Abstract
To have the sparsity of deep neural networks is crucial, which can improve the learning ability of them, especially for application to high-dimensional data with small sample size. Commonly used regularization terms for keeping the sparsity of deep neural networks are based on L1-norm or L2-norm; however, they are not the most reasonable substitutes of L0-norm. In this paper, based on the fact that the minimization of a log-sum function is one effective approximation to that of L0-norm, the sparse penalty term on the connection weights with the log-sum function is introduced. By embedding the corresponding iterative re-weighted-L1 minimization algorithm with k-step contrastive divergence, the connections of deep belief networks can be updated in a way of sparse self-adaption. Experiments on two kinds of biomedical datasets which are two typical small sample size datasets with a large number of variables, i.e., brain functional magnetic resonance imaging data and single nucleotide polymorphism data, show that the proposed deep belief networks with self-adaptive sparsity can learn the layer-wise sparse features effectively. And results demonstrate better performances including the identification accuracy and sparsity capability than several typical learning machines.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
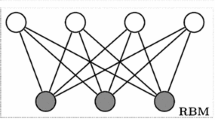
Restricted Boltzmann machine (RBM) which is a stochastic artificial neural network with two layers: one is a visible input layer, and the other is a hidden layer, can learn the probability distribution over the raw features [1,2,3]. According to the distribution of visible units, two different RBMs have been proposed, i.e., Bernoulli-Bernoulli RBM (BBRBM) [4] and Gaussian-Bernoulli RBM (GBRBM) [5, 6]. BBRBM assumes that the visible unit is of binary data, and GBRBM assumes that the visible unit is Gaussian distribution. In practice, the appropriate RBM type can be selected based on the dataset. Deep-belief network (DBN) is a probability generation model, which can be viewed as a composition of simple, unsupervised networks such as RBMs [1, 4, 7]. DBN which is widely regarded as one of the effective deep learning models, can obtain the multi-layer nonlinear representation of the data by greedy layer-wise training [8,9,10]. DBN possesses inherent power for unsupervised feature learning [11], and it has been widely used in many fields, e.g., image classification, document processing, object segmentation, social network, automatic control [12, 13].
For DBN models, it consists of one layer of visible units and multiple layers of hidden units, and neurons between neighboring layers can have a widely interconnected structure [1]. However, studies from the brain’s nervous system have shown that such a system employs a highly sparse mechanism. In brain organization and processing information, there exist two kinds of sparse properties: one is connection sparseness and the other is response sparseness [14]. The connection sparseness is also known as the sparse structure of the network or sparse topology. And the response sparseness is the sparse representation or sparse coding of neurons. These two kinds of sparseness guarantee the high promotion ability of human neural systems. Compared with the distributed information representation, sparsity can provide a higher quality of storage and better generalization capabilities [14]. By a combination of experimental, computational, and theoretical studies, it has been verified the existence of an underlying principle involved in sensory information processing. Namely, the information is represented by a relatively small number of simultaneously active neurons out of a large population, commonly referred to as sparse coding [15]. In [14], it confirmed the existence of sparse connectivity within the basal ganglia. DeepMind generated a function of grid cells to achieve automatic tracking just like humans recently, where the use of regularization to obtain sparsity is critical to grid-like representations [16].
At present, there are many approaches to get the topology sparseness as well as representation sparseness of DBNs, and introducing regularization terms into the parameters updating procedure are most commonly used to get the sparsity. For regularization algorithm, the purpose is to solve a constrained optimization issue and to remove unnecessary connections in the training process. In this way, it can both reduce the complexity of the model and improve the generalization ability of the network. The methods of weighting constraints include Gaussian regularizer [17], Laplace regularizer [18], weight elimination [19] and soft weight sharing [20]. Among them, the most widely used methods are the Gaussian regularizer and Laplace regularizer. And they use L2-norm or L1-norm on the connections as their penalty functions. Moreover, some other techniques, such as Dropout, DropConnect, pruning, low-rank matrix decomposition, knowledge distillation, have also been used to achieve the topology sparseness or reduce the complexity of neural networks [17,18,19,20,21,22,23,24].
In addition to adding regularization on the connections to achieve the sparse topology, simulating the sparse response of the hidden neurons is another important way to improve the generalization ability. Hinton et al. [25] showed that networks with low response frequency neurons have a better explanation and generalization ability. Lee and Ng penalized the mean of the hidden neuron’s response to get the response sparsity of the hidden layer by using Kullback-Leibler (KL) divergence [26, 27]. Ranzato et al. [28] used the energy-based method to add a coding pattern with sparse representation. By adding the L1/L2 penalty to the response probability of the hidden neurons, a network was proposed to obtain the local correlation of the hidden neurons [29]. The L1-norm constraint was applied directly to the response of the hidden neurons in the fine-tuning process [30]. Srivastava and Hinton et al. used Dropout to prevent neural networks from over-fitting [31]. Almost all of the above methods to obtain the sparseness of DBN are based on penalization with L1-norm, L2-norm or KL divergence.
For controlling the sparsity, the best way is straightforward to use L0-norm since it directly measures the number of non-zero elements. However, since it is a NP-hard problem to solve the optimization problem based on L0-norm, many methods for obtaining the spareness are based on L1-norm, L2-norm or KL divergence instead. Although L1-norm is generally used as a substitution of L0-norm, there still exists a key difference between L1-norm and L0-norm, namely, the dependence on magnitude [24]. For L0-norm, the penalization on the components is democratic, while for L1-norm, the penalization on larger components is penalized more heavily than the small components. For L2-norm, it is not possible to give a sparse solution. The KL divergence is used for measuring how different two distributions are, and it is usually applied to control the average activation values of hidden neurons. Finding a better way to address the issue of sparsity in DBN is still a challenging topic. In this paper, by introducing a log-sum minimization problem and its corresponding iterative re-weighted-L1 minimization algorithm, we dedicate to seek the approximative solution of the optimum problem with L0-norm and further, sparse connections of DBNs.
