Abstract
A bacterial strain, py1294T, isolated from a root of Paris polyphylla Smith var. yunnanensis collected from Yunnan province, southwest China, was characterised by using a polyphasic approach to clarify its taxonomic position. Strain py1294T was found to be Gram-positive, aerobic, spore-forming, peritrichous flagella and rod shaped. Growth was found to occur in the presence of 0–8 % (w/v) NaCl (optimum 1–3 %), at pH 6.5–9.5 (optimum 8.0) and at 10–42 °C (optimum 30 °C). The major cellular fatty acids were identified as anteiso-C15:0, anteiso-C17:0, iso-C16:0 and iso-C14:0. The predominant quinone was identified as MK-7 and a minor amount of MK-6 was detected. The diagnostic polar lipids were diphosphatidylglycerol, phosphatidylglycerol and phosphatidylethanolamine. The cell wall peptidoglycan was found to contain meso-diaminopimelic acid. Phylogenetic analysis of the 16S rRNA gene sequence showed that strain py1294T forms a well-supported clade with Oceanobacillus damuensis PT-20T (97.9 % sequence similarity) within the genus Oceanobacillus, although it also shares a high sequence similarity with Ornithinibacillus contaminans (97.5 %). Crucially, the DNA–DNA relatedness value between strain py1294T and O. damuensis PT-20T was 29.7 ± 3.2 %. The G+C content was determined to be 42.3 mol%. On the basis of the phylogenetic and phenotypic data, a novel species Oceanobacillus endoradicis sp. nov. is proposed, with py1294T (=DSM 100726T = KCTC 33731T) as the type strain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Paris polyphylla Smith var. yunnanensis (Franch.) Hand.-Mazz. is a perennial medicinal plant mainly distributed in the southwest of China. The rhizomes of this plant are used in traditional Chinese medicine as a haemostatic, immunity adjustment and antimicrobial agent (Yu et al. 2010; Zhou et al. 2003). Steroidal saponins are the main active ingredient, which have important physiological activities (Waller and Yamasaki 1996). Plant endophytic microorganisms are an important source of natural bioactive compounds with great potential applications in medicine, agriculture and industry (Strobel and Daisy 2003; Strobel et al. 2004). Therefore, in previous work, we attempted to collect bacterial isolates from the endophytic habitat of P. polyphylla var. yunnanensis, and expect to find some useful resources, which could produce steroidal saponins or participate in the metabolism of steroidal saponins. Interestingly, amongst more than three hundred endophytic isolates, we identified an Oceanobacillus-like strain, py1294T, which was isolated from a rhizome sample from Chuxiong, Yunnan province, southwest China.
The genus Oceanobacillus was established by Lu et al. (2001) and the description of the genus was emended by Yumoto et al. (2005), Lee et al. (2006) and Hirota et al. (2013) on the basis of chemotaxonomic and phenotypic data. At the time of writing, this genus comprises 21 validly named species, including six recently described species, Oceanobacillus limi (Amoozegar et al. 2014), Oceanobacillus luteolus (Wu et al. 2014), Oceanobacillus arenosus (Kim et al. 2015), Oceanobacillus bengalensis (Yongchang et al. 2015), Oceanobacillus rekensis and Oceanobacillus damuensis (Long et al. 2015). Members of the genus Oceanobacillus have been isolated from a wide variety of environments. Oceanobacillus iheyensis, Oceanobacillus profundus, Oceanobacillus pacificus, O. arenosus, Oceanobacillus locisalsi, O. limi and O. bengalensis were isolated from marine environments or salt lake; Oceanobacillus kimchii, Oceanobacillus kapialis, Oceanobacillus indicireducens and Oceanobacillus polygoni were isolated from fermented products (food or liquor); O. rekensis and O. damuensis were isolated from saline alkali soil; other species were isolated from fish, insect, activated sludge, mural paintings, wastewater treatment, sand dune, algal mat and soil. However, there is not yet any reports of Oceanobacillus species distributed in plant endophytic environments. Here, we describe an endophytic bacterial strain, py1294T, which belongs to the genus Oceanobacillus.
