplasms" (Supp 1). To increase the comprehensiveness of our searches, we manually screened the references mentioned in the previous reviews and included articles. Searches were conducted in the English Language without any published date limitation.
Study Selection
Two researchers (SS and MM) carried out an independent review of the literature search results to select relevant studies based on our inclusion. It is worth noting that any disagreement was resolved through consensus and discussion. The studies that met the following inclusion criteria were selected for use in the meta-analysis: studies that investigated the association between MTHFR polymorphisms (including C677T and/or A1298C) and BC susceptibility were conducted with a non-familial, case–control design, reported sufficient genotype data or possessed the information to calculate the pooled OR and the corresponding 95% confidence intervals (CIs), and included participants who were from an Asian population. So, studies without usable data, with participants other than those of an Asian population, or with combined ethnicities, having cases not confirmed by other approaches, and those lacking full texts were excluded. When there were overlapping published studies, only a recent study with the most complete data for our meta-analysis was included.
Data Extraction
To achieve more precision, two researchers separately performed the extraction of data for all the eligible studies (SS and MM). The following items were extracted: first author’s name, year of publication, study location, ethnicity, participants' descriptive characteristic, type of polymorphism, type of control, total sample size, and the number of alleles and genotypes in both case and control groups. A standardized data abstraction of Excel forms was used to extract the pertinent data.
Quality Assessment
The Newcastle–Ottawa Scale (NOS) was used to assess the quality of included studies. The following three domains were applied to critically evaluate each study: the selection of case/control, comparability, and outcome. It is noteworthy that NOS scores ranged from 0 to 9 and that studies with scores of seven or more were considered as having a good quality.
Statistical Analysis
All the statistical analyses were conducted using STATA (version 11.0; Stata Corporation, College Station, TX). Crude odds ratios (ORs) with corresponding 95% CIs were estimated to investigate the strength of association between the MTHFR polymorphisms (includes C677T and/or A1298C) and BC susceptibility. With respect to C677T variant, genotypes were analyzed using genetic models such as the allelic (T vs. C), recessive (TT vs. CT + CC), dominant (TT + CT vs. CC), homozygote (TT vs. CC), heterozygous (CT vs. CC), and codominant models (CT vs. TT + CC). Likewise, A1298C genotypes were analyzed using the following genetic models: the allelic (C vs. A), recessive (CC vs. AC + AA), dominant (CC + AC vs. AA), homozygote (CC vs. AA), heterozygous (AC vs. AA), and codominant models (AC vs. CC + AA). Hardy–Weinberg Equilibrium (HWE) was used to assess the distribution of genotypes in the control group of each included study which has not deviated from HWE by using the Chi-square test. Inter-study heterogeneity was evaluated using the Cochran Q test and I2 statistic. It should be noted that a P < 0.1 for Q test and I2 greater than 50% showed a substantial heterogeneity. In the presence of heterogeneity, the random-effects model was used to pool the ORs and their 95% CIs, otherwise, the fixed-effect model was applied (P > 0.1 with I2 < 50%). Subgroup analyses were conducted to stratify the pooled findings based on the potential suspected variables including ethnicity (Western Asia vs. Southern Asia vs. Eastern Asia), source of control (population base vs. hospital base vs. NI), genotyping (other techniques vs. PCR–RFLP), agreement with HWE (yes vs. no), and quality status (high vs. low) to explore the source of heterogeneity. Sensitivity analyses as an additional procedure were performed to assess the stability of the pooled ORs after excluding each study using the leave-one-out method and also to find the source of heterogeneity. Evidence of potential publication bias across included studies was quantitatively examined using Egger’s and Begg’s tests and funnel plot asymmetry was assessed visually.
