Abstract
Two moderately halophilic strains, PT-11T and PT-20T, were isolated from saline alkali soil samples collected in Shache County, Xinjiang Province, China. Both strains are aerobic, Gram-positive, motile rods. Strain PT-11T grows at 15–40 °C and at pH 6.5–10.0, while PT-20T grows at 15–40 °C and at pH 6.5–11.0. The major cellular fatty acids in both strains include anteiso-C15:0, anteiso-C17:0 and iso-C15:0. For both strains, the polar lipids consist of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, an unidentified phospholipid and several unidentified lipids. In addition, strain PT-20T also contains phosphatidylcholine. The major isoprenoid quinone for both strains is MK-7. The genomic G+C content is 36.7 % for PT-11T and 39.2 % for PT-20T. Phylogenetic analyses of 16S rRNA gene sequences indicated that these two isolates are members of the genus Oceanobacillus. DNA–DNA hybridization indicated that strains PT-11T and PT-20T should be considered two distinct species. On the basis of both phylogenetic and chemotaxonomic data analyses, therefore, we conclude that PT-11T and PT-20T represent two novel species within the genus Oceanobacillus, for which we propose the names Oceanobacillus rekensis sp. nov. and Oceanobacillus damuensis sp. nov., respectively. The type strains are PT-11T (=KCTC 33144T = DSM 26900T) and PT-20T (=KCTC 33146T = DSM 26901T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Oceanobacillus is composed of obligately aerobic, Gram-positive, motile, spore-forming, rod-shaped bacteria. The genus was first proposed by Lu et al. (2001) with the description of Oceanobacillus iheyensis, isolated from a deep-sea mud sample, as the type species (Lu et al. 2001). The description of Oceanobacillus was later amended upon isolation of Oceanobacillus oncorhynchi from the skin of a rainbow trout (Yumoto et al. 2005). Virgibacillus picturae (Heyrman et al. 2003), isolated from samples of a biofilm formation on mural paintings, was later reclassified as Oceanobacillus picturae (Lee et al. 2006). Members of Oceanobacillus have been isolated from deep-sea sediment, chironomid egg mass, sludge, soil, salt lake, marine solar saltern, sand dune, marine sand, the skin of a rainbow trout, algal mat, painting, wastewater-treatment system, fermented shrimp paste, fermented food, soy sauce production equipment, and indigo fermentation fluid. At the time of this writing, eighteen Oceanobacillus species have been described: O. iheyensis (Lu et al. 2001), O. picturae (Heyrman et al. 2003; Lee et al. 2006), O. oncorhynchi (including two subspecies; Yumoto et al. 2005), O. chironomi (Raats and Halpern 2007), O. profundus (Kim et al. 2007), O. caeni (Nam et al. 2008), O. kapialis (Namwong et al. 2009), O. sojae (Tominaga et al. 2009), O. locisalsi (Lee et al. 2010), O. neutriphilus (Yang et al. 2010), O. kimchii (Whon et al. 2010), O. chungangensis (Lee et al. 2013), O. indicireducens (Hirota et al. 2013a, b), O. polygoni (Hirota et al. 2013a, b), O. limi (Amoozegar et al. 2014), O. luteolus (Wu et al. 2014), O. pacificus (Yu et al. 2014) and O. arenosus (Kim et al. 2015).
In this study, two moderately halophilic bacterial strains, PT-11T and PT-20T, were isolated from saline alkali soil samples and were investigated using phylogenetic and chemotaxonomic methods. We determine these strains to be two novel species of the genus Oceanobacillus.