On the other hand, for DBNs, sampling method based on Markov Chain Monte Carlo (MCMC) strategy is often used. In such a sampling process, each variable is sampled from a posterior distribution given the current state of other variables. Then the unbiased estimate of the position parameter is obtained by using the logistic likelihood method with multiple alternating Gibbs sampling. However, this procedure is time-consuming and requires a large number of samples for an accurate estimate. Hinton proposed a fast learning algorithm, i.e., the k-step contrastive divergence (CD), to overcome the complication of Gibbs sampling [32]. It is known that maximizing the log likelihood of the data is equivalent to minimizing the KL divergence (denoted by \(KL(P^{0}\|{P}_{\theta }^{\infty })\)) between the initial data distribution P0 and the equilibrium distribution of the visible variables, \({P}_{\theta }^{\infty }\), where 𝜃 refers to the coefficients of the learning machine, and \(P^{\infty }_{\theta }\) is generated by the sustained Gibbs sampling from the generation model. For CD algorithm with k steps, it minimizes the difference between \(KL(P^{0}\|P^{\infty }_{\theta })\) and \(KL({P}_{\theta }^{k}\|{P}_{\theta }^{\infty })\) instead of minimizing \(KL(P^{0}\|{P}_{\theta }^{\infty })\), where \(P^{k}_{\theta }\) denotes the reconstruction distribution of the data generated by k steps Gibbs sampling. The CD algorithm with k steps is an effective alternative to maximum likelihood learning, which can relieve high computational demands for samples acquired from the equilibrium distribution. As Hinton pointed out, the intuitive motivation for using the k-step CD algorithm is to leave the initial distribution over the visible variables unaltered when implementing Gibbs sampling in Markov chain. Rather than running the Markov chain to the equilibrium state and comparing the difference between the initial state and the final state, one can simply run the Markov chain with k steps and update the parameters of the model to reduce the tendency of the chain to deviate from the initial distribution.
In this paper, we aim to obtain a DBN with sparsity learning capability by embedding the k-step CD algorithm with the iterative re-weighted-L1 minimization algorithm, and further apply it to biomedical data, such as brain magnetic resonance imaging (MRI) data and single nucleotide polymorphism (SNP) data. For the MRI data, it contains the concentration of grey matter in different brain regions and the correlation values of different regions of interest (ROI), which do not have any spatial topological structure. Similarly, the SNP data contains the DNA polymorphism data caused by single nucleotide variation, and it doesn’t possess spatial topological structure either. Thus, other deep learning models, such as the convolution operation of convolutional neural networks which considers domain relationships of the data are no longer applicable. Additionally, it should be noticed that for biomedical datasets, they are usually of small sample size but a large number of features. Although nowadays deep networks have been constantly shown to be one of the most powerful tools in big data analysis, its application to biomedical data is still limited. That is because deep learning models are very easy to fall into overfitting for data with n ≪ p, where n is the sample size and p is the number of features [33, 34]. Seeking a deep network with strong sparse self-learning ability is just an effective solution to this issue. It will be shown in Section 2 that the re-weighted-L1 function keeps more approximation ability of L0-norm than L1-norm or L2-norm. Thus by combining a iterative re-weighted-L1 algorithm, the connections of a DBN can be updated adaptively in a sparse way by the k-step CD algorithm, and such a network is called a self-adaptive sparse DBN. In this way, the deviation of the Markov chain from the initial distribution can be reduced, and it thus efficaciously minimize the probability that the model distorts the data. Experiments on the biomedical data show that the proposed self-adaptive sparse DBN is valid to avoid over-fitting, and it can achieve good identification ability by learning the sparse structure of the feature space.
2 Iterative algorithm for re-weighted L 1 minimization
In this section, the iterative re-weighted-L1 minimization algorithm will be presented; it is a good approximation to the sparse optimum solution of minimization problem with L0-norm.
In [35], it was proved that minimization of \({\sum }_{i=1}^{n}\log |x_{i}|\) is consistent with minimization of \({\sum }_{i=1}^{n}|x_{i}|^{p}\) when \(p\rightarrow 0\). Further, because
where x is a n-dimensional vector with xi being the i-th element, [24, 35, 36] put forward that the minimization of \({\sum }_{i=1}^{n}\log |x_{i}|\) is an effective approximation to that of ∥x∥0. Furthermore, in order to ensure the existence of \(\log |x_{i}|\) when \(|x_{i}|\rightarrow 0\), a small ε is usually added to make the minimization problem meaningful, and the solution of
is proposed as an alternative to get better approximate solution of minimization problem with L0-norm than the often used L1-norm [24, 36]. The objective function of (2) is called as the log-sum function.
In order to get the iterative solution of (2) for self-adaptive sparse CD algorithm with k steps, we introduce the absolute value optimization problem
where ci ≥ 0. Noting that |xi| is the minimum of zi with conditions |xi|≤ zi, (3) has the following equivalent form
By (4), the optimization problem (2) can be rewritten in its equivalent form
That is, if x∗ is the optimum solution of (2), then u∗ is the optimum solution of (5) when u∗ = |x∗|. Alternatively, if u∗ is the optimum solution of (5), then x∗ is the optimum solution of (2).
A general form of (5) is
where g is a concave function with C being a convex set. The solution to the problem (6) can be improved step by step by minimizing a linearized g:
The solution obtained by the iteration algorithm is the approximation of convex optimization. In particular, for the problem (5), one can get that
Thus, by the equivalent relationship between the optimum solutions of (2) and (5), we have
Let
then (8) can be rewritten as
Thus, the solution of (2) can be approximated by the iterative solutions of (10).
