Abstract
In the present work we studied the expression of genes from nitrogen central metabolism in the yeast Dekkera bruxellensis and under regulation by the Nitrogen Catabolite Repression mechanism (NCR). These analyses could shed some light on the biological mechanisms involved in the adaptation and survival of this yeast in the sugarcane fermentation process for ethanol production. Nitrogen sources (N-sources) in the form of ammonium, nitrate, glutamate or glutamine were investigated with or without the addition of methionine sulfoximine, which inhibits the activity of the enzyme glutamine synthetase and releases cells from NCR. The results showed that glutamine might act as an intracellular sensor for nitrogen availability in D. bruxellensis, by activating NCR. Gene expression analyses indicated the existence of two different GATA-dependent NCR pathways, identified as glutamine-dependent and glutamine-independent mechanisms. Moreover, nitrate is sensed as a non-preferential N-source and releases NCR to its higher level. After grouping genes according to their regulation pattern, we showed that genes for ammonium assimilation represent a regulon with almost constitutive expression, while permease encoding genes are mostly affected by the nitrogen sensor mechanism. On the other hand, nitrate assimilation genes constitute a regulon that is primarily subjected to induction by nitrate and, to a lesser extent, to a repressive mechanism by preferential N-sources. This observation explains our previous reports showing that nitrate is co-consumed with ammonium, a trait that enables D. bruxellensis cells to scavenge limiting N-sources in the industrial substrate and, therefore, to compete with Saccharomyces cerevisiae in this environment
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since we first showed that Dekkera bruxellensis is present at high counts in yeast populations from sugarcane industrial fermentation in Brazil (De Souza Liberal et al. 2007; Basílio et al. 2008), efforts have been made to understand the mechanisms enabling this yeast to adapt and even surpass the population of Saccharomyces cerevisiae in this process. In this regard, two main branches of investigation have been pursued: tolerance to industrial stresses and ability to scavenge limiting growth factors from the industrial substrate.
In the context of a putative nutritional advantage, D. bruxellensis uses ammonium or urea as nitrogen source (N-source) for the basal metabolism, as do most yeasts and filamentous fungi (Siverio 2002). Amino acids are also suitable as N-sources, especially in anaerobiosis (Blomqvist et al. 2012), oxygen limitation (De Barros et al. 2013a) and under wine fermentation (Crauwels et al. 2015). Moreover, this yeast can also use nitrate in the central metabolism through its conversion to ammonium and further assimilation into glutamate (De Barros et al. 2011). When nitrate is used as N-source, yeast cells turn their metabolism to respiration to the detriment of fermentation (De Barros et al. 2013b). Additionally, the use of this N-source could solve the problem of redox imbalance caused by a limitation in the ability to reoxidise anabolic NADH in oxygen limitation, which leads to a decrease in the ethanol production, the so-called Custer effect (De Barros et al. 2011; Galafassi et al. 2013). Since the use of nitrate by yeasts found in ethanol fermentation using sugarcane juice is limited to some species (Basílio et al. 2008; Silva et al. 2016), it might represent a clear advantage for D. bruxellensis over S. cerevisiae, considering that the latter does not assimilate this N-source (De Barros et al. 2011). However, regardless of the type of nitrogen present in the fermentation substrate, it will always be found in limiting conditions, as ammonium or nitrate (De Barros et al. 2011) or even as amino acids (Walford 1996). Therefore, the metabolism of nitrogen is an important area of investigation in order to understand the adaptation of this yeast in the industrial process.
In S. cerevisiae the metabolism of nitrogen is regulated by two interplaying general mechanisms governing the regulation of gene expression and enzyme functioning: the General Amino Acids Control (GAAC) and the Nitrogen Catabolite Repression (NCR) (Ter Schure et al. 2000; Magasanik and Kaiser 2002). The latter acts by repressing the expression of genes involved in the use of a variety of alternative N-sources when ammonium, glutamate or glutamine are available in the medium (Magasanik and Kaiser 2002). In general terms, the NCR mechanism is composed of two negative regulator proteins, Dal80p and Gzf3p, and two positive regulator proteins, Gln3p and Gat1p. The common feature in these transcription factors (TF) is the zinc-finger DNA binding motif that recognises a GATAAG element (GATA motif) in the promoter of target genes (Magasanik and Kaiser 2002). Additionally, Ure2p is a central player in the NCR mechanism by controlling the cytoplasmic-nuclear location of transcription factors (Georis et al. 2011). In the presence of ammonium, Ure2p binds to Gln3p and impairs its migration to the nucleus while Dal80p and Gzf3p bind to GATAAG elements, which results in the repression of NCR-sensitive genes. When nitrogen is scarce or in the presence of non-preferential sources, Gln3p is released from Ure2p and migrates to nucleus to activate the expression of GAT1 gene, and together Gln3p and Gat1p activate the expression of NCR-sensitive genes (Georis et al. 2011).