Materials and methods
Bacterial strains and culture conditions
Strains PT-11T and PT-20T were isolated from saline alkali soil samples collected in Shache County, Xinjiang Province, in northwestern China. The typical inland arid climate and unique geographical conditions at this site cause salt to accumulate on the surface of the soil profile. The pH value of the saline soil at sampling sites varied from 7.41 to 7.97. Twenty soil samples were collected at a depth of 10-30 cm at each sampling site (Shache Town, Reke Town, Damu Town) and stored in 50-ml sterile Falcon centrifuge tubes (Shanghai Sangon, China). Each 1 g soil sample was thoroughly shaken in 25 ml PT medium (pH 8.2–8.5) containing, per liter, 7.5 g casein peptone, 100 g NaCl, 10 g yeast extract, 0.016 mol phenol and 1.5 ml MS mixture (0.05 g each of betaine, proline, glycine, d-sorbitol and glutamate, added to 1000 ml H2O) at 28 °C for a week. The suspension, following dilution, was spread onto TSA plates [1.5 % (w/v) tryptone, 0.5 % (w/v) soya peptone, 2 % (w/v) agar, pH adjusted to 7.5] with a total NaCl concentration of 10 % (w/v) and then incubated at 28 °C for a week. Single colonies on the plates were purified by transferring them onto fresh plates and reincubating them. Strains PT-11T and PT-20T were selected and preserved as glycerol stocks at −80 °C before lyophilization. The two strains have been deposited at the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ; German Collection of Microorganisms and Cell Cultures) and the Korean Collection for Type Cultures (KCTC).
Phylogenetic analyses
Methods used for extraction of genomic DNA from strains PT-11T and PT-20T, as well as methods for PCR amplification, primers used, and DNA sequencing conditions of the 16S rRNA gene have been previously described by Li et al. (2007). We obtained nearly complete 16S rRNA gene sequences (1592 bp for PT-11T and 1455 bp for PT-20T). Identification of phylogenetic neighbours and calculation of pairwise 16S rRNA gene sequence similarities were achieved using the NCBI’s BLAST search (Altschul et al. 1990) and the EzTaxon-e server (http://ezbiocloud.net/eztaxon; Kim et al. 2012). Phylogenetic trees were constructed using the neighbour-joining, maximum-likelihood and minimum-evolution methods with the program MEGA 5.2 (Tamura et al. 2011). Evolutionary distances were calculated using Kimura’s two-parameter model (Kimura 1980, 1984). The resultant tree topologies were evaluated by bootstrap analysis with 1000 resamplings. The G+C contents of PT-11T and PT-20T genomic DNA were determined by reversed-phase HPLC according to the method described by Mesbah et al. (1989). DNA–DNA hybridizations were performed between strains PT-11T and PT-20T, between PT-11T and Oceanobacillus profundus, between PT-11T and Oceanobacillus polygoni, and between PT-11T and Ornithinibacillus contaminans using the microplate method as reported by Ezaki et al. (1989).
Morphological, physiological, and chemotaxonomic tests
For phenotypic and chemotaxonomic analysis, strains PT-11T and PT-20T were cultivated on TSB medium with a total NaCl concentration of 10 % (w/v). Cell morphology and size were observed under a scanning electron microscope (SEM) (JSM-7500F, JEOL) using cells in the exponential growth phase. For observation of negatively stained cells by transmission electron microscopy (TEM; Hitachi, H-600IV, Japan), cells were cultivated on TSB medium with 10 % NaCl (w/v). TEM preparation and observation were performed as described previously (Yumoto et al. 2001). Gram-staining was performed as described by Smibert and Krieg (1994). The endospores were detected by using the Schaeffer-Fulton staining method using cells grown for 2 days (Murray et al. 1994). Salt tolerance tests were carried out by growing the cells in TSB medium with different concentrations of NaCl [0, 3, 5, 7, 10, 12, 15, 17, 20, 23, and 25 % (w/v)] while keeping pH value constant at 8.0 and temperature at 30 °C. We determined the optimal pH for each strain by incubating cells in TSB medium adjusted to different pH levels (pH 3.0, 4.0, 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, 9.0, 10.0, 11.0, 11.5, and 12.0) while keeping salt concentration constant at 10 % (w/v) NaCl and temperature constant at 30 °C. We determined growth temperatures for strains PT-11T and PT-20T by incubating cells in TSB medium (pH 8.0) with 10 % (w/v) NaCl at 5, 10, 15, 20, 25, 28, 30, 35, 37, 40, 45, and 50 °C. Nitrate reduction and hydrolysis of gelatin, starch, Tween 80, cellulose and urea were carried out according to the methods described by Cappuccino and Sherman (2011). Catalase ability was tested with 3 % H2O2 and oxidase ability was determined according to the protocols described by Barrow and Feltham (1993). Other enzyme activities were assayed using the API ZYM system according to the manufacturer’s instructions. Carbon-source utilisation was determined using a Biolog Gen III microplate according to the manufacturer’s instructions.