The following iterative re-weighted-L1 minimization algorithm, i.e., Algorithm 1, gives the iterative process of solving minimization problem (2). Based on it, the solution of (2) can be dynamically updated, and it is a good approximation to the sparse optimum solution of minimization problem with L0-norm.

Furthermore, in order to show the advantage of the re-weighted-L1 function (i.e., \(f_{R,\epsilon }\triangleq \eta \cdot |x|=\frac {1}{|x|+\epsilon }\cdot |x|\)) in achieving sparsity, we compare it with the following penalty functions: f0(x) = 1{x≠ 0} and f1(x) = |x|. From Fig. 1, it shows that fR,𝜖 can well approximate to f0 as \(\epsilon \rightarrow 0\).
As can be shown, by combining the iterative re-weighted-L1 minimization algorithm with the k-step CD algorithm, one can obtain more sparse solution of the connective weights than using the L1-norm or the CD algorithm with just one step. In this way, we can avoid over-fitting problem and improve the generalization of DBNs.
3 Self-adaptive sparse DBNs (SAS-DBNs)
In this section, by embedding the iterative re-weighted-L1 minimization algorithm, i.e., Algorithm 1, we will present the sparse self-adaptively learning for k-step CD, and get the sparse architecture for each component of DBN, namely, RBM. As a result, the unsupervised sparse learning solution of the whole DBN can be achieved.
A DBN is stacked and trained by multi-layer RBMs, and each RBM can find a compressed feature representation of the current input data. In an RBM, there are two layers: one is the visible input layer, and the other is a hidden layer. There are connections between the layers but no connections between units within each layer. Assume v is the Nv-dim visible units, h is the Nh-dim hidden units, α is the Nv-dim bias of the visible units, β is the Nh-dim bias of the hidden units, and \(W=\{W_{ij}\}_{N_{v}\times N_{h}}\) with each Wij is the connection weight between vi and hj. The likelihood function P𝜃(v) is defined as the distribution of the observed data v𝜃, where 𝜃 = {W,α,β} is the set of all parameters. Maximizing P𝜃(v) is the main task of training an RBM, and that is equal to determining the parameters to fit the given training samples, i.e., to find a 𝜃∗, such that
where \({\mathscr{L}}(\theta ) = \log P_{\theta }(v) \). \({\mathscr{L}}(\theta )\) is just the objective function to be optimized for training RBM. By performing stochastic steepest ascent in the log probability on the training data, the weights can be updated based on
in which, ζ is the learning rate, and 〈⋅〉 is the operator of expectation with the corresponding distribution denoted by the subscript.
Due to the difficulty of calculating Z𝜃, it is a quite hard task to obtain an unbiased sample of 〈vihj〉model. A much faster learning method is the k-step CD algorithm. The k-step CD algorithm is a fast learning algorithm of RBM which was proposed by Hinton [32]. Different from the Gibbs sampling method, k-step CD algorithm initializes the system states, i.e., neurons in the visible layer, with the training data. In this way, as Hinton pointed out, only a few state transitions are needed to obtain a good approximation of the model distribution. In the k-step CD algorithm, firstly, the initial states of the visible layer units, i.e., v(0), are assigned as the values of the training data, and based on the conditional probability p(h = 1|v(0)), the initial states of the hidden units h(0) can be sampled. Then, the conditional probability p(v = vr|h(0)) is utilized to obtain v(1), where vr is 1 and real-value for BBRBM and GBRBM respectively. The above processes are executed alternately until both v(k) and h(k) are obtained, which are used to approximately estimate 〈vihj〉model. Specially, for BBRBM,
For GBRBM,
in which, f(⋅) is the sigmoid activation function, σi is the standard deviation associated with Guassian visible units vi, and \(\mathcal {N}(\cdot |\mu ,\sigma ^{2})\) denotes the Guassion probability density function with mean μ and standard deviation σ.
For each RBM, if there is no restriction on the responses of hidden neurons and connections between layers, an unstructured network will be generated, which leads to the poor interpretability, high computational complexity and large space resource consumption of the network [37]. Adding sparse regularization terms on the objective function of training RBM is more likely to learn the structural characteristics of the data [38]. In Section 2, it has already been shown that the re-weighted-L1 function is an effective approximation to the sparse solution of minimization problem with L0-norm, thus, by introducing the sparse penalty term on the connection weights with the re-weighted-L1 function, and embedding its corresponding iterative re-weighted-L1 minimization algorithm with k-step CD, the sparse connection architecture of the RBM will be obtained. At the same time, since L2-norm penalty term can reduce the noise or variation in the data, we also introduce L2-norm term in the connection weights. Additionally, the KL divergence on the average responses of hidden neurons is also utilized to achieve the sparse responses of the hidden layer neurons. The improved objective function to be optimized for training RBM is
where λ1, λ2 and λ3 are penalty parameters, ε is a given constant. The KL divergence, \(KL(\rho \parallel p_{j})=\rho \log \frac {\rho }{p_{j}}+(1-\rho )\log \frac {1-\rho }{1-p_{j}}\) is the relative entropy between the two random variables with the mean ρ and the mean pj. It is used to measure the difference between two different distributions. The purpose of introducing the KL divergence is to force the average response value of the hidden neurons to be approximately equal to the default initial value ρ. Here we take \(p_{j}=\frac {1}{N_{s}}{\sum }^{N_{s}}_{q=1}\frac {1}{1+e^{-{\sum }^{N_{v}}_{i=1}v^{(q)}_{i}W_{ij}-\beta _{j}}}\) as the average activation degree of the j-neuron in the hidden layer with Ns samples, and Nv is the number of nodes in the current visual layer. The deviations of the KL divergence for the parameters are
where \(\sigma ^{(q)}_{j}=\sigma \left ({\sum }_{i=1}^{N_{v}}v^{(q)}_{i}W_{ij}+\beta _{j}\right )=\frac {1}{1+e^{-{\sum }^{N_{v}}_{i=1}v^{(q)}_{i}W_{ij}-\beta _{j}}}\). Thus, by combining the iterative re-weighted-L1 minimization algorithm with the k-step CD sampling process, the parameters of an unsupervised RBM can be updated in a sparse self-adaptive way by Algorithm 2.