Upstream of this regulatory process is the TOR (Target of Rapamycin) pathway, which is a kinase cascade transducer mechanism connecting the availability of nitrogen in the environment and cell growth control (Magasanik and Kaiser 2002, Georis et al. 2011). TOR may use ammonium and/or glutamine as an intracellular sensor for nitrogen, and, by doing so, it is directly connected to the central pathway of ammonium assimilation (Magasanik and Kaiser 2002, Georis et al. 2011). In this regard, ammonium is mainly assimilated by the amination of 2-oxoglutarate into glutamate, by NADPH-dependent glutamate dehydrogenase (Gdh1p), and the further amination of glutamate into glutamine, by ATP-dependent glutamine synthetase (GS or Gln1p). In the excess of glutamine in the medium, this amino acid can be used to transaminate 2-oxoglutarate to produce two molecules of glutamate by NADH-dependent glutamine/2-oxoglutarate amino transferase (GOGAT or Glt1p) (Magasanik and Kaiser 2002). Inhibition of both TOR by rapamycin and GS by methionine sulfoximine (MSX) mimics, in different ways, nitrogen limitation or shortage in the environment and may account for the release of expression in NCR-sensitive genes. The expression of these genes enables cells to scavenge any possible N-source available (Crespo and Hall 2002; Georis et al. 2011). In S. cerevisiae, ammonium is a strong inducer of NCR by impairing the use of other N-sources. However, it does not seem to inhibit nitrate usage by D. bruxellensis as both N-sources are co-consumed in industrial (De Barros et al. 2011) and laboratory (De Barros et al. 2013b) media. This indicates that the regulatory mechanisms governing the use of N-source might be different between these yeasts. Therefore, an understanding of the regulatory network of nitrogen metabolism may uncover the mechanisms allowing this yeast to survive and grow in a nitrogen-limited substrate, such as sugarcane juice. This is rather important, especially in the context of the nutritional advantage observed for D. bruxellensis due to the assimilation of nitrate.
In the present work we used the inhibition of GS by MSX in order to study the regulation of key genes in the central metabolism of ammonium and nitrate in D. bruxellensis. The results showed that MSX is effective in releasing the NCR mechanism in this yeast and indicated that glutamine is an intracellular sensor molecule of NCR in this yeast. The presence of nitrate itself was enough to achieve this response, independently of MSX treatment. The relative expression profiles of target genes showed the presence of glutamine-dependent and glutamine-independent GATA mechanisms for gene regulation, and genes could be grouped according to their response to NCR. Finally, we confirmed previous results showing that nitrate genes are more subject to an inductive mechanism by nitrate than a repressive mechanism by ammonium or glutamate/glutamine.
Materials and methods
Strain and culture conditions
The strain D. bruxellensis GDB 248 was used as it has been a model for physiological and genetic studies of this yeast isolated from fuel-ethanol fermentation processes (De Barros et al. 2011; Leite et al. 2012). Cells were maintained in YPD plates (yeast extract: 10 g/l; peptone: 20 g/l; glucose: 20 g/l; agar: 20 g/l). Seed cultures were prepared by inoculating fresh yeast colonies in liquid complete synthetic defined (SD) medium (yeast nitrogen base: 1.7 g/l; ammonium sulphate: 5 g/l; glucose: 20 g/l) and cultivating the cells at 30 °C and 180 rpm for 48 h. Afterwards, yeast cells were transferred to fresh synthetic media to initial 0.1–0.2 OD600nm. The synthetic media contained either ammonium (sulphate salt), nitrate (sodium salt), glutamate or glutamine as nitrogen sources, added to medium to achieve an initial nitrogen concentration of 75 mM. Methionine sulfoximine (Sigma Co., St Louis, USA) was added at different concentrations (0–20 mM) in the experiments of NCR inhibition. Cultivations were performed in 150 μl volume sterile microtitre plates (in 200 μl wells) in multireader Synergy HT (Biotek, Swissland) at 30 °C with automatic recording of O.D. at 600 nm. Yeast growth rates were calculated from the slope of the exponential growth phase (Leite et al. 2012). Experiments were performed in biological triplicates and the data represent the mean arithmetic value.