Cellular fatty acids of strains PT-11T and PT-20T were assayed together with O. profundus DSM 18246T in order to examine differences between the novel strains and a closely related species. The isolates were cultured aerobically on TSA plates with 10 % (w/v) NaCl at 37 °C and cells were harvested during the exponential growth phase. Fatty acid methyl esters were prepared and identified with a MIDI Sherlock Microbial Identification System (Sherlock license CD version 6.1). We analysed the polar lipids profile by extracting polar lipids with methanol/chloroform/saline (2:1:0.8 by vol) from 1 g freeze-dried cells, as described by Kates et al. (1972). Separation and identification of lipids was done by two-dimensional chromatography on a silica gel TLC plate (10 × 10 cm), as described previously (Raj et al. 2013). The major quinone of strains PT-11T and PT-20T was determined by HPLC after extraction with a chloroform/methanol (2:1 v/v) mixture and purified by TLC (Imhoff 1984; Hiraishi and Hoshino 1984; Hiraishi et al. 1984).
Results and discussion
Two novel isolates, PT-11T and PT-20T, were found to be aerobic, Gram -positive, spore-forming rods that are motile by means of a peritrichous flagellum (Fig. 1). The cell size of each is 1.0–3.0 μm in length and 0.4–1.2 μm in width. Colonies are circular, smooth and creamy white. Growth of strain PT-11T was found to occur at 5–20 % (w/v) NaCl (optimum 5–12 %), pH 6.5–10.0 (optimum 8.0–9.0), and 15–40 °C (optimum 30–37 °C). Growth of strain PT-20T was found to occur at 3–15 % (w/v) NaCl (optimum 10–15 %), pH 6.5–11.0 (optimum 7.5–9.0), and 15–40 °C (optimum 30–37 °C) (Table 1). Both strains can hydrolyse gelatin and Tween 80 but not starch or cellulose. Oxidase and catalase reactions are positive for both strains. Nitrate (NO3) is reduced to nitrite (NO2) by both strains. Both PT-11T and PT-20T are negative for urease, methyl red, Voges–Proskauer, H2S and indole production tests. Strain PT-11T was found to be sensitive to aztreonam, minocycline and nalidixic acid. Strain PT-20T was found to be sensitive to aztreonam, vancomycin, fusidic acid, troleandomycin, rifamycin SV and nalidixic acid.
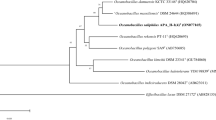
On the basis of pairwise 16S rRNA gene sequence comparisons, strain PT-11T was found to be 97.7 % identical to O. profundus DSM 18246T, 97.3 % identical to Ornithinibacillus contaminans CCUG 53201T, and 97.1 % identical to O. polygoni SA9T. For PT-20T, the highest similarities were found to O. contaminans CCUG 53201T (97.6 % identity), Ornithinibacillus bavariensis WSBC24001T (97.4 %), O. profundus DSM 18246T (97.3 %), and O. polygoni SA9T (97.1 %). A neighbour-joining tree based on 16S rRNA gene sequences indicated that strains PT-11T and PT-20T are closely related to O. profundus DSM 18246T and O. polygoni SA9T (Fig. 2). Essentially the same tree topology was obtained with the maximum-likelihood and minimum-evolution algorithms. These phylogenetic analyses revealed that PT-11T and PT-20T form a separate cluster within a subgroup of the genus Oceanobacillus.