Based on Algorithm 2, each RBM can be trained to have sparse connections adaptively, and this process is called as an RBM reconstructed with SAS-k-CD. Thus, the unsupervised process of a DBN with self-adaptive sparsity (SAS-DBN) can be obtained by those stacking sparse RBMs. The following Fig. 2 presents the whole learning process of SAS-DBN with stacked sparse RBMs.
Furthermore, the learned sparse DBN can be further fine-tuned with some fine-tuning methods, e.g., the back propagation (BP) algorithm. On noting that the fine-tuning process will redistribute the weights as well as the representations of the entire network, i.e., the learned sparsity of SAS-DBN will be destroyed. To ensure that the sparse structure of the learned SAS-DBN is still held during fine-tuning, two methods are generally utilized. One is that we can retain the sparse structure of SAS-DBN, and then carry out fine-tuning. Another one is bringing sparse learning into fine-tuning. In our experiments, we have compared both methods, namely, fine-tuning based on the retained sparse structure learned by SAS-DBN, and fine-tuning adding sparsity regularization terms, such as KL divergence, L1-norm and log-sum function. It indicates that there exists no significant difference between them. Thus, we only considered the first method here.
4 Experiments
One of the main purpose of implementing sparseness on deep learning is to avoid over-fitting and improve the identification ability, especially for high-dimensional data with small sample size. In this section, we will use both the brain functional magnetic resonance imaging data and single nucleotide polymorphism data to demonstrate the validation of the proposed DBN with self-adaptive sparsity. For the MRI data, adding the fine-tuning process does improve its accuracy rate, but there exists nearly no improvement on the accuracy rate of the SNP data with or without fine-tuning. Considering the time complexity caused by the fine-tuning process, we only utilized fine-tuning for the MRI data and didn’t use it for the SNP data. Moreover, the experiments show that although these datas are with a large number of features but low samples, SAS-DBN can better represent data in a sparse architecture, and it has the best classification accuracy among many classifiers we have tested.
4.1 Experiments on MRI database
This dataset is the MLSP 2014 Schizophrenia Classification Challenge data, which can be downloaded from https://www.kaggle.com/c/mlsp-2014-mri. The data are collected on a 3T MRI scanner at the Mind Research Network. Image preprocessing was performed using Statistical Parametric Mapping software, and further feature extraction was performed using independent component analysis, yielding features from different imaging modalities, i.e., source-based morphometric (SBM) loadings and functional network connectivity (FNC) features for structural magnetic resonance images (sMRI) and resting state functional MRI (rs-fMRI), respectively. The data consist of 40 patients with schizophrenia and schizoaffective disorders (both are denoted by SZs) and 46 healthy controls (HCs). A diagnosis of SZs was made by using the Structured Clinical Interview for DSM-IV (DSM: Diagnostic and Statistical Manual of Mental Disorders) [39]. 410 features (32 for SBM and 378 for FNC) are extracted for each sample. SBM loadings are weights of brain maps obtained from the application of ICA, which indicate the concentration of grey matter in different regions of the subject’s brain. The concentration of gray-matter is indicative of the computational power in a particular region of the brain [40]. FNC features are the pair-wise correlation values between the time courses of 28 ICA brain maps, which can be seen as features describing the subject’s overall level of synchronicity between brain areas [41]. We denote the data as \(D=\{(x_{i},y_{i})\}^{n}_{i=1}\), where n = 86 (40 SZs and 46 HCs);  (32 from the SBM and 378 from the FNC); yi ∈{0,1} (0 = ’Healthy Control’, 1 = ’Schizophrenic Patient’). 86 subjects were randomly divided into two subsets: 70% subjects for training and the rest 30% ones for testing. For the sake of obtaining a stable classification, we performed random sampling from 86 subjects repeatedly for 30 times.
(32 from the SBM and 378 from the FNC); yi ∈{0,1} (0 = ’Healthy Control’, 1 = ’Schizophrenic Patient’). 86 subjects were randomly divided into two subsets: 70% subjects for training and the rest 30% ones for testing. For the sake of obtaining a stable classification, we performed random sampling from 86 subjects repeatedly for 30 times.
The deep architecture of SAS-DBN we used here contains 5 hidden layers with 200, 100, 50, 50, and 50 units respectively. In such a stacked DBN, we adopt a GBRBM as the first RBM and BBRBM as others. Because the grid search method can simply make a complete search over a given parameters space, easily be parallelized and find the optimal parameter for deep learning with a much more stable solution, it was used to find the optimal parameters in this paper. Specifically, one of the parameters is selected by the grid search method when other parameters are fixed. And all parameters are optimized by repeating the above process. Based on the grid search method, the optimal parameters corresponding to GBRBM and BBRBM can be obtained. Corresponding to GBRBM and BBRBM respectively, the learning rate ζ are 0.001 and 0.1, the penalty parameter λ1 are 0.03 and 0.0002, the re-weight L1-norm penalty parameter λ2 are 0.001 and 0.001, 𝜖 are 0.1 and 0.1, the KL divergence penalty parameter λ3 are 0.1 and 0.1, and the sparse parameter ρ are 0.01 and 0.01, respectively. For simplifying, the steps of CD for each RBM is selected as the same k.