RNA extraction and cDNA synthesis
For gene expression analyses, yeast cells were cultivated in flasks with SD medium containing ammonium sulphate until reaching 1.0 OD600nm. Cells were collected by centrifugation, washed with sterile water and re-suspended in the same volume of SD medium containing different N-sources, with or without MSX at 20 mM, and incubated for 1 h at 30 °C and 180 rpm. After incubation, cells were immediately stored at −80 °C for RNA extraction.
For RNA extraction, cells were thawed, centrifuged at 4 °C and re-suspended in 250 µl of AE buffer (50 mM sodium acetate and 10 mM EDTA, pH 5.3) plus 60 µl 10% SDS. Cells were lysed by vortexing followed by incubation at 65 °C for 10 min. The lysates were centrifuged at 9520 RCF for 5 min at 4 °C and the supernatants were transferred to new tubes. Total RNA was purified with NucleoSpin® RNA II kit (Macherey–Nagel, Germany) according to manufacturer instructions. Quantification of RNA was done by using a Nanodrop (GE Health Care, EUA), with purity evaluated by 260/280 nm ratio, and integrity was evaluated in 1% agarose gel in DEPC-treated TAE buffer, with staining with ethidium bromide. cDNA was produced by using the ImProm-II™ Reverse Transcription System Promega II kit (Promega, USA), following manufacturer’s instructions.
Primer design and qPCR
Nucleotide sequences of D. bruxellensis genes orthologous to S. cerevisiae were recovered from the genome database deposited at the NCBI. Primers for qPCR were designed using the tool Primer Express® of Applied Biosystems (http://www.appliedbiosystems.com/support/apptech/) with the following criteria: length of 19–21 nucleotides, GC content around 50%, average Tm of 60 °C and amplicons with 70–150 bp. Information regarding the primers used in the present study were previously reported as supplementary material by De Barros Pita et al. (De Barros et al. 2013a, b). All parameters of quality, efficiency and validation were evaluated according to conditions previously established (De Barros et al. 2012). Amplifications were performed in ABI Prism 7300 (Applied Biosystems, Foster City, CA, USA) using SYBR Green PCR master Mix kit (Applied Biosystems). Reference genes data normalisation and analysis by geNorm algorithm as well as relative quantity determination followed the MIQE Guidelines as recommended for D. bruxellensis by De Barros et al. (2012).
Relative expression profiles of each gene were transformed in a binary matrix with the criterion of 0 (fold change ≤2) and 1 (fold change >2) for each condition to produce a distance matrix clustered by UPGMA using the online software freely available at http://genomes.urv.cat/UPGMA/(Garcia-Vallve et al. 1999). The nodes represent the parameters for each sub-network dichotomized according to the framing of the genes to each parameter (Y = yes; N = no).
Gene promoter analysis
Nucleotide sequences comprising 1000 bp upstream from the transcription start point of each gene were collected from the D. bruxellensis CBS 2499 v2.0 genome (Piskur et al. 2012) deposited at JGI database (http://genome.jgi.doe.gov/Dekbr2/Dekbr2.info.html). Those sequences were analysed for the presence of TF binding motifs by the tool “find TF binding site” of the Yeastract database (http://www.yeastract.com/index.php) (Teixeira et al. 2014). The presence of NCR-related regulatory elements and their positions were recorded.
Results and discussion
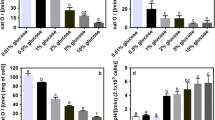
In the present work we used MSX in order to release NCR in the yeast D. bruxellensis and evaluate its consequences to the regulation of key genes in nitrogen central metabolism. The first approach was to test the effectiveness of this compound in the yeast growth, as a selective inhibitor of GS. This would enforce a glutamine shortage, mimicking nitrogen starvation and cell metabolic reprogramming. Growth on SD medium with ammonium as nitrogen source showed a characteristic lag phase, lasting for 5–6 h, followed by an exponential growth at rate of 0.14 h−1. On the other hand, cells exposed to MSX did not grow when ammonium was the N-source (Fig. 1a), indicating that MSX is also active in D. bruxellensis and should be blocking glutamine biosynthesis. The results showed that D. bruxellensis was more tolerant to MSX than S. cerevisiae. Currently, the literature reports the use of 2 mM of MSX to inhibit growth of S. cerevisiae, while our experiments showed that D. bruxellensis grows normally at this concentration. Full inhibition of cell growth was achieved only with 20 mM (data not shown). Glutamine was used as N-source by this yeast as shown by the high growth rate at 0.15 h−1 with a very short lag phase (Fig. 1c). It was observed that 20 mM of MSX decreased growth rate to 0.13 h−1, rather than stalling it, as observed for ammonium. Therefore, the toxic effect of MSX is not effective when using glutamine as N-source. Additional evidence came from a cultivation in which glutamate was the N-source. This amino acid is also used by the yeast cells, which grew at 0.12 h−1 following a very short lag phase (Fig. 1b). The presence of MSX did not stop yeast growth, but significantly reduced it to 0.03 h−1. This indicated that an unknown alternative low-active pathway for glutamine biosynthesis from glutamate was in operation.