According to Rossello-Mora (2006), DNA–DNA hybridization is “the gold standard method” for defining a bacterial species. The DNA–DNA relatedness value between strain PT-11T and PT-20T is 54.5 ± 2.3 %, while it is 48.6 ± 4.5 % between PT-11T and O. profundus, 46.5 ± 2.0 % between PT-11T and O. polygoni, and 32.1 ± 2.9 % between PT-11T and Orn. contaminans. These relatedness values are all lower than 70 %, which was recommended as the value for delineating separate prokaryotic species by Wayne et al. (1987). The genomic G+C content of strains PT-11T and PT-20T were determined to be 36.7 and 39.2 % respectively, which is within the range for other members of the genus Oceanobacillus.
Whole-cell fatty acids analysis revealed that anteiso-C15:0 (52.3 %), iso-C15:0 (14.4 %), summed feature 4 (iso I/anteiso B C17:1, 7.6 %), anteiso-C17:0 (6.7 %), iso-C16:0 (3.5 %) and iso-C14:0 (3.1 %) are the predominant fatty acids in strain PT-11T. The relative proportions of the predominant fatty acids in strain PT-20 T are somewhat different, as follows: anteiso-C15:0 (36.5 %), anteiso-C17:0 (14.5 %), iso-C16:0 (8.7 %), iso-C15:0 (7.4 %), C8:1 w9c (6.5 %), C16:0 (5.0 %) and iso-C14:0 (3.0 %). The major cellular fatty acids for members of the genus Oceanobacillus are anteiso-C15:0, iso-C15:0 and iso-C14:0, consistent with the results for PT-11T and PT-20T. MK-7 was found to be the major quinone of both strains PT-11T and PT-20T.
For both strains PT-11T and PT-20T, the polar lipids were found to consist of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, an unidentified phospholipid and several unidentified lipids. For PT-20T, phosphatidylcholine is also present (Fig. 3).
Polar lipids profile of strain PT-20T, PT-11T and Oceanobacillus profundus DSM 18246T separated by two-dimensional TLC. DPG diphosphatidylglycerol, PG phosphatidylglycerol, PE phosphatidylethanolamine, PC phosphatidylcholine, PL unidentified phospholipid, AL unidentified aminolipid, L unidentified polar lipid
Phylogenetic analysis of 16S rRNA gene sequences indicates that strains PT-11T and PT-20T are members of the genus Oceanobacillus. The chemotaxonomic results (cellular fatty acids, polar lipids, quinone, etc.) are also consistent with this conclusion. Although the two strains have similar properties, several differences exist between them such as differences in growth conditions; utilisation of glycerol, d-mannose, d-mannitol and d-turanose; enzyme activities; and cellular fatty acids composition. On the basis of these differences and DNA–DNA hybridization results, we propose two novel species: Oceanobacillus rekensis sp. nov. represented by strain PT-11T and Oceanobacillus damuensis sp. nov. represented by strain PT-20T.
Description of Oceanobacillus rekensis sp. nov
Oceanobacillus rekensis (rek.en’sis. N.L. masc. adj. rekensis pertaining to Reke Town, Shache County, Xinjiang Province, China, where the type strain was isolated).