4.1.1 Model selection for the steps of CD
We first discuss how to select the steps of CD, and perform SAS-DBNs with different k which are chosen from 1 to 7. Other parameters of SAS-DBNs are chosen as mentioned above. Figure 3 shows the classification accuracy rates (AR) and the non-zero ratio of the network connections with different k, where AR is defined as
in which, Corrp, Corrc, InCorrc and InCorrp denote the number of correctly predicted schizophrenic patients, correctly predicted healthy subjects, healthy subjects incorrectly classified as schizophrenic patients, and schizophrenic patients incorrectly classified as healthy subjects, respectively.
a shows AR of SAS-DBNs with different k steps CD and the numbers 1 to 7 refer to the different selections of k. b displays that with different k, the combinatorial graph with a bar graph for the average non-zero ratio of connections in the first RBM and a line chart for AR. The horizontal axis denotes the different selections of k. For the vertical axis, the primary axis on the left is denoted as the average non-zero ratio of connections and the secondary axis on the right is denoted as the AR. Here shows the connection rate of the first RBM, and the connections of other RBMs also have the same tendencies
Figure 3 shows the RBM has its best AR with k = 5. Meanwhile, the non-zero ratio of the network connections keeps down with the increasing of k, and the decline tends to be gentle after k = 5. Thus, we choose 5 as the steps of CD.
4.1.2 Weight sparsity
Weight sparsity is an important index to demonstrate the validation of a deep learning machine in overcoming the over-fitting problem. In order to show the sparse learning ability of the proposed SAS-DBN, we compare SAS-DBN with six different deep learning models, namely, stacked RBM adding a SVM classifier (denoted by SR-SVM), DBN with L2-norm penalty on the connection weights (denoted by DBN), DBN with L1-norm penalty on the connection weights and KL divergence penalty on the hidden neurons (denoted by sDBN), DBN with Dropout (denoted by DP-DBN), DBN with DropConnect (denoted by DPC-DBN), DBN with singular value decomposition (denoted by SVD-DBN), and DBN with pruning method (denoted by Pr-DBN). Each network is well trained and especially, for DP-DBNs, when the dropout ratio is selected as 0.1, 0.2, 0.3, 0.4 and 0.5, the average accuracy of 30 times is 0.709, 0.6833, 0.6795, 0.6795 and 0.6859, respectively. Thus, we choose the DP-DBN model with the dropout ratio being 0.1. Similarly, for DPC-DBNs, the average accuracy of 30 times is 0.7358, 0.7385, 0.7321, 0.7346 and 0.7410, respectively, with the drop connect ratio being 0.1, 0.2, 0.3, 0.4 and 0.5. So we choose the DPC-DBN with the drop connect ratio being 0.5. Similarly, for Pr-DBNs, we select the Pr-DBN with the pruning ratio of weights being 0.001. For each network, we perform the learning process for 30 times. For each time, the training data are sampled randomly 70% from the original data, and the rest data are the testing data.
In what follows, we will show the weight sparsity of SAS-DBN in comparison with other DBNs. The learning capability of weight sparsity can then strongly indicate why the SAS-DBN is valid for such kind of high-dimensional small sample size data.
For a quantifiable comparison, we list the weights sparsity of SAS-DBN and other six deep learning methods with two different indicators. The first indicator is the non-zero ratio of connections, and the results are shown in Table 1. Additionally, in Fig. 4a, we further compare SAS-DBN with two models, i.e., DPC-DBN and Pr-DBN, which have the highest average classification accuracy among the other six deep networks. In this figure, the different 5 colors, i.e., red, yellow, green, pink and blue refer to the non-zero ratio of each w(i) (i = 1,⋯ ,5) respectively.
There is another normalized indicator to measure the sparsity of connections within the network, i.e., the Hoyer sparseness measure (HSM). It is based on the relationship between the L1-norm and the L2-norm of the weights [42, 43]. The HSM of the weight matrix Wm×n is defined as follows:
The value of HSM is in [0,1]. When HSM closes to 1, the connection of the network becomes much sparser, i.e., most of the values in connections are zeros and only a few have significant values. The average HSM with random 30 learning times for three deep networks are shown in Table 2 and Fig. 4b.
Further, Fig. 5 shows that the learning ability for weight sparseness of SAS-DBN, DPC-DBN and Pr-DBN. It shows that SAS-DBN has more sparse connections than DPC-DBN and Pr-DBN, which is an important reason that SAS-DBN owns better identification and extraction abilities of sparse features than the other two deep learning models.
From Tables 1 and 2, Figs. 4 and 5, one can draw the conclusion that the proposed SAS-DBN has the sparsest connections than the others deep networks, and especially has the sparse learning ability of w(1). It should be noticed that the learning of w(1) is quite crucial, since it is the first and basic step to extract essential features and remove redundancy and noise from the original data.
4.1.3 Classification accuracy rates of fifteen methods
In order to show the classification results, we compare SAS-DBN with other several typical classifiers, i.e., linear SVM (denoted by SVM-I), SVM with RBF kernel (denoted by SVM-II), KNN, Naive Bayes (NB), Random Forest (RF), Sparse Representation Based Classifier (SRC), discriminant analysis classifier (DAC), and other six different deep learning models [44,45,46]. For each classifier, we perform classifications for 30 times. For each time, the training data are sampled randomly 70% from the original data, and the rest data are testing data.
The convergence performances of the proposed method here in the training and testing data are shown in Fig. 6. In which, there is a sharp fall between epochs 20 and 40. That is because the first 30 epochs were the training of SAS-DBN, and then was the fine-tuning. Moreover, the average results are shown in Table 3 and Fig. 7. Among these fifteen methods, the proposed method here has the best performance. Especially in comparing with other methods, the AR has been increased as 11.92%, 13.22%, 9.55%, 15.23%, 11.93%, 23.83%, 13.70%, 6.93%, 2.55%, 1.56%, 5.97%, 1.39%, 3.36%, 0.68%, respectively. From the above two subsections, we know SAS-DBN do have the best sparse learning ability and discrimination capability of the fMRI data.