Results
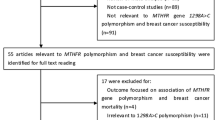
A total of 1097 studies were identified through comprehensive database searches. Out of all the screening studies, only a number of 36 qualified studies (Akilzhanova et al. 2006; Akram et al. 2012; Chou et al. 2006; DELIGEZER et al. 2005; Ergul et al. 2003; He and Shen 2017; Hedayatizadeh-Omran et al. 2017; Hekim et al. 2007; Hesari et al. 2019; Hosseini et al. 2011; Huang et al. 2014; Jiang-hua et al. 2014; Kalyankumar and Jamil 2006; Kaya et al. 2016; Lajin et al. 2012; Lee et al. 2004; Lin et al. 2004; Liu et al. 2013; Lu et al. 2015; Mir et al. 2008; Mohammadzadeh et al. 2016; Naushad et al. 2011; Niu et al. 2017; Ozen et al. 2013; Pooja et al. 2015; Prasad and Wilkhoo 2011; Rahimi et al. 2019; Sangrajrang et al. 2010; Shrubsole et al. 2004; Song et al. 2016; Suzuki et al. 2008; Waseem et al. 2016; Weiner et al. 2010; Weiwei et al. 2014; Yu et al. 2007; Zhang et al. 2015) were found to be eligible based on inclusion criteria for the current meta-analysis. Figure 1 shows the flowchart of how the studies were identified and then selected step by step. A total of 1097 studies were identified through comprehensive database searches.
The association between C677T polymorphism and BC was reported by 36 studies (containing 11,261 cases and 13,318 controls) and the association between A1298C polymorphism and BC was demonstrated by 19 studies (including 7424 cases and 8204 controls). As the basic characteristics of the included studies are shown in Table 1, it was found that the genotype distributions of the controls were not in line with those of HWE in studies conducted by the He and Shen ( 2017), Weiwei et al. (2014), Hosseini et al. (2011), and Jiang-hua et al. (2014) with studies on C677T polymorphism and also by Akram et al. (2012), Ozen et al. (2013), Mir et al. (2008), Sangrajrang et al. (2010), and studies on A1298C polymorphism. The included studies were published between 2003 (Ergul et al. 2003) and 2019 (Rahimi et al. 2019).
Association of the MTHFR C677T Polymorphism with Breast Cancer Risk for Asians
The forest plots which represent the association of the C677T polymorphism with BC risk among an Asian population are shown in Fig. 2. The use of the random-effects model based on 36 qualified studies on the pooled results indicated that there was a significant association between genetic models including the allelic (OR 1.21, 95% CI 1.09–1.33, P < 0.01, I2 = 78.9%), the dominant (OR 1.17, 95% CI 1.05–1.30, P < 0.01, I2 = 71.8%), the recessive (OR 1.43, 95% CI 1.23–1.67, P < 0.01, I2 = 55.8%), and the homozygous models (OR 1.48, 95% CI 1.25–1.75, P < 0.01, I2 = 59.9%) with an increased risk of BC among an Asian population, whereas the heterozygous (OR 1.08, 95% CI 0.98–1.19, P = 0.11, I2 = 57.5%) and the codominant models (OR 0.98, 95% CI 0.90–1.08, P = 0.72, I2 = 59.6%) did not reveal such a significant association.
According to the evidence of inter-study heterogeneity across the qualified studies (P < 0.01 with I2 > 50%), subgroup analyses were carried out according to moderator variables including ethnicity, source of control, genotyping, agreement with HWE, and quality status. With reference to Table 2, the results of the subgroups indicated the reduced heterogeneity in some of the strata. Meanwhile, it was found that there was a significant association between the all genetic models and the risk of BC in participants of Eastern Asia and hospital-based controls. Yet, the association between the codominant models and BC risk remained non-significant in different strata.
Following sensitivity analyses, the findings showed that after each study was removed, the pooled ORs for the allelic, dominant, recessive, homozygous, and codominant models maintained their stability. However, the lower and higher pooled ORs in sensitivity analyses for these genetic models are presented in Table 3. But, for the heterozygous model, the sensitivity analysis results revealed that removal of a study conducted by Aklizhanova et al. (2013) changed the pooled OR (OR 1.10, 95% CI 1.00–1.20).
The funnel plot shapes of the genetic models were almost symmetrical, showing the lack of any evidence for publication bias (Supp 1 Fig. S1–S6). However, significant potential publication biases were statistically found using Begg and Egger’s tests for the recessive (P Begg's test = 0.01, P Egger's test = 0.04), and the homozygous models (P Begg's test = 0.02, P Egger's test = 0.06). After including the findings of censored studies using the trimming estimator with the linear method, there were no significant differences for these two genetic models in the pooled ORs between before and after the inclusion of censored studies.