Materials and methods
Strains and culture conditions
Healthy root samples of P. polyphylla. Smith var. yunnanensis, a traditional Chinese medicinal plant, were collected from Chuxiong, Yunnan province, southwest China (101°55.573′E 25°38.235′N), and used as sources for the isolation of endophytic bacteria. Root pieces were washed several times with running tap water and surface sterilised by stepwise washing in 5 % NaOCl for 5 min, 70 % ethanol for 8 min and finally washed several times with sterile distilled water. The dried root was pulverised in a blender, distributed on tenfold-diluted nutrient agar (10−1 NA) medium and incubated at 28 °C for 2 weeks. Since the growth of strain py1294T was better on Luria–Bertani (LB) medium, for characterising this strain, it was routinely grown aerobically on LB medium for 3 days at 30 °C and pH 7.5, except where indicated otherwise. The purified strain was maintained at −80 °C on LB medium without agar and supplemented with 20 % (v/v) glycerol.
Lysinibacillus mangiferahumi M-GX18T was obtained from Dr. Ming-He Mo (Yunnan University) for use as a reference strain during the detection of the predominant quinone. The type strain O. damuensis PT-20T was kindly provided by Dr. Yongqiang Tian (Sichuan University) and cultured under comparable conditions as a reference strain.
Phenotypic characterisation
Cell morphology was examined by using phase-contrast microscopy (Leica, DM2000), transmission electron microscopy (JEOL, JEM-2100) and scanning electron microscopy (XL30 ESEM-TMP, Philips-FEI). Gram-staining was carried out according to Smibert and Krieg (1994) combined with a KOH lysis test (Gregersen 1978). Tolerance of NaCl was tested on LB plates at different NaCl concentrations (0–10 %, w/v, at intervals of 1 %). The temperature range for growth was determined at 0, 4, 10, 15, 20, 30, 37, 42 and 50 °C. Growth at various pH (4.0–10.0, at intervals of 0.5 pH units) was examined in LB liquid by using the following buffer system: pH 4.0–5.0, 0.1 mol/L citric acid/0.1 mol/L sodium citrate; pH 6.0–8.0, 0.1 mol/L KH2PO4/0.1 mol/L NaOH; pH 9.0–10.0, 0.1 mol/L NaHCO3/0.1 mol/L Na2CO3 (Tang et al. 2010). The pH value was measured and readjusted after autoclaving (OHAUS, STARTER3100). A spectroscopic method of monitoring turbidity at OD660 (Lambda 35 UV/Vis spectrometer, PerkinElmer) was used to assess the growth at various pH. Catalase activity was measured by using 3 % H2O2 solution. Oxidase activity was detected using API oxidase reagent according to the manufacturer’s instructions. The utilisation of carbon sources was performed using the Biolog GENIII system at 30 °C for 24 h. To determine steroid production, Liebermann-Burchard and Salkowski reactions were performed after strain py1294T was cultured in Erlenmeyer flasks of LB liquid at 30 °C for 5 days (Zhang et al. 2007). Other physiological and biochemical characteristics, including acid production from carbohydrates were tested using the API ZYM, API 20NE and API 50CH (bioMérieux) following the manufacturers’ instructions.
Chemotaxonomic characterisation
For analysis of fatty acids, cell mass of strain py1294T and O. damuensis PT-20T was harvested from tryptic soy agar (TSA; Difco) after incubation for 3 days at 30 °C. Cellular fatty acids were extracted and analysed by using the standard MIDI Sherlock Microbial Identification System (version 6.1; MIDI database: TSBA6) according to the manufacturer’s instructions (Sasser 1990) on an Agilent Technologies 7890A GC. The respiratory quinones were extracted and analysed by HPLC (Agilent Technologies 1260 Infinity) (Collins et al. 1977; Groth et al. 1997), and identified by comparison with known quinones from strain L. mangiferahumi M-GX18T as a reference. Total polar lipids were extracted and identified by two-dimensional TLC using the method of Minnikin et al. (1984) after staining with molybdatophosphoric acid, molybdenum blue, ninhydrin and α-naphthol. Diaminopimelic acid isomers was analysed according to the procedures developed by Hasegawa et al. (1983).