Aerobic, Gram-positive, motile, spore-forming rods (0.4–1.2 × 1.0–3.0 μm). Endospores are ellipsoid and terminally positioned. Colonies are circular, smooth and creamy white, with diameter 0.5–2.0 mm. Growth occurs at 5–20 % (w/v) NaCl (optimum 5–12 %), at pH 6.5–10.0 (optimum 8.0–9.0) and at 15–40 °C (optimum 30–37 °C). Can hydrolyse gelatin and Tween 80 but not starch or cellulose. Oxidase and catalase reactions are positive. Able to reduce nitrate (NO3) to nitrite (NO2). Negative for urease, egg yolk reaction, methyl red, Voges-Proskauer, H2S and indole production tests. Positive for Biolog Gen III MicroStation substrates dextrin, d-maltose, d-trehalose, d-sucrose, d-turanose, stachyose, d-raffinose, α-d-glucose, d-fructose, d-glucose-6-phosphate, d-fructose-6-phosphate, l-alanine, l-glutamic acid, l-serine, pectin, d-gluconic acid, d-glucuronic acid, d-glucuronamide, d-lactic acid methyl ester, l-malic acid, bromo-succinic acid, Tween 40, γ-amino-butyric acid, α-hydroxy-butyric acid, acetoacetic acid, propionic acid, acetic acid. In API-ZYM assays, esterase (C4), esterase lipase (C8) and naphthol-AS-B1-phosphohydrolase are present. Alkaline phosphatase, lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, α-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase are absent. The predominant polar lipids are diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, an unidentified phospholipid and several unidentified lipids. The major isoprenoid quinone is MK-7. The major cellular fatty acids are anteiso-C15:0, iso-C15:0, summed feature 4 (iso I/anteiso B C17:1), anteiso-C17:0, iso-C16:0 and iso-C14:0. The genomic G+C content of the type strain is 36.7 %.
The type strain is PT-11T (=KCTC 33144 = DSM 26900), isolated from a saline alkali soil sample collected in Reke Town, Shache County, Xinjiang Province, China. The 16S rRNA sequence of strain PT-11T has been deposited in Genbank under accession number HQ620695.
Description of Oceanobacillus damuensis sp. nov
Oceanobacillus damuensis (da.mu.en’sis. N.L. masc. adj. damuensis pertaining to Damu Town, Shache County, Xinjiang Province, China, where the type strain was isolated).
Aerobic, Gram-positive, motile, spore-forming rods (0.4–1.2 × 1.0–3.0 μm). Endospores are ellipsoid and terminally positioned. Colonies are circular, smooth, and creamy white, with diameter 0.5–1.5 mm. Growth occurs at 3–15 % (w/v) NaCl (optimum 10–15 %), at pH 6.5–11.0 (optimum 7.5–9.0), and at 15–40 °C (optimum 30–37 °C). Can hydrolyse gelatin and Tween 80 but not starch or cellulose. Oxidase and catalase reactions are positive. Able to reduce nitrate (NO3) to nitrite (NO2). Negative for urease, egg yolk reaction, methyl red, Voges–Proskauer, H2S and indole production tests. Positive for Biolog Gen III MicroStation substrates dextrin, d-maltose, d-trehalose, N-acetyl-d-glucosamine, N-acetyl-β-d-mannosamine, α-d-glucose, l-fucose, 1 % sodium lactate, d-glucose-6-phosphate, d-fructose-6-phosphate, l-alanine, l-glutamic acid, lincomycin, pectin, d-galacturonic acid, d-glucuronic acid, glucuronamide, d-lactic acid methyl ester, citric acid, α-keto-glutaric acid, l-malic acid, bromo-succinic acid, Tween 40, α-hydroxy-butyric acid, β-hydroxy-d,l-butyric acid, α-keto-butyric acid, and acetoacetic acid. In API-ZYM assays, esterase lipase (C8), naphthol-AS-B1-phosphohydrolase, β-glucuronidase, α-glucosidase, and β-glucosidase are present. Alkaline phosphatase, esterase (C4), lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase are absent. The predominant polar lipids are diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholine, an unidentified phospholipid and several unidentified lipids. The major isoprenoid quinone is MK-7. The major cellular fatty acids are anteiso-C15:0, anteiso-C17:0, iso-C16:0, iso-C15:0, C8:1 w9c, C16:0 and iso-C14:0. The genomic G+C content of the type strain is 39.2 %.