Association of the MTHFR A1298C Polymorphism with Breast Cancer Risk for Asians
A number of 19 qualified studies examined the association between A1298C polymorphism and BC risk. It was found, using random-effects model, that the allelic (OR 1.15, 95% CI 1.04–1.28, P < 0.01, I2 = 70.4%) and the homozygous models (OR 1.38, 95% CI 1.15–1.66, P < 0.01, I2 = 44.2%) were associated with the risk of BC among BC patients compared to controls; however, no such significant association was seen between the dominant (OR 1.11, 95% CI 0.99–1.25, P = 0.068, I2 = 60.3%), recessive (OR 1.23, 95% CI 0.99–1.52, P = 0.065, I2 = 65.5%), heterozygous (OR 1.23, 95% CI 0.97–1.57, P = 0.085, I2 = 89.5%), and the codominant models (OR 0.97, 95% CI 0.87–1.08, P = 0.57, I2 = 55.4%) (Fig. 3).
Following subgroup analyses, inter-study heterogeneity was changed among some of the strata. Table 2 shows more detailed results of subgroup analyses. It is worth noting that, in all the investigated genetic models except for the codominant mode, the participants in Western Asia were significantly associated with the risk of BC.
Moreover, after the step-by-step removal of the included studies, no statistical change was witnessed between pre- and post-sensitivity pooled ORs for the allele, the heterozygous, the homozygotes, and the codominant models. However, the pooled OR for the dominant model was sensitive to the study by Chou et al. (2006) (OR 1.13, 95% CI 1.02–1.26) and the pooled OR for the recessive model was sensitive to that of Akram et al. (2012) (OR 1.31, 95% CI 1.11–1.55) (Table 3).
In spite of the fact that the included studies were visually and quantitatively assessed, no significant evidence of publication bias was found, indicating a low risk of publication bias; except for the homozygous model (P Begg's test = 0.02, P Egger's test = 0.01) (Supp 1 Fig. S7–S12). After using a nonparametric method (Duval and Tweedie), it was found that the summary effect size did not change before and after the included studies were censored to pool the association between the homozygous model with the risk of BC.
Discussion
BC is a multifactorial disease that is not only developed due to environmental factors but also by genetic variants that affect the expression and function of proteins. The MTHFR enzyme plays a very important role in folate metabolism, hence its involvement in the process of DNA methylation (Botezatu et al. 2013; Cheng et al. 2012). DNA methylation may lead to suppression of tumor suppressor genes as well as activation of proto-oncogenes (Meneses-Sanchez et al. 2019). This is the first systematic review and meta-analysis which has incorporated a considerable number of studies (36 studies) on the association of RNLS MTHFR C677T and A1298C variants with susceptibility to BC among Asians. It was found that the MTHFR C677T polymorphism was associated with BC. Also, concurrently, this association was more documented through subgroup analyses in terms of ethnicity, source of control, genotyping, and agreement with HWE. In the case of A1298C polymorphism, allelic and homozygous models were associated with increased risk of BC; moreover, Western Asia populations showed the same association in the heterogeneous models which are considered as the first report in this regard.
Located in exon 4, the C677T (rs1801133) polymorphism is folate binding site of the MTHFR gene and affects the process of protein expression (Teng et al. 2013), thereby playing an important role in relation to cancers. The results of the current meta-analysis study revealed that although the C677T variant, in 24,579 subjects and 36 studies, significantly increased the risk of BC in homozygous, recessive, dominant, and allelic models, no such a significant difference was found in heterozygous and overdominant models. Consistent with these results, Hi and Shen in a meta-analysis study on the MTHFR C677T polymorphism and BC demonstrated that this variant may be a risk factor for BC in 14,299 Asian populations (He and Shen 2017). Despite the results achieved in the subgroup analyses, we could find a significant association between the heterozygous models with the risk of BC in participants of Eastern Asia and hospital-based controls. Hence, it may be hypothesized that the controls and ethnicity may have been the source of the biased results. Furthermore, the results of the sensitivity analysis in this model showed that removing a study conducted by Aklizhanova et al. (2013) changed the pooled OR (OR 1.10, 95% CI 1.00–1.20). It has been reported that folate deficiency is related to aberrant DNA methylation, increased mutations, and breaks in chromosomes that can be associated with an enhanced risk of cancer (Choi and Mason 2000; Tomita 2016). Interestingly, several studies have shown that the modification of DNA methylation by MTHFR C677T and A1298C variants may only occur under conditions of limited folate content and that folate stabilizes the MTHFR C677T polymorphism (Del Gobbo et al. 2018). The study by Aklizhanova et al. was performed on the Kazakhstan population where traditional food is meat, which leads to the deficiency of folate, thus promoting the risk of BC development (Akilzhanova et al. 2006). Additionally, they found low levels of serum folate not only in BC cases but also in normal subjects, bringing about the hypothesis that low levels of serum folate in participants may be the possible causes of this discrepancy.