4.2 Experiments on SNP database
In this section, subject recruitment and data collection were conducted by the Mind Clinical Imaging Consortium (MCIC). The SNPs data were collected from 208 subjects including 92 schizophrenia patients (age: 34 ± 11, 22 females) and 116 healthy controls (age: 32 ± 11, 44 females). All of them were provided written informed consent. Healthy participants were free of any medical, neurological or psychiatric illnesses and had no history of substance abuse. By the clinical interview of patients for DSM IV-TR disorders [47] or the comprehensive assessment of symptoms and history, patients met criteria for DSM-IV-TR schizophrenia [48, 49]. Antipsychotic history was collected as part of the psychiatric assessment.
A blood sample was obtained for each participant and DNA was extracted. Genotyping for all participants was performed at the Mind Research Network using the Illumina Infinium HumanOmni1-Quad assay covering 1,140,419 SNP loci. Bead Studio was used to make the final genotype calls. The PLINK software package http : //pngu.mgh.harvard.edu/ purcell/plink) was used to perform a series of standard quality control procedures, resulting in the final data set spanning 777,635 SNP loci. Each SNP was categorized into three clusters based on their genotype and was represented with discrete numbers: 0 for \('BB^{\prime }\) (no minor allele), 1 for \('AB^{\prime }\) (one minor allele) and 2 for \('AA^{\prime }\) (two minor alleles). Further, SNP with > 20% missing data was deleted, and missing data were further imputed. SNPs with minor allele frequency < 1% were removed. For genetic diseases, because pathways organize genes into biologically functional groups and model their interactions that capture correlation between genes, the genes involving in the same pathway process have similar effects and act together in regulating a biological system [50]. Therefore, using the pathway information can improve not only the predictive performance but also the interpretability of the selected features. In this paper, we select the SNPs falling into the schizophrenia genes’ pathway for analysis. After the selection, the dimension for each sample is 12513.
The deep architecture of SAS-DNN used here contains three hidden layers with 2000, 1000, and 500 units respectively. We use three BBRBM modules as the stacked SAS-DNN. Based on the grid search method, the learning rate ζ is 0.1, the penalty parameter λ1 is 0.0002, the re-weight L1-norm penalty coefficients λ2 is 0.0002, 𝜖 is 0.1, the KL divergence penalty coefficients λ3 is 0.05, the sparse parameters ρ is 0.05, and the steps of CD is 5. 208 subjects were randomly divided into two subsets: 148 subjects for training and the rest 60 ones for testing. We perform the SAS-DNN 30 times to obtain the average accuracy rate. The convergence performances of the proposed method here in the training and testing data are shown in Fig. 8. It indicated that the proposed method here can converge quickly in few iterations.
4.2.1 Weight sparsity
We compare the weights sparsity of SAS-DBN with the other six deep networks mentioned in Section 4.1.2. In the experiments, each network is well trained separately, and especially, for DP-DNNs, when the dropout ratio is selected as 0.1, 0.2, 0.3, 0.4 and 0.5, the average accuracy of 30 times is 0.8483, 0.8517, 0.765, 0.825 and 0.8717, respectively. Thus, we choose the DP-DNN model with the dropout ratio being 0.5. Similarly, for DPC-DNNs, the average accuracy of 30 times is 0.8717, 0.8633, 0.8783, 0.82 and 0.8717, respectively, with the drop connect ratio being 0.1, 0.2, 0.3, 0.4 and 0.5. So we choose the DPC-DNN with the drop connect ratio being 0.3. Similarly, for Pr-DNNs, we select the Pr-DNN with the pruning ratio of weights being 0.001. we perform the learning process 30 times. At each time, the training data are sampled from 70% of the original data.
The ratio of connection weights with non-zeros values between different layers on SNP data is shown in Table 4a. The HSM is presented in Table 4b. From them, we can see that SAS-DBN has the lowest non-zeros connection ratio and the highest HSM among the deep networks. Additionally, in Fig. 9a and b, we further compare SAS-DBN with two models, i.e., sDBN and SVD-DBN, which have the highest average classification accuracy among the other six deep networks.
4.2.2 Classification results on the SNP database
The performance of each classifier is quantified by using AR, Sensitivity (SS), Specificity (SC), positive predictive value (PPV) and negative predictive value (NPV). And the last four are defined as follows
AR, SS, SC, PPV and NPV provide different assessments of the classifier’s performance from five aspects, i.e., the proportion of all samples that are correctly predicted, the proportion of patients correctly predicted among all the predicted patients, the proportion of healthy subjects correctly predicted among all the predicted healthy subjects, the proportion of correctly predicted patients among the true patients, and the proportion of healthy subjects correctly predicted among those true healthy subjects.
We compare the proposed method, i.e., SAS-DBN with several classifiers that have fairly good performances. The classification results on the testing data set for different classifiers are summarized in Table 5. In Table 5, CLASSIFIER I refers to linear SVM, II for SVM with multi-layer perceptron kernel, III for sparse representations based classifier, IV for DBN, V for sDBN, VI for DP-DBN, VII for SVD-DBN, VIII for DPC-DBN, IX for Pr-DBN and X for SAS-DBN. For each classifier, we perform the classification process 30 times. At each time, the training data are sampled from 70% of the original data, and the rest data are testing data. The average results are shown in Table 5. We can see that the SAS-DBN performs the best among the classifiers. The AR has been increased as 7.59%, 22.93%, 5.90%, 11.29%, 6.91%, 11.80%, 6.40%, 11.13% and 13.99% respectively. Figure 10 further shows the classification results of all methods, where the horizontal axis represents different classifiers. The primary vertical axis on the left is denoted as the AR and the secondary axis on the right denotes other classification results, i.e., the SS, SC, PPV and NPV. Through the comparison, we can conclude that SAS-DBN has the best performance on the classification accuracy.