Growth kinetics of Dekkera bruxellensis GDB 248 strain in YNB media containing glucose and different nitrogen sources as ammonium (a), glutamate (b), glutamine (c) and nitrate (d) in the absence (open circle) or presence (filled circle) of methionine sulfoximine at 20 mM. The results represent the mean value of triplicates
Nitrate is used as an N-source by D. bruxellensis after its reductive conversion to ammonium using NAD(P)H as reduced co-factor. It provides nutritional advantage for this yeast over S. cerevisiae in industrial processes, such as fermentation of sugarcane juice (De Barros et al. 2011) and could also be used as an alternative electron acceptor to overcome the Custer effect (De Barros et al. 2011; Galafassi et al. 2013). The regulation of nitrate genes has been studied by our group and in the present context we intended to test the influence of the NCR mechanism over their expression. The yeast grew at a rate of 0.10 h−1 in the presence of nitrate, preceded by a 5–6 h lag phase (Fig. 1d), a characteristic of this strain. The presence of MSX, and consequent inhibition of glutamine biosynthesis extended that lag phase to 14 h, while only decreasing the exponential growth rate to 0.08 h−1 (Fig. 1d). This was somewhat surprising, since nitrate is reduced to ammonium and one could expect to observe the same behaviour observed in ammonium-containing medium. Once again, since GS should be inactivated by MSX, this suggested the presence of an alternative low-active pathway for glutamine biosynthesis in this yeast.
The results above led us to speculate that such a putative and alternative glutamine biosynthetic pathway, if it truly exists, should be under strong repression by ammonium. In this case, in ammonium-based medium the presence of MSX blocks the GS activity. Then, the presence of ammonium itself inhibits that alternative pathway and no glutamine is produced whatsoever. When nitrate is used as N-source, the presence of MSX blocks the conventional pathway for glutamine biosynthesis. However, as the conversion to ammonium flows at slow rates, as indicated by the reduced growth (Fig. 1d), ammonium should not accumulate to repress that alternative glutamine biosynthetic pathway. When glutamate is used as N-source, the alternative pathway should not be repressed as well. We simulated the reaction by GOGAT for the interconversion glutamine-glutamate using online eQuilibrator software (Flamholz et al 2012). The result showed the estimated ΔG° of −49.4 kJ/mol in favour of the conversion of glutamine to glutamate. Thus, this reverse reaction seems unlikely. Nonetheless, the identification of this putative pathway is beyond the subject of this report.
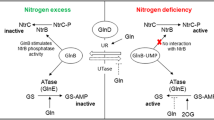
In addition to physiological data, the present study evaluated the expression of genes involved in the central metabolism of nitrogen, which were divided into three groups (Fig. 2). In group 1, we positioned genes encoding transporters for organic and inorganic nitrogen compounds, such as MEP1 (ammonium permease), GAP1 (general amino acids permease), YNT1 (nitrate permease) and PUT4 (proline permease). This last gene was used because it is a typical marker for the NCR mechanism in S. cerevisiae (Ter Schure et al. 2000). The second group comprises genes coding for enzymes involved in the conversion of the inorganic substrates (ammonium or nitrate) to glutamate and glutamine, such as GDH1 (NADPH-dependent glutamate dehydrogenase), GDH2 (NAD+-dependent glutamate dehydrogenase), GLT1 (NAD+-dependent glutamate synthase), YNR1 (nitrate reductase) and YNI1 (nitrite reductase) (Fig. 2). The third group includes genes encoding the main regulatory proteins involved in ammonium and nitrate assimilation (YNA1 and GAT1). Their expressions in different media were relative to ammonium-based medium. Expression of genes in glutamine medium was not included due to lack of physiological effect of MSX in this condition. Moreover, the promoter region of each gene was scanned for the presence of DNA-binding motifs for the transcription factors involved in NCR (Table 1).