The type strain is PT-20T (=KCTC 33146 = DSM 26901), isolated from a saline alkali soil sample collected in Damu Town, Shache County, Xinjiang Province, China. The 16S rRNA sequence of strain PT-20T has been deposited in Genbank under accession number HQ620704.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Amoozegar MA, Bagheri M, Makhdoumi-Kakhki A, Didari M, Schumann P, Spröer C, Sánchez-Porro C, Ventosa A (2014) Oceanobacillus limi sp. nov., a moderately halophilic bacterium from a salt lake. Int J Syst Evol Microbiol 64:1284–1289
Barrow G I, Feltham RKA (1993). Cowan and Steel’s Manual for the identification of Medical bacteria, 3rd edn. Cowan & Steels Manual for the Identification of Medical Bacteria
Cappuccino JG, Sherman N (2011) Microbiology: A Laboratory Manual. Pearson/Benjamin Cummings, San Francisco
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Heyrman J, Logan NA, Busse HJ, Balcaen A, Lebbe L, Rodriguez-Diaz M, Swings J, De Vos P (2003) Virgibacillus carmonensis sp. nov., Virgibacillus necropolis sp. nov. and Virgibacillus picturae sp. nov., three novel species isolated from deteriorated mural paintings, transfer of the species of the genus Salibacillus to Virgibacillus, as Virgibacillus marismortui comb. nov. and Virgibacillus salexigens comb. nov., and emended description of the genus Virgibacillus. Int J Syst Evol Microbiol 53:501–511
Hiraishi A, Hoshino Y (1984) Distribution of rhodoquinone in Rhodospirillaceae and its taxonomic implications. J Gen Appl Microbiol 30:435–448
Hiraishi A, Hoshino Y, Kitamura H (1984) Isoprenoid quinone composition in the classification of Rhodospirillaceae. J Gen Appl Microbiol 30:197–210
Hirota K, Aino K, Nodasaka Y, Yumoto I (2013a) Oceanobacillus indicireducens sp. nov., a facultative alkaliphile that reduces an indigo dye. Int J Syst Evol Microbiol 63:1437–1442
Hirota K, Hanaoka Y, Nodasaka Y, Yumoto I (2013b) Oceanobacillus polygoni sp. nov., a facultatively alkaliphile isolated from indigo fermentation fluid. Int J Syst Evol Microbiol 63:3307–3312
Imhoff JF (1984) Quinones of phototrophic purple bacteria. FEMS Microbiol Lett 25:85–89
Kämpfer P, Falsen E, Lodders N, Langer S, Busse HJ, Schumann P (2010) Ornithinibacillus contaminans sp. nov., an endospore-forming species. Int J Syst Evol Microbiol 60:2930–2934
Kates M, Work TS, Work E (1972). Techniques of lipidology: isolation, analysis and identification of lipids. Lab Tech Biochem Mol Biol 267
Kim YG, Choi DH, Hyun S, Cho BC (2007) Oceanobacillus profundus sp. nov., isolated from a deep-sea sediment core. Int J Syst Evol Microbiol 57:409–413
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Lee JH, Yi H, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kim W, Chatuphon S, Kim JH, Sukhoom A (2015) Oceanobacillus arenosus sp.nov., a moderate halophilic bacterium isolated from marine sand. Int J Syst Evol Microbiol 63:3307–3312
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kimura M (1984) The neutral theory of molecular evolution. Cambridge University Press, New York
Lee JS, Lim JM, Lee KC, Lee JC, Park YH, Kim CJ (2006) Virgibacillus koreensis sp. nov., a novel bacterium from a salt field, and transfer of Virgibacillus picturae to the genus Oceanobacillus as Oceanobacillus picturae comb. nov. with emended descriptions. Int J Syst Evol Microbiol 56:251–257
Lee SY, Oh TK, Kim W, Yoon JH (2010) Oceanobacillus locisalsi sp. nov., isolated from a marine solar saltern. Int J Syst Evol Microbiol 60:2758–2762