Compared with [47, 51] (the data here are the same as ours), we use 148 samples as the training and 60 samples as the testing. In these papers, they use 90% subjects for training and only the rest 10% are used for testing. The average accuracy here is over 98%, while the accuracy of classification in these papers are around 85%, and the best accuracy of classification is 65%, and the average accuracy is about 55% by Li et al. [52].
4.3 Remark
For two kinds of high-dimensional data with small sample size, the above two experiments show that by the iterative re-weighted-L1 minimization algorithm combining with k-step CD, the proposed deep belief networks with adaptive sparsity have the best classification performance than other typical classifiers including several deep networks. A crucial reason for the good classification results of SAS-DBN lies in that the iterative re-weighted-L1 minimization algorithm can effectively obtain the approximate solution of L0 minimization problem, especially by combining it with k-step CD algorithm, the parameters of the network can be adjusted adaptively in a quite sparse way. Furthermore, the learned sparse deep network can effectively remove the noise and redundant features from the original data. By the sparse structure, discriminative features of the data can be extracted in a sparse layer-wise way, which is critical for preventing overfitting and improving identification accuracy.
5 Conclusion
This work is motivated by the sparsity mechanism of brain neural networks. By exploring substitutions of the approximating solution of minimization problem with L0-norm, the k-step CD sampling process embedding the re-weighted-L1 minimization algorithm, and further the self-adaptive sparse DBNs are proposed. In this way, layer-wise sparse features of data can be effectively extracted, which are vital for learning the true distribution of the data, thereby it can enhance the generalization capacity as well as the discernment performance of the network. It is promising that the self-adaptive sparse DBNs will be a practical solution to the small number of samples but a large number of features problem. Numerical experiments of applying to brain imaging and genomics data give a good demonstration of their effectiveness, and it shows that the proposed methods can not only get a sparse deep network architecture in a self-adaptive way, but also have the potential to avoid over-fitting problem.
References
Qiao C, Gao B, Shi Y (2020) SRS-DNN: a deep neural network with strengthening response sparsity. Neural Comput Applic 32:8127–8142
Liu K, Wu J, Liu H, Sun M, Wang Y (2021) Reliability analysis of thermal error model based on DBN and Monte Carlo method. Mech Syst Signal Process 146:107020
Yan X, Liu Y, Jia M (2020) Multiscale cascading deep belief network for fault identification of rotating machinery under various working conditions. Knowl Based Syst 193:105484
Chen CLP, Feng S (2020) Generative and discriminative fuzzy restricted Boltzmann machine learning for text and image classification. IEEE Trans Cybern 50(5):2237–2248
Chu J, Wang H, Meng H, Jin P, Li T (2019) Restricted Boltzmann machines with Gaussian visible units guided by pairwise constraints. IEEE Trans Cybern 49(12):4321–4334
Zhang J, Wang H, Chu J, Huang S, Li T, Zhao Q (2019) Improved Gaussian–Bernoulli restricted Boltzmann machine for learning discriminative representations. Knowl Based Syst 185:104911
Qiao J, Wang L (2021) Nonlinear system modeling and application based on restricted Boltzmann machine and improved BP neural network. Appl Intell 51:37–50
Bengio Y (2009) Learning deep architectures for ai. Found Trends Mach Learn 2(1):1–127
LeCun Y, Bengio Y, Hinton GE (2015) Deep learning. Nature 521(7553):436–444
Gu L, Huang J, Yang L (2019) On the representational power of restricted boltzmann machines for symmetric functions and boolean functions. IEEE T Neur Net Lear 30(5):1335–1347
Chen Y, Zhao X, Jia X (2015) Spectral-spatial classification of hyperspectral data based on deep belief network. IEEE J Select Topics Appl Earth Observ Remote Sens 8(6):2381–2392
Hinton GE, Salakhutdinov RR (2006) Reducing the dimensionality of data with neural networks. Science 313(5786):504–507
Salakhutdinov R, Hinton G (2009) Replicated softmax: An undirected topic model. In: Proceedings of the 22nd International Conference on Neural Information Processing Systems, Curran Associates Inc., Red Hook, NY, USA, NIPS’09, pp 1607–1614
Morris G, Nevet A, Bergman H (2003) Anatomical funneling, sparse connectivity and redundancy reduction in the neural networks of the basal ganglia. J Physiol-Paris 97(4-6):581–589
Olshausen BA, Field DJ (2004) Sparse coding of sensory inputs. Current Opin Neurobiol 14 (4):481–487
Banino A, Barry C, Uria B, Blundell C, Lillicrap T, Mirowski P, Pritzel A, Chadwick M, Degris T, Modayil J, Wayne G, Soyer H, Viola F, Zhang B, Goroshin R, Rabinowitz N, Pascanu R, Beattie C, Petersen S, Kumaran D (2018) Vector-based navigation using grid-like representations in artificial agents. Nature 557, 429–433
Girosi F, Poggio T (1995) Regularization theory and neural networks architectures. Neural Comput 7(2):219–269
Williams P (1995) Bayesian regularization and pruning using a laplace prior. Neural Comput 7 (1):117–143
Weigend AS, Rumelhart DE, Huberman BA (1990) Generalization by weight-elimination with application to forecasting.. In: Proceedings of the 1990 conference on advances in neural information processing systems 3, Morgan Kaufmann Publishers Inc., San Francisco, CA, USA, NIPS-3, pp 875–882
Nowlan SJ, Hinton GE (1992) Simplifying neural networks by soft weight-sharing. Neural Comput 4(4):473–493