Scheme of the metabolic routes involved in the uptake and metabolisation of different nitrogen sources in yeast. Permeases and enzymes are underlined. Black arrows represent the transport of metabolites from outside the cells. Thick gray arrows represent enzyme reactions. Black line with flattened tip represents the inhibition of glutamine synthetase by methionine sulfoximine (MSX)
The presence of MSX in medium containing ammonium promoted a slight induction of permease-encoding GAP1, PUT4 and YNT1, while transcriptional activity of MEP1 remained unaltered (Fig. 3). In S. cerevisiae, GAP1 is under NCR when in ammonium or nitrogen-rich conditions, in which its transcription is repressed by Ure2p. Under nitrogen starvation or nitrogen-poor condition it is induced by the Gat1p and Gln3p (Magasanik and Kaiser 2002). PUT4 encodes a permease for proline than can also transport alanine and glycine. Its expression is tightly regulated by ammonium and follows the pattern presented for GAP1 (Ter Schure et al. 2000). In D. bruxellensis GAP1, PUT4 and MEP1 were up-regulated in low nitrogen concentration or when proline was the N-source, irrespective of sugar concentration (De Barros et al. 2013a). On the other hand, MEP1 was not up-regulated in the present report for MSX-treated cells (Fig. 3). In S. cerevisiae, two mechanisms of NCR are in operation: (i) a TorC1-dependent pathway acting through a GATA regulator Gat1p and (ii) a TorC1-independent Gln3p pathway. Treatment with MSX relocates Gln3p to the nucleus for the up-regulation of its target genes. On the other hand, the location of Gat1p is not changed by MSX and consequently it does not influence the expression of genes under Gat1p control (Georis et al. 2011). In S. cerevisiae, this gene encodes a transcriptional factor that induces the expression of NCR-sensitive genes in parallel to Gln3p-regulated mechanism. However, transcription of this gene is activated by Gln3p in response to nitrogen-poor condition and repressed by Dal80p in nitrogen-rich medium (Magasanik and Kaiser 2002; Siverio 2002). We observed a significant up-regulation of GAT1 by MSX (Fig. 3). Therefore, we concluded that in D. bruxellensis the orthologue of GAT1 is also regulated by the Gln3p-dependent/glutamine-dependent mechanism like GAP1 and PUT4 above. Therefore, from our previous results (De Barros et al. 2013a) and the results of the present work, we propose that two GATA pathways are also operating in D. bruxellensis: (i) GATA-dependent/glutamine-dependent regulation of GAP1 and PUT4 and (ii) GATA-dependent/glutamine-independent regulation of MEP1. Analysis of the promoter region of MEP1 revealed the presence of three DNA binding motifs for Gcn4p TF (Table 1), which makes its expression tightly regulated by ammonium. The presence of this motif in the promoter of GAP1 and PUT4 also indicated an ammonium-dependent regulation (Table 1). Gcn4p induces the expression of target genes under amino acids limitation, and its stability in the cells depends on the ubiquitin-related mechanism that responds to the nutrient availability in the medium (Meimoun et al. 2000). This means that under adequate supply of glutamine, or in ammonium-rich medium, Gcn4p is degraded and its target genes are down-regulated. The lack of induction of MEP1 by MSX, as observed in ammonium medium (Fig. 3) together with the presence of Gcn4p motifs in its promoter (Table 1) revealed that the strength of ammonium repression is higher than the activation by glutamine starvation. The fourth permease-encoding gene YNT1 showed up-regulation by sixfold in the presence of MSX, higher than the other two permease genes (Fig. 3). This indicates that this gene is under a tight repressive control of glutamine-dependent GATA pathway. We previously showed that YNT1 is highly expressed by nitrate and also that this induction was slightly suppressed by the presence of ammonium in nitrate-based medium (De Barros et al. 2011, 2013b). Altogether, these results showed that YNT1 is subject to repressive regulation by glutamine, which is relieved upon MSX treatment. This was corroborated by the presence of GATA binding motifs and absence of a Gcn4p binding motif in its promoter region (Table 1).