Molecular analysis
The genomic DNA was extracted and the 16S rRNA gene was amplified by PCR according to the method of Li et al. (2007). The identification of phylogenetic neighbours was achieved by using the EzTaxon-e server (http://www.ezbiocloud.net/eztaxon; Kim et al. 2012). The 16S rRNA gene sequence was aligned and compared with reference sequences retrieved from the GenBank database by using CLUSTAL X software (Thompson et al. 1997). The phylogenetic trees were constructed using neighbor-joining (Saitou and Nei 1987), maximum-likelihood (Felsenstein 1981) and maximum parsimony (Fitch 1971) tree-making algorithms by using the software package MEGA version 6 (Tamura et al. 2013), while the positions with gaps were completely excluded. Distances were calculated using distance options according to Kimura’s two-parameter model (Kimura 1980). The topologies of the phylogenetic trees were evaluated by using the bootstrap resampling method of Felsenstein (1985) with 1000 replicates. The G+C content of genomic DNA was determined using the HPLC method as described by Mesbah et al. (1989), with DNA prepared according to the method of Marmur (1961). The DNA–DNA relatedness was determined with five replications using the fluorometric micro-well method (Ezaki et al. 1989) with consideration of the modifications described by Goris et al. (1998). Fluorescence was measured by using a microplate spectrofluorometer (Gemini XPS; Molecular Devices).
Results and discussion
Phenotypic characterisation
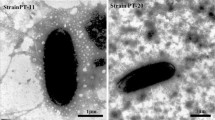
Strain py1294T shows a range of phenotypic properties typical of members of the genus Oceanobacillus. It was observed to be Gram-positive, strictly aerobic, catalase and oxidase positive, and to produce terminal endospores and peritrichous flagella (Supplementary Fig. S1). Colonies were observed to be yellow, round and convex. Cells were observed to be rods, 0.3–0.6 μm wide and 2–6 μm long. Strain py1294T was found to be able to grow at 10–42 °C, in 0–8 % (w/v) NaCl and at pH 6.5–9.5. Optimal growth was observed at 30 °C, in 1–3 % (w/v) NaCl and at pH 8.0. The strain is clearly distinct from its close neighbour O. damuensis, which was described as halophilic (optimum 10–15 % NaCl) and alkaliphilic (optimum pH 7.5–9.0). This difference was also found with the other closely related species listed in Table 1. Negative results from both Liebermann-Burchard and Salkowski reactions indicated that the fermentation broth of strain py1294T did not contain steroids.
In the API 50CH strip, acid was found to be produced from d-ribose, d-fructose, N-acetylglucosamine, aesculin, salicin, cellobiose, starch, glycogen, d-mannitol and d-xylose. In Biolog GENIII plates, the following substrates are utilised: dextrin, d-maltose, d-trehalose, d-cellobiose, gentiobiose, sucrose, d-turanose, alpha-d-lactose, d-melibiose, d-salicin, N-acetyl-d-glucosamine, N-acetyl-beta-d-mannosamine, N-acetyl-d-galactosamine, alpha-d-glucose, d-fructose, d-galactose, 3-methyl glucose, l-rhamnose, inosine, d-mannitol, d-arabitol, glycerol, l-alanine, pectin, l-galactonic acid lactone, d-gluconic acid, d-glucuronic acid, alpha-keto-glutaric acid, Tween 40, beta-hydroxy-dl-butyric acid, acetoacetic acid, acetic acid. Alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, trypsin, alpha-chymotrypsin, naphthol-AS-BI-phosphohydrolase, beta-galactosidase (ONPG), hydrolysis of aesculin and gelatin are positive. Esterase lipase (C14), valine arylamidase, cystine arylamidase, acid phosphatase, alpha-galactosidase, beta-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, N-acetyl-beta-glucosaminidase, alpha-mannosidase, beta-fucosidase, nitrate reduction, indole production, urease and arginine dihydrolase are negative. Phenotypic features that differentiate strain py1294T from closely related species are summarised in Table 1.