In a total of 19 studies on 15,628 subjects, it was revealed that another polymorphism, rs1801131 (A1298C), located in exon 7 of the MTHFR gene (Castiglia et al. 2019), was significantly associated with BC in homozygous and allelic models, even though no significant difference was found in recessive, dominant, heterozygous, and overdominant models. Interestingly, the subgroups analysis seemed to suggest that genetic models of recessive, dominant, heterozygous, homozygous, and allelic in participants from Western Asia were significantly associated with the risk of BC. Additionally, a number of other studies have shown that this polymorphism was not significantly related to the risk of BC (Rai 2014; Zhang et al. 2016). Nonetheless, the results of subgroup analysis also revealed that this polymorphism was significantly associated with the risk of BC in participants from Western Asia in all the investigated genetic models, except for the codominant model. Therefore, it has been speculated that the discrepancy between the findings was most likely due to the racial ethnic differences, ethnicity, patient selection criteria, sources of control, and sample size. A meta-analysis conducted by He and shen (2017) investigated only the MTHFR C677T variant, while our meta-analysis assessed both MTHFR C677T and A1298C SNPs. Moreover, they included 19 Asian studies and concluded no significant association between MTHFR C677T and breast cancer among Asians. However, we included 36 Asian studies (almost twofold compared to the study by He and Shen (2017), indicating the association of MTHFR C677T and breast cancer. In addition, in the case of the A1298C variant, allelic and homozygous genetic models were associated with increased risk of BC; moreover, Western Asia populations showed the same association in the heterogeneous models which are considered as the first report in this regard.
Although this is an up-to-date meta-analysis study, there were a number of limitations that need to be mentioned. First, given the high rate of heterogeneity in a number of findings, it is likely that the results are negatively affected, suggesting further studies are required. Second, subgroup analysis was not performed according to the levels of serum folate. Third, this analysis was carried out only on researches published in English. However, the present study also had some highlights which include more reliable results due to the increased sample size, as well as the use of all the possible genetic models in examining the association between these two polymorphisms and the risk of BC.
Conclusion
In conclusion, the present meta-analysis showed that the MTHFR C677T variants might be a promising factor that predisposes BC in an Asian population. In contrast to other studies reporting a non-significant association between the A1298C variant and BC, we found that the A1298C variant acts as a risk factor for BC, particularly in the Western Asian population. Nevertheless, further studies are required to show the association of MTHFR C677T and A1298C gene polymorphisms with BC in an Asian population in the presence of other factors such as folate levels.