Expression of genes coding for proteins of the ammonium central pathway (GDH1, GLT1 and GDH2) was not changed by MSX in ammonium-based medium. Actually, under ammonium limitation these genes were down-regulated (De Barros et al. 2013a), indicating their almost exclusive regulation by ammonium. Moreover, the lack of up-regulation of GDH1 and GLT1 genes seems to be the consequence of two mechanisms as indicated in Table 1. First, GDH1 did not present any NCR-related motif, corroborating its almost constitutive expression. Second, GLT1 contains motifs for Gcn4p and Dal80p, indicating its tight negative regulation by ammonium. On the other hand, genes for the nitrate metabolism were only slightly up-regulated (Fig. 3). Since this was observed independently of YNA1 mRNA levels (Fig. 3), it seems that nitrate genes are mainly controlled by the activation of Yna1p. In our previous work (De Barros et al. 2011, 2013b) we concluded that nitrate genes are mostly regulated by the presence of nitrate and less subjected to NCR, which is confirmed in the present study.
In glutamate-based medium, we observed a slight up-regulation of the three permease encoding genes and down-regulation of GDH1, GDH2 and GLT1 relative to ammonium-based medium (Fig. 4a). This overall picture was practically the same for glutamate-growing cells treated with MSX, with an increase only in scale of fold change (Fig. 4b). Our previous work showed that GLT1 was down-regulated in glutamate-rich medium relative to ammonium, while GDH2 was up-regulated in this condition (De Barros et al. 2013a). This profile was reproduced also in glutamate-limited medium (De Barros et al. 2013a). The explanation follows that presented for ammonium. In this case, the excess of assimilable N in the form of glutamate not only activates a repressive mechanism that impairs Gcn4p function, but also might induce migration of Gap1p from the cytoplasm. GAT1 was up-regulated independently of presence of MSX (Fig. 4b). These results indicated a hierarchical regulation of GAT1 as a consequence of the nitrogen sensing mechanisms. First, GAT1 expression seems to be maximally achieved in the presence of MSX (tenfold), i.e., absence of glutamine, and progressively decreases in the presence of glutamine (threefold) and ammonium (onefold). Glutamine biosynthesis occurs via amination of glutamate by GS. In this case, ammonium is obtained directly from the medium (in ammonium-based medium) or is produced by deamination of glutamate by action of Gdh2p activity (in glutamate-based medium). It seems that in glutamate as sole N-source, glutamine synthesis is limited by the low rate of Gdh2p activity, as suggested by the observed low expression of GDH2 (Fig. 4). Cultivation in glutamate or glutamine showed significant differences in only two genes (De Barros et al. 2013b). GDH2 was up-regulated in glutamate, as expected from the catabolic activity of its enzyme. On the other hand, GLT1 was up-regulated in glutamine, since glutamate must be produced by the transaminase activity encoded by this gene. In addition, PUT4 was only slightly down-regulated in glutamine relative to glutamate. Second, full induction is achieved in ammonium medium containing MSX (16-fold), supporting the statement above that glutamine is a sensor that helps the negative regulation of GAT1, as shown by the growth curves (Fig. 1). Transcription of nitrate genes was unchanged in glutamate medium relative to ammonium medium (Fig. 4a). Actually, the results indicated a down-regulation of YNT1 and YNR1 in such conditions. On the other hand, in MSX, a significant up-regulation of the three nitrate genes was observed (Fig. 4b). These genes seem to behave as a regulon, with the same co-regulation pattern. Different from GAT1, their first regulatory mechanism seems to be the relief of NCR controlled by the Gln3p-dependent mechanism followed by repression triggered by the nitrogen-rich condition of glutamine.
Expression of yeast genes in Dekkera bruxellensis GDB 248 cells incubated in YNB medium containing glutamate (a) or glutamate in the presence of methionine sulfoximine (b) relative to background expression in YNB medium containing ammonium. Error bars represent the variation among biological triplicates
When nitrate was used as N-source, MEP1, GAP1, GDH1, GDH2 and GLT1 were up-regulated relative to ammonium (Fig. 5a) in a higher level than that achieved in glutamate (Fig. 4a). On the other hand, the presence of MSX did not change their expression profile (Fig. 5b). For MEP1, GAP1 and GLT1, the up-regulation can be explained by the dependence of Gcn4p binding to their promoters in the absence of ammonium. GAT1 transcription level was higher in nitrate than in ammonium (Fig. 5a), although it showed down-regulation when MSX was added to the medium (Fig. 5b). As expected, nitrate genes were up-regulated by nitrate irrespective to the presence of MSX (Fig. 5a, b). By comparing the expression level of nitrate and nitrate + MSX condition it became clear that nitrate itself was enough to fully release D. bruxellensis cells from NCR.