Chemotaxonomic characterisation
Strain py1294T was found to contain meso-diaminopimelic acid as the diagnostic diamino acid in the cell wall, which is significantly different from that of members of another closely related genus, Ornithinibacillus, which contain l-Orn–d-Asp (Mayr et al. 2006). Fatty acid composition was compared under the same experimental conditions using the close phylogenetic neighbour based on the 16S rRNA gene sequence, O. damuensis PT-20T. Major fatty acids (>5 % of the total fatty acids) in strain py1294T were identified as anteiso-C15:0 (64.4 %), anteiso-C17:0 (13.9 %), iso-C16:0 (7.0 %) and iso-C14:0 (5.2 %). C16:0 (2.5 %) and iso-C15:0 (4.5 %) were also detected. The presence of a large amount of anteiso-C15:0 and the absence of C18:1 ω9c distinguishes strain py1294T from its close neighbour O. damuensis (Table 2). This fatty acid profile is similar to the description for the genus Oceanobacillus, but the content of each component and the absence of iso-C17:0 were distinct from the isolate’s closely related species. Polar lipid extracts were found to contain diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, two unidentified phospholipids and three unidentified polar lipids (Supplementary Fig. S2). The major isoprenoid quinone was found to be MK-7 (96.7 %), with MK-6 (3.3 %) present in a minor amount. Strain py1294T exhibits chemotaxonomic profiles similar to members of the genus Oceanobacillus. However, the isolate could be distinguished from the closely related species of the genus Oceanobacillus by its fatty acid profile (Table 2) and polar lipids (Table 1).
Molecular analysis
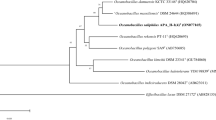
The almost complete 16S rRNA gene sequence of strain py1294T (1492 bp, GenBank/EMBL/DDBJ accession number KU189324) was determined. The 16S rRNA gene sequence displayed high levels of similarity to members of both the genera Oceanobacillus (between 97.9 and 93.6 %) (1354–1492 bp) and Ornithinibacillus (between 97.5 and 93.4 %) (1254–1491 bp) calculated by using CLUSTAL X software. High similarities are exhibited with O. damuensis (97.9 %, 1452 bp), O. picturae (96.9 %, 1492 bp), O. kapialis (96.8 %, 1445 bp), O. profundus (96.7 %, 1354 bp), O. polygoni (96.7 %, 1491 bp) and Ornithinibacillus contaminans (97.5 %, 1254 bp) respectively. Except for O. damuensis and Orn. contaminans, the similarity of 16S rRNA genes between py1294T and other Oceanobacillus species are lower than 97 %. In the phylogenetic tree reconstructed using the neighbour-joining algorithm, strain py1294T clustered with the type strain of O. damuensis with 98 % bootstrap support (Fig. 1). This relationship was also supported by the maximum-like and maximum-parsimony methods with high bootstrap supports of 93 and 94 % (Supplementary Fig. S3). Although strain py1294T shows high sequence similarity with Orn. contaminans CCUG 53201T, they distributed in different clades (Fig. 1a). In addition, given the close phylogenetic relationship between both genera, we generated two sequences sub-datasets, which included strain py1294T and the members of the genera Oceanobacillus and Ornithinibacillus respectively (in each case the same outgroup, Bacillus crassostreae, was used). Interestingly, the phylogenetic relationship between py1294T and the genus Ornithinibacillus was maintained (Fig. 1b) whilst it located outside of the genus Ornithinibacillus (Fig. 1c and Supplementary Fig. S3 C/F). However, based on the sequence dataset that only included the sequences of genus Oceanobacillus, py1294T still clustered with O. damuensis, with higher bootstrapping support (Fig. 1b and Supplementary Fig. S3 B/E). These results indicated that strain py1294T belongs to the genus Oceanbacillus.
Neighbour-joining tree based on 16S rRNA gene sequences, showing the phylogenetic relationships of strain py1294T. Bootstrap values (expressed as percentages of 1000 replications, and ≥50) are given at nodes. B. crassostreae is used as outgroup. Bar 0.005 substitutions per nucleotide position. Oce., Oceanobacillus; Orn., Ornithinibacillus; B., Bacillus
The DNA–DNA relatedness value between strain py1294T and O. damuensis PT-20T was 29.7 ± 3.2 %, a value that is well-below the threshold value (70 %) recommended by Wayne et al. (1987) for the definition of novel prokaryotic species.
The DNA G+C content of strain py1294T was determined to be 42.3 mol%. Strain py1294T shares similar chemotaxonomic characteristics with members of the genus Oceanobacillus in terms of the peptidoglycan type, polar lipids, predominant quinone and major fatty acids. However, strain py1294T can be distinguished from closely related species in some chemotaxonomic and phenotypic features, such as NaCl and pH ranges for growth, and the absence of iso-C17:0. Therefore, based on the phylogenetic analyses and the chemotaxonomic characteristics, strain py1294T should be classified as the representative of a novel species within the genus Oceanobacillus, for which the name Oceanobacillus endoradicis sp. nov. is proposed.