References
Akilzhanova A et al (2013) Genetic profile and determinants of homocysteine levels in Kazakhstan patients with breast cancer. Anticancer Res 33:4049–4059
Akilzhanova A, Takamura N, Zhaojia Y, Aoyagi K, Karazhanova L, Yamashita S (2006) Kazakhstan: a folate-deficient area? Eur J Clin Nutr 60:1141–1143
Akram M, Malik F, Kayani MA (2012) Mutational analysis of the MTHFR gene in breast cancer patients of Pakistani population. Asian Pac J Cancer Prev 13:1599–1603
Antonaros F et al (2019) MTHFR C677T polymorphism analysis: a simple, effective restriction enzyme-based method improving previous protocols. Mol Genet Genom Med 7:e628
Botezatu A, Socolov D, Iancu IV, Huica I, Plesa A, Ungureanu C, Anton G (2013) Methylenetetrahydrofolate reductase (MTHFR) polymorphisms and promoter methylation in cervical oncogenic lesions and cancer. J Cell Mol Med 17:543–549
Castiglia P et al (2019) Methylenetetrahydrofolate reductase (MTHFR) C677T and A1298C polymorphisms in breast cancer: a Sardinian preliminary case-control study. Int J Med Sci 16:1089
Cheng H, Deng Z, Wang Z, Zhang W, Su J (2012) MTHFR C677T polymorphisms are associated with aberrant methylation of the IGF-2 gene in transitional cell carcinoma of the bladder. J Biomed Res 26:77–83
Choi S-W, Mason JB (2000) Folate and carcinogenesis: an integrated scheme. J Nutr 130:129–132
Chou Y-C et al (2006) Genetic polymorphisms of the methylenetetrahydrofolate reductase gene, plasma folate levels and breast cancer susceptibility: a case–control study in Taiwan. Carcinogenesis 27:2295–2300
Del Gobbo GF, Price EM, Hanna CW, Robinson WP (2018) No evidence for association of MTHFR 677C> T and 1298A> C variants with placental DNA methylation. Clin Epigenet 10:34
Deligezer U, Akisik EE, Dalay N (2005) Homozygosity at the C677T of the MTHFR gene is associated with increased breast cancer risk in the Turkish population. In Vivo 19:889–893
Ergul E, Sazci A, Utkan Z, Canturk NZ (2003) Polymorphisms in the MTHFR gene are associated with breast cancer. Tumor Biol 24:286–290
Ferreira MA et al (2019) Genome-wide association and transcriptome studies identify target genes and risk loci for breast cancer. Nat Commun 10:1–18
Floris M et al (2020) MTHFR, XRCC1 and OGG1 genetic polymorphisms in breast cancer: a case-control study in a population from North Sardinia. BMC Cancer 20:1–15
Ghoncheh M, Mahdavifar N, Darvishi E, Salehiniya H (2016) Epidemiology, incidence and mortality of breast cancer in Asia Asian Pacific. J Cancer Prev 17:47–52
He J et al (2014) Association between dietary intake of folate and MTHFR and MTR genotype with risk of breast cancer. Genet Mol Res 13:8925–8931
He L, Shen Y (2017) MTHFR C677T polymorphism and breast, ovarian cancer risk: a meta-analysis of 19,260 patients and 26,364 controls. OncoTargets Ther 10:227–238. https://doi.org/10.2147/ott.s121472
Hedayatizadeh-Omran A, Alizadeh-Navaei R, Toghani-Hulari F, Amjadi O (2017) Association between MTHFR (C677T) gene polymorphism with breast cancer in Northern Iran. WCRJ 4:e876
Hekim N, Ergen A, Yaylım İ, Yılmaz H, Zeybek Ü, Öztürk O, İsbir T (2007) No association between methylenetetrahydrofolate reductase C677T polymorphism and breast cancer. Cell Biochem Funct 25:115–117
Hesari A et al (2019) Evaluation of the two polymorphisms rs1801133 in MTHFR and rs10811661 in CDKN2A/B in breast cancer. J Cell Biochem 120:2090–2097
Hosseini M, Houshmand M, Ebrahimi A (2011) MTHFR polymorphisms and breast cancer risk. Arch Med Sci: AMS 7:134
Huang C-Y et al (2014) Evaluation of the contribution of methylenetetrahydrofolate reductase genotypes to Taiwan breast cancer. Anticancer Res 34:4109–4115
Jiang-hua Q, De-chuang J, Zhen-duo L, Shu-de C, Zhenzhen L (2014) Association of methylenetetrahydrofolate reductase and methionine synthase polymorphisms with breast cancer risk and interaction with folate, vitamin B 6, and vitamin B 12 intakes. Tumor Biol 35:11895–11901