Expression of yeast genes in Dekkera bruxellensis GDB 248 cells incubated in YNB medium containing nitrate (a) or nitrate in the presence of methionine sulfoximine (b) relative to background expression in YNB medium containing ammonium. Error bars represent the variation among biological triplicates
Based on the expression profiles from the genes analysed in the present study, we clustered the data and graphically represented them as a dendrogram (Fig. 6). Note that all genes involved in nitrate assimilation, except YNA1, behave as a regulon, meaning that they responded in exactly the same way i.e. repressed by ammonium and induced by nitrate, with the following differences. YNT1 was the most up-regulated among them due to the presence of three GATA motifs in its promoter and presented no repressive response by ammonium. YNI1 was the second most up-regulated gene due to the presence of one GATA motif and two Gcn4p binding motifs. However, these two Gcn4 motifs make YNI1 repressible by ammonium. Finally, the presence of only one GATA motif explained the weak regulatory response observed for YNR1. YNA1 and GDH2 also formed a separate regulon that showed a positive response exclusively to the presence of nitrate, perceived by the cells as nitrogen limitation or starvation. Unfortunately, there is no data available for their promoter elements. On the other hand, GDH1 did not show any differential expression in the conditions relative to ammonium as reference. This gene does not present any NCR element in its promoter (Table 1), but only elements relevant to carbon control (data not shown). GDH1 expression responds to both nitrogen and carbon source in S. cerevisiae, being rather dependent on the control exerted by Ada3p and Gcn4/5p involved in repression by glucose. With a respirable carbon source, such as ethanol, this gene is controlled by Snf2p (Riego et al. 2002). In a parallel study, we observed that GDH1 expression changes in anaerobiosis, in which fermentation demands high glycolytic flux to sustain adequate ATP supply for growth (manuscript in preparation). PUT4 gene of D. bruxellensis showed only mild regulation by NCR and did not seem to be an efficient marker to study this regulatory mechanism, as reported for S. cerevisiae. This set of data represents the first picture on the regulatory circuit of NCR in the yeast D. bruxellensis, studies of which are still very limited by the shortage of molecular tools for gene modification and strain construction via sporulation/breeding classical methods. Nevertheless, it presents the first clues on the regulation of genes involved in nitrate assimilation, a physiological trait that might provide adaptation to industrial processes, such as ethanol fermentation of sugarcane substrates.
Clustering by UPGMA of genes of Dekkera bruxellensis GDB 248 according to the profile of relative expression in all conditions showed in Figs. 3, 4 and 5 as described in “Materials and methods”. The nodes represent the parameters for each sub-network dichotomized according to the framing of the genes to each parameter (Y = yes; N = no)
References
Basílio ACM, Araújo PRL, Morais JOF, da Silva Filho EA, de Morais Jr MA, Simões DA (2008) Detection and identification of wild yeast contaminants of the industrial fuel ethanol fermentation process. Curr Microbiol 5:322–326
Blomqvist J, Nogué VS, Gorwa-Grauslund M, Passoth V (2012) Physiological requirements for growth and competitiveness of Dekkera bruxellensis under oxygen-limited or anaerobic conditions. Yeast 29:265–274
Crauwels S, Van Assche A, de Jonge R, Borneman AR, Verreth C, Troels P, De Samblanx G, Marchal K, Van de Peer Y, Willems KA, Verstrepen KJ, Curtin CD, Lievens B (2015) Comparative phenomics and targeted use of genomics reveals variation in carbon and nitrogen assimilation among different Brettanomyces bruxellensis strains. Appl Microbiol Biotechnol 99:9123–9134
Crespo JL, Hall MN (2002) Elucidating TOR signalling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol Mol Biol Rev 66:579
Da Silva TC, Leite FC, De Morais Jr MA (2016) Distribution of Dekkera bruxellensis in a sugarcane-based fuel-ethanol fermentation plant. Lett Appl Microbiol 62:354–358