Description of Oceanobacillus endoradicis sp. nov
Oceanobacillus endoradicis (en.do.ra′di.cis. Gr. prep. endo in, within; L. n. radix—icis a root; N.L. gen. n. endoradicis of the inside of a root).
Cells are Gram-positive, aerobic, catalase and oxidase positive, rod-shaped, 0.3–0.6 μm wide and 2–6 μm long. Cells produce terminal endospores and peritrichous flagella. Colonies are yellow, round, convex and 0.5–1.5 mm in diameter after incubation at 30 °C on LB for 3 days. Growth occurs at 10–42 °C (optimum 30 °C), pH 6.5–9.5 (optimum at pH 8.0) and in the presence of 0–8 % (w/v) NaCl (optimum 1–3 %). The cell wall peptidoglycan contains meso-diaminopimelic acid. The major fatty acids (>5 % of the total) are anteiso-C15:0, anteiso-C17:0, iso-C16:0 and iso-C14:0. Polar lipids are diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, unidentified phospholipids and unidentified polar lipids. The predominant isoprenoid quinone is MK-7. The DNA G+C content of the type strain is 42.3 mol%.
The type strain, py1294T (=DSM 100726T = KCTC 33731T) was isolated from the root of P. polyphylla. Smith var. yunnanensis, collected from Chuxiong, Yunnan province, China. The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain py1294T is KU189324.
References
Amoozegar MA, Bagheri M, Makhdoumi-Kakhki A, Didari M, Schumann P, Spröer C, Sánchez-Porro C, Ventosa A (2014) Oceanobacillus limi sp. nov., a moderately halophilic bacterium from a salt lake. Int J Syst Evol Microbiol 64:1284–1289
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–789
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Goris J, Suzuki KI, De Vos P, Nakase T, Kersters K (1998) Evaluation of a microplate DNA:DNA hybridization method compared with the initial renaturation method. Can J Microbiol 44:1148–1153
Gregersen T (1978) Rapid method for distinction of Gram negative from Gram-positive bacteria. Eur J Appl Microbiol Biotechnol 5:123–127
Groth I, Schumann P, Rainey FA, Martin K, Schuetze B, Augsten K (1997) Demetria terragena gen. nov., sp. nov., a new genus of actinomycetes isolated from compost soil. Int J Syst Bacteriol 47:1129–1133
Hasegawa T, Takizawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322
Heyrman J, Logan NA, Busse H-J, Balcaen A, Lebbe L, Rodriguez-Diaz M, Swings J, De Vos P (2003) Virgibacillus carmonensis sp. nov., Virgibacillus necropolis sp. nov. and Virgibacillus picturae sp. nov., three novel species isolated from deteriorated mural paintings, transfer of the species of the genus Salibacillus to Virgibacillus, as Virgibacillus marismortui comb. nov. and Virgibacillus salexigens comb. nov., and emended description of the genus Virgibacillus. Int J Syst Evol Microbiol 53:501–511
Hirota K, Hanaoka Y, Nodasaka Y, Yumoto I (2013) Oceanobacillus polygoni sp. nov., a facultatively alkaliphile isolated from indigo fermentation fluid. Int J Syst Evol Microbiol 63:3307–3312
Kim Y-G, Choi DH, Hyun S, Cho BC (2007) Oceanobacillus profundus sp. nov., isolated from a deep-sea sediment core. Int J Syst Evol Microbiol 57:409–413
Kim O-S, Cho Y-J, Lee K, Yoon S-H, Kim M, Na H, Park S-C, Jeon YS, Lee J-H, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kim W, Siamphan C, Kim J-H, Sukhoom A (2015) Oceanobacillus arenosus sp. nov., a moderate halophilic bacterium isolated from marine sand. Int J Syst Evol Microbiol 65:2943–2948
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Lee J-S, Lim J-M, Lee KC, Lee J-C, Park Y-H, Kim C-J (2006) Virgibacillus koreensis sp. nov., a novel bacterium from a salt field, and transfer of Virgibacillus picturae to the genus Oceanobacillus as Oceanobacillus picturae comb. nov. with emended descriptions. Int J Syst Evol Microbiol 56:251–257
Li W-J, Xu P, Schumann P, Zhang Y-Q, Pukall R, Xu L-H, Stackebrandt E, Jiang C-L (2007) Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China) and emended description of the genus Georgenia. Int J Syst Evol Microbiol 57:1424–1428