Kalyankumar C, Jamil K (2006) Methylene tetrahydofolate reductase (MTHFR) C677T and A1298C polymorphisms and breast cancer in South Indian population. Int J Cancer Res 2:143–151
Kaya EF, Karakus N, Ulusoy AN, Özaslan C, Kara N (2016) Association of the MTHFR gene C677T polymorphism with breast cancer in a Turkish population. Oncol Res Treat 39:534–538
Kumar P, Yadav U, Rai V (2015) Methylenetetrahydrofolate reductase gene C677T polymorphism and breast cancer risk: evidence for genetic susceptibility. Meta Gene 6:72–84
Lajin B, Sakur AA, Ghabreau L, Alachkar A (2012) Association of polymorphisms in one-carbon metabolizing genes with breast cancer risk in Syrian women. Tumor Biol 33:1133–1139
Lee S-A et al (2004) Methylenetetrahydrofolate reductase polymorphism, diet, and breast cancer in Korean women. Exp Mol Med 36:116–121
Lin W-Y et al (2004) The MTHFR C677T polymorphism, estrogen exposure and breast cancer risk: a nested case-control study in Taiwan. Anticancer Res 24:3863–3868
Liu Y, Zhou L-S, Xu X-M, Deng L-Q, Xiao Q-K (2013) Association of dietary intake of folate, vitamin B 6 and B 12 and MTHFR genotype with breast cancer risk Asian Pacific. J Cancer Prev 14:5189–5192
Lu Q, Jiang K, Li Q, Ji Y-J, Chen W-L, Xue X-H (2015) Polymorphisms in the MTHFR gene are associated with breast cancer risk and prognosis in a Chinese population. Tumor Biol 36:3757–3762
Meneses-Sanchez P, Garcia-Hernandez SC, Porchia LM, Pérez-Fuentes R, Torres-Rasgado E, Soto ADA, Gonzalez-Mejia ME (2019) C677T and A1298C methylenetetrahydrofolate reductase polymorphisms and breast cancer susceptibility among Latinos: a meta-analysis. Breast Cancer 26:602–611
Mir MM, Dar JA, Dar NA, Dar MS, Salam I, Lone MM, Chowdary NA (2008) Combined impact of polymorphism of folate metabolism genes; glutamate carboxypeptidase, methylene tetrahydrofolate reductase and methionine synthase reductase on breast cancer susceptibility in Kashmiri women. Int J Health Sci 2:3
Mohammadzadeh G, Karimi M, Bazyar M, Hosseini S-M (2016) Lack of association between MTHFR C677T polymorphism and breast cancer risk in Ahvaz, west south-Iran. Adv Biomed Res. https://doi.org/10.4103/2277-9175.176352
Momenimovahed Z, Salehiniya H (2019) Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer: Targets Ther 11:151
Mubarik S, Malik SS, Wang Z, Li C, Fawad M, Yu C (2019) Recent insights into breast cancer incidence trends among four Asian countries using age-period-cohort model. Cancer Manage Res 11:8145
Naushad SM, Pavani A, Digumarti RR, Gottumukkala SR, Kutala VK (2011) Epistatic interactions between loci of one-carbon metabolism modulate susceptibility to breast cancer. Mol Biol Rep 38:4893–4901
Niu Z, Zhao H, Hou X (2017) Association of MTHFR, MTRR and MTR polymorphisms with breast cancer risk: a study in Chinese females. Int J Clin Exp Pathol 10:7059–7066
Odin E, Wettergren Y, Carlsson G, Danenberg PV, Termini A, Willén R, Gustavsson B (2006) Expression and clinical significance of methylenetetrahydrofolate reductase in patients with colorectal cancer. Clin Colorectal Cancer 5:344–349
Ozen F, Erdis E, Sik E, Silan F, Uludag A, Ozdemir O (2013) Germ-line MTHFR C677T, FV H1299R and PAI-1 5G/4G variations in breast carcinoma. Asian Pac J Cancer Prev 14:2903–2908
Paydar P et al (2019) Epigenetic modulation of BRCA-1 and MGMT genes, and histones H4 and H3 are associated with breast tumors. J Cell Biochem 120:13726–13736
Pooja S et al (2015) MTHFR 677C> T polymorphism and the risk of breast cancer: evidence from an original study and pooled data for 28031 cases and 31880 controls. PLoS ONE 10:e0120654
Prasad VV, Wilkhoo H (2011) Association of the functional polymorphism C677T in the methylenetetrahydrofolate reductase gene with colorectal, thyroid, breast, ovarian, and cervical cancers. Oncol Res Treat 34:422–426