De Barros Pita W, Leite FCB, de Souza Liberal AT, Simões DA, de Morais Jr MA (2011) The ability to use nitrate confers advantage to Dekkera bruxellensis over S cerevisiae and can explain its adaptation to industrial fermentation processes. Antonie Van Leeuwenhoek 100:99–107
De Barros Pita W, Leite FCB, de Souza Liberal AT, Pereira LF, Carazzolle MF, Pereira GA, De Morais Jr MA (2012) A new set of reference genes for RT-qPCR assays in the yeast Dekkera bruxellensis. Can J Microbiol 58:1362–1367
De Barros Pita W, Castro Parente D, Simões DA, Passoth V, de Morais Jr MA (2013a) Physiology and gene expression profiles of Dekkera bruxellensis in response to carbon and nitrogen availability. Antonie Van Leeuwenhoek 104:855–868
De Barros Pita W, Tiukova I, Leite FC, Passoth V, Simões DA, de Morais Jr MA (2013b) The influence of nitrate on the physiology of the yeast Dekkera bruxellensis grown under oxygen limitation. Yeast 30:111–117
De Souza Liberal AT, Basílio ACM, Brasileiro BTRV, da Silva Filho EA, Simões DA, de Morais Jr MA (2007) Identification of the yeast Dekkera bruxellensis as major contaminant in continuous fuel ethanol fermentation. J Appl Microbiol 102:538–547
Flamholz A, Noor E, Bar-Even A, Milo R (2012) eQuilibrator—the biochemical thermodynamics calculator. Nucl Acids Res 40:D770–775
Galafassi S, Capusoni C, Moktaduzzaman M, Compagno C (2013) Utilization of nitrate abolishes the “Custers effect” in Dekkera bruxellensis and determines a different pattern of fermentation products. J Ind Microbiol Biotechnol 40:297–303
Garcia-Vallve S, Palau J, Romeu A (1999) Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol Biol Evol 16:1125–1134
Georis I, Tate JJ, Cooper TG, Dubois E (2011) Nitrogen-responsive regulation of GATA protein family activators Gln3 and Gat1 occurs by two distinct pathways, one inhibited by rapamycin and the other by methionine sulfoximine. J Biol Chem 286:44897–44912
Leite FCB, Basso TO, De Barros Pita WB, Gombert A, Simões DA, de Morais Jr MA (2012) Quantitative aerobic physiology of the yeast Dekkera bruxellensis, a major contaminant in bioethanol production plants. FEMS Yeast Res 13:34–43
Magasanik B, Kaiser CA (2002) Nitrogen regulation in Saccharomyces cerevisiae. Gene 290:1–18
Meimoun A, Holtzman T, Weissman Z, McBride HJ, Stillman DJ, Fink GR, Kornitzer D (2000) Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex. Mol Biol Cell 11:915–927
Piskur J, Ling Z, Marcet-Houben M, Ishchuk OP, Aerts A, LaButti K, Copeland A, Lindquist E, Barry K, Compagno C, Bisson L, Grigoriev IV, Gabaldon T, Phister T (2012) The genome of wine yeast Dekkera bruxellensis provides a tool to explore its food-related properties. Int J Food Microbiol 157:202–209
Riego L, Avendano A, DeLuna A, Rodriguez E, Gonzalez A (2002) GDH1 expression is regulated by GLN3, GCN4, and HAP4 under respiratory growth. Biochem Biophys Res Commun 293:79–85
Siverio JM (2002) Assimilation of nitrate by yeasts. FEMS Microbiol Rev 26:277–284
Teixeira MC, Monteiro PT, Guerreiro JF, Gonçalves JP, Mira NP, dos Santos SC, Cabrito TR, Palma M, Costa C, Francisco AP, Madeira SC, Oliveira AL, Freitas AT, Sá-Correia I (2014) The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucl Acids Res 42:D161–D166
ter Schure EG, van Rielb NAW, Verripsa CT (2000) The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol Rev 24:67–83
Walford SN (1996) Composition of Cane Juice. Proc S Afr Sug Technol Ass 70:265–266
Funding
This work was sponsored by the Bioethanol Research Network of the State of Pernambuco (CNPq-FACEPE/PRONEM program, Grant No APQ-1452-2.01/10) and by Grants of the National Council of Science and Technology (CNPq No 472533/2013-4 and 474847/2013-6).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Cajueiro, D.B.B., Parente, D.C., Leite, F.C.B. et al. Glutamine: a major player in nitrogen catabolite repression in the yeast Dekkera bruxellensis . Antonie van Leeuwenhoek 110, 1157–1168 (2017). https://doi.org/10.1007/s10482-017-0888-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-017-0888-5