Long X, Ye R, Zhang S, Liu B, Zhang Y, Zeng Z, Tian Y (2015) Oceanobacillus damuensis sp. nov. and Oceanobacillus rekensis sp. nov., isolated from saline alkali soil samples. Antonie Van Leeuwenhoek 108:731–739
Lu J, Nogi Y, Takami H (2001) Oceanobacillus iheyensis gen. nov., sp. nov., a deep-sea extremely halotolerant and alkaliphilic species isolated from a depth of 1050 m on the Iheya Ridge. FEMS Microbiol Lett 205:291–297
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218
Mayr R, Busse HJ, Worliczek HL, Ehling-Schulz M, Scherer S (2006) Ornithinibacillus gen. nov., with the species Ornithinibacillus bavariensis sp. nov., and Ornithinibacillus californiensis sp. nov. Int J Syst Evol Microbiol 56:1383–1389
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Namwong S, Tanasupawat S, Lee KC, Lee J-S (2009) Oceanobacillus kapialis sp. nov., from fermented shrimp paste in Thailand. Int J Syst Evol Microbiol 59:2254–2259
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic tree. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. MIDI Inc, Newark
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 607–654
Strobel G, Daisy B (2003) Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev 67:491–502
Strobel G, Daisy B, Castillo U, Harper J (2004) Natural products from endophytic microorganisms. J Nat Prod 67:257–268
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tang S-K, Wang Y, Guan T-W, Lee J-C, Kim C-J, Li W-J (2010) Amycolatopsis halophila sp. nov., a halophilic actinomycete isolated from a salt lake. Int J Syst Evol Microbiol 60:1073–1078
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Waller GR, Yamasaki K (1996) Saponins used in traditional and modern medicine. Plenum Press, NewYork
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE et al (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation ofapproaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Wu M, Yang G, Yu Z, Zhuang L, Jin Y, Zhou S (2014) Oceanobacillus luteolus sp. nov., isolated from soil. Int J Syst Evol Microbiol 64:1495–1500
Yongchang O, Xiang W, Wang G (2015) Oceanobacillus bengalensis sp. nov., a bacterium isolated from seawater of the Bay of Bengal. Antonie Van Leeuwenhoek 108:1189–1196
Yu H, Zhang L, Li L, Zheng C, Guo L, Li W, Sun P, Qin L (2010) Recent developments and future prospects of antimicrobial metabolites produced by endophytes. Mycrobiol Res 165:437–449
Yumoto I, Hirota K, Nodasaka Y, Nakajima K (2005) Oceanobacillus oncorhynchi sp. nov., a halotolerant obligate alkaliphile isolated from the skin of a rainbow trout (Oncorhynchus mykiss), and emended description of the genus Oceanobacillus. Int J Syst Evol Microbiol 55:1521–1524
Zhang X-J, Tang L-L, Wang Y-D (2007) Isolation and screening of steroidal saponins-producing enophytic fungi, actinomycete from paris polyphylla var. chinensis Franch. Prog Mod Biomed 7:358–360 (in Chinese)
Zhou L, Yang C, Li J, Wang S, Wu J (2003) Heptasaccharide and octasaccharide isolated from Paris polyphylla var. yunnanensis and their plant growth-regulatory activity. Plant Sci 165:571–575
Acknowledgments
This research was supported by the National Natural Science Foundation of China (Grant Numbers: 31500007 and 31560309), and the opening project of the State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences (Grant Number: SKLMR-20140601). We are grateful to Dr. Yongqiang Tian of Sichuan University for providing the type strain of O. damuensis PT-20T.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, LL., Tang, SK., Chu, X. et al. Oceanobacillus endoradicis sp. nov., an endophytic bacterial species isolated from the root of Paris polyphylla Smith var. yunnanensis . Antonie van Leeuwenhoek 109, 957–964 (2016). https://doi.org/10.1007/s10482-016-0695-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0695-4