Hinton GE, Osindero S, Teh YW (2006) A fast learning algorithm for deep belief nets. Neural comput 18:1527–54
Denton E, Zaremba W, Bruna J, LeCun Y, Fergus R (2014) Exploiting linear structure within convolutional networks for efficient evaluation. In: Proceedings of the 27th international conference on neural information processing systems - volume 1, MIT Press, Cambridge, MA, USA, NIPS’14, pp 1269–1277
Wang E, Davis JJ, Zhao R, Ng HC, Niu X, Luk W, Cheung PYK, Constantinides GA (2019) Deep neural network approximation for custom hardware: Where we’ve been, where we’re going. ACM Comput Surv 52(2):1–39
J Candès E, Wakin MB, Boyd SP (2007) Enhancing sparsity by reweighted l1 minimization. J Fourier Anal Appl 14:877–905
Nair V, Hinton GE (2009) 3d object recognition with deep belief nets. In: Bengio Y, Schuurmans D, Lafferty JD, Williams CKI, Culotta A (eds) Advances in neural information processing systems 22, Curran Associates, Inc. pp 1339–1347
Lee H, Ekanadham C, Ng AY (2008) Sparse deep belief net model for visual area v2. In: Platt JC, Koller D, Singer Y, Roweis ST (eds) Advances in neural information processing systems 20 curran associates Inc. pp 873–880
Lee H, Grosse R, Ranganath R, Ng AY (2011) Unsupervised learning of hierarchical representations with convolutional deep belief networks. Commun Acm 54(10):95–103
Ranzato M, Poultney C, Chopra S, LeCun Y (2007) Efficient learning of sparse representations with an energy-based model, MIT Press 1137–1144
Luo H, Shen R, Niu C, Ullrich C (2011) Sparse group restricted boltzmann machines. In: Proceedings of the twenty-fifth AAAI conference on artificial intelligence, AAAI Press, AAAI’11, pp 429–434
Zhang J, Ji N, Liu J, Pan J, Meng D (2015) Enhancing performance of the backpropagation algorithm via sparse response regularization. Neurocomputing 153:20–40
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: A simple way to prevent neural networks from overfitting. J Mach Learn Res 15:1929–1958
Hinton GE (2002) Training product of expert by minimizing contrastive divergence. Neural Comput 14(8):1771–1800
Kong Y, Yu T (2018) A graph-embedded deep feedforward network for disease outcome classification and feature selection using gene expression data. Bioinformatics 34(21):3727–3737
Min S, Lee B, Yoon S (2016) Deep learning in bioinformatics. Brief Bioinform 18(5):851–869
Rao B, Kreutz-Delgado K (1999) An affine scaling methodology for best basis selection. IEEE T Signal Proces 47(1):187–200
Wipf D, Nagarajan S (2010) Iterative reweighted ℓ1 and ℓ2 methods for finding sparse solutions. IEEE J Sel Top Signal Process 4(2):317–329
Hinton GE (2010) A practical guide to training restricted boltzmann machines. Momentum 9:926–947
Fischer A, Igel C (2014) Training restricted boltzmann machines: An introduction. Pattern Recogn 47(1):25–39
Segal D (2015) Diagnostic and statistical manual of mental disorders (5th ed.), American Cancer Society 101–105
Segall J, Allen E, Jung R, Erhardt E, Arja S, Kiehl K, Calhoun V (2012) Correspondence between structure and function in the human brain at rest. Front Neuroinform 6:10
Allen E, Erik BE (2011) A baseline for the multivariate comparison of resting-state networks. Syst Neurosci 5:2
Hoyer PO (2004) Non-negative matrix factorization with sparseness constraints. J Mach Learn Res 5:1457–1469
Thom M, Palm G (2013) Sparse activity and sparse connectivity in supervised learning, JMLR 14
Hu J, Li T, Wang H, Fujita H (2016) Hierarchical cluster ensemble model based on knowledge granulation. Knowl Based Syst 91:179–188
Lan L, Wang Z, Zhe S, Cheng W, Wang J, Zhang K (2019) Scaling up kernel SVM on limited resources: a low-rank linearization approach. IEEE Trans Neural Netw Learn Syst 30(2):369–378
Guo X, Zhang C, Luo W, Yang J, Yang M (2020) Urban impervious surface extraction based on multi-features and random forest. IEEE Access 8:226609–226623
Cao H, Duan J, Lin D, Calhoun V, Wang YP (2013) Integrating fmri and snp data for biomarker identification for schizophrenia with a sparse representation based variable selection method. BMC Med Genomics 6(3):S2
Meier L, Geer S, Bühlmann P (2008) The group lasso for logistic regression, group lasso for logistic regression. J R Stat Soc Ser B 70(1):53–71
Onitsuka T, Shenton ME, Salisbury DF, Dickey CC, Kasai K, Toner SK, Frumin M, Kikinis R, Jolesz FA, MR W (2004) Middle and inferior temporal gyrus gray matter volume abnormalities in chronic schizophrenia: An mri study. Am J Psychiatry 161(9):1603–1611
Tyekucheva S, Marchionni L, Karchin R, Parmigiani G (2011) Integrating diverse genomic data using gene sets. Genome Biol 12(10):R105
Cao H, Lin D, Duan J, Calhoun V, Wang YP (2012) Biomarker identification for diagnosis of schizophrenia with integrated analysis of fmri and snps. In: 2012 IEEE Int C Bioinform, pp 1–6
Li Y, Namburi P, Yu Z, Guan C, Feng J, Gu Z (2009) Voxel selection in fmri data analysis based on sparse representation. IEEE T Bio-Med Eng 56(10):2439–2451
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This research was supported by the National Natural Science Foundation of China (No. 12090020 and No.12090021), the National Key Research and Development Program of China (No. 2020AAA0106302), the Science and Technology Innovation Plan of Xian (No. 2019421315KYPT004JC006) and was partly Supported by HPC Platform, Xi’an Jiaotong University.
Rights and permissions
About this article
Cite this article
Qiao, C., Yang, L., Shi, Y. et al. Deep belief networks with self-adaptive sparsity. Appl Intell 52, 237–253 (2022). https://doi.org/10.1007/s10489-021-02361-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-021-02361-y