Lee DC, Kang H, Weerawongwiwat V, Kim B, Choi YW, Kim W (2013) Oceanobacillus chungangensis sp. nov., isolated from a sand dune. Int J Syst Evol Microbiol 63:3666–3671
Li WJ, Xu P, Schumann P, Zhang YQ, Pukall R, Xu LH, Stackebrandt E, Jiang CL (2007) Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int J Syst Evol Microbiol 57:1424–1428
Lu J, Nogi Y, Takami H (2001) Oceanobacillus iheyensis gen. nov., sp. nov., a deep-sea extremely halotolerant and alkaliphilic species isolated from a depth of 1050 m on the Iheya Ridge. FEMS Microbiol Lett 205:291–297
Mayr R, Busse HJ, Worliczek HL, Ehling-Schulz M, Scherer S (2006) Ornithinibacillus gen. nov., with the species Ornithinibacillus bavariensis sp. nov. and Ornithinibacillus californiensis sp. nov. Int J Syst Evol Microbiol 56:1383–1389
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Murray RGE, Doetsch RN, Robinow CF (1994) Determinative and cytological light microscopy. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 21–41
Nam JH, Bae W, Lee DH (2008) Oceanobacillus caeni sp. nov., isolated from a Bacillus-dominated wastewater treatment system in Korea. Int J Syst Evol Microbiol 58:1109–1113
Namwong S, Tanasupawat S, Lee KC, Lee JS (2009) Oceanobacillus kapialis sp. nov., from fermented shrimp paste in Thailand. Int J Syst Evol Microbiol 59:2254–2259
Raats D, Halpern M (2007) Oceanobacillus chironomi sp. nov., a halotolerant and facultatively alkaliphilic species isolated from a chironomid egg mass. Int J Syst Evol Microbiol 57:255–259
Raj PS, Ramaprasad EVV, Vaseef S, Sasikala C, Ramana CV (2013) Rhodobacter viridis sp. nov., a phototrophic bacterium isolated from mud of a stream. Int J Syst Evol Microbiol 63:181–186
Rosselló-Mora R (2006) DNA-DNA reassociation methods applied to microbial taxonomy and their critical evaluation. Molecular identification, systematics, and population structure of prokaryotes. Springer Berlin, Heidelberg, pp 23–50
Smibert RM, Krieg NR (1994) Phenotypic characteristics. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Manual of Methods for General and Molecular Bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tominaga T, An S-Y, Oyaizu H, Yokota A (2009) Oceanobacillus soja sp. nov. isolated from soy sauce production equipment in Japan. J Gen Appl Microbiol 55:225–232
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Murray RGE et al (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Whon TW, Jung MJ, Roh SW, Nam YD, Park EJ, Shin KS, Bea JW (2010) Oceanobacillus kimchii sp. nov. isolated from a traditional Korean fermented food. J Microbiol 48(6):862–866
Wu M, Yang GQ, Yu Z, Zhuang L, Jin YQ, Zhou SG (2014) Oceanobacillus luteolus sp nov., isolated from soil. Int J Syst Evol Microbiol 64:1495–1500
Yang JY, Huo YY, Xu XW, Meng FX, Wu M, Wang CS (2010) Oceanobacillus neutriphilus sp. nov., isolated from activated sludge in a bioreactor. Int J Syst Evol Microbiol 60:2409–2414
Yu C, Yu S, Zhang Z, Li Z, Zhang XH (2014) Oceanobacillus pacificus sp. nov., isolated from a deep-sea sediment. Int J Syst Evol Microbiol 64:1278–1283
Yumoto I, Yamazaki K, Hishinuma M, Nodasaka Y, Suemori A, Nakajima K, Inoue N, Kawasaki K (2001) Pseudomonas alcaliphila sp. Nov., a novel facultatively psychrophilic alkaliphile isolated from seawater. Int J Syst Evol Microbiol 51:349–355
Yumoto I, Hirota K, Nodasaka Y, Nakajima K (2005) Oceanobacillus oncorhynchi sp. nov., a halotolerant obligate alkaliphile isolated from the skin of a rainbow trout (Oncorhynchus mykiss), and emended description of the