Rahimi Z, Bozorgi M, Rahimi Z, Shakiba E, Yari K, Jalilian N, Vaisi-Raygani A (2019) MTHFR C677T polymorphism is associated with the risk of breast cancer among Kurdish population from Western Iran. Int J Cancer Manage. https://doi.org/10.5812/ijcm.67895
Rai V (2014) Methylenetetrahydrofolate reductase A1298C polymorphism and breast cancer risk: a meta analysis of 33 studies. Ann Med Health Sci Res 4:841–851
Sangrajrang S, Sato Y, Sakamoto H, Ohnami S, Khuhaprema T, Yoshida T (2010) Genetic polymorphisms in folate and alcohol metabolism and breast cancer risk: a case–control study in Thai women. Breast Cancer Res Treat 123:885–893
Shrubsole MJ et al (2004) MTHFR polymorphisms, dietary folate intake, and breast cancer risk: results from the Shanghai Breast Cancer Study. Cancer Epidemiol Prev Biomark 13:190–196
Song A, Zhao L, Li Y, Wu L, Li Y, Liu X, Lan S (2016) Haplotypes of the MTHFR gene are associated with an increased risk of breast cancer in a Han Chinese population in Gansu province. IUBMB Life 68:526–534
Suzuki T et al (2008) One-carbon metabolism-related gene polymorphisms and risk of breast cancer. Carcinogenesis 29:356–362
Teng Z, Wang L, Cai S, Yu P, Wang J, Gong J, Liu Y (2013) The 677C> T (rs1801133) polymorphism in the MTHFR gene contributes to colorectal cancer risk: a meta-analysis based on 71 research studies. PLoS ONE 8:e55332
Tomita LY (2016) Folate and cancer: is there any association? J Inborn Errors Metab Screen 4:2326409816661357
Wan L, Li Y, Zhang Z, Sun Z, He Y, Li R (2018) Methylenetetrahydrofolate reductase and psychiatric diseases. Transl Psychiatry 8:1–12
Waseem M et al (2016) Association of MTHFR (C677T) gene polymorphism with breast cancer in North India. Biomark Cancer. https://doi.org/10.4137/BIC.S40446
Weiner A et al (2010) Polymorphic variants of folate metabolizing genes (C677T and A1298C MTHFR and C1420T SHMT1 and G1958A MTHFD) are not associated with the risk of breast cancer in the West Siberian Region of Russia. Mol Biol 44:720–727
Weiwei Z, Liping C, Dequan L (2014) Association between dietary intake of folate, vitamin B6, B12 & MTHFR, MTR Genotype and breast cancer risk. Pak J Med Sci 30:106
Yip C-H (2009) Breast cancer in Asia. Cancer epidemiology. Humana Press, Totowa, pp 51–64
Yu C-P, Wu M-H, Chou Y-C, Yang T, You S-L, Chen C-J, Sun C-A (2007) Breast cancer risk associated with multigenotypic polymorphisms in folate-metabolizing genes: a nested case-control study in Taiwan. Anticancer Res 27:1727–1732
Zhang X, Liu T, Li Y, Li S (2015) Association between MTHFR 677C/T and 1298A/C gene polymorphisms and breast cancer risk. Genet Mol Res 14:230
Zhang J, Zhang L, Li G (2016) Association between MTHFR gene 1298A> C polymorphism and breast cancer susceptibility: a meta-analysis based on 38 case-control studies with 40,985 subjects. World J Surg Oncol 14:230
Zhu B, Wu X, Zhi X, Liu L, Zheng Q, Sun G (2014) Methylenetetrahydrofolate reductase C677T polymorphism and type 2 diabetes mellitus in Chinese population: a meta-analysis of 29 case-control studies. PLoS ONE 9:e102443
Acknowledgements
The current study was funded by a Grant Number (#22963) from the Vice-chancellor for Research, Shiraz University of Medical Sciences, Shiraz, Iran.
Funding
No funding was received.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no conflicts of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rezaee, M., Akbari, H., Momeni-Moghaddam, M.A. et al. Association of C677T (rs1081133) and A1298C (rs1801131) Methylenetetrahydrofolate Reductase Variants with Breast Cancer Susceptibility Among Asians: A Systematic Review and Meta-Analysis. Biochem Genet 59, 367–397 (2021). https://doi.org/10.1007/s10528-020-10020-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-020-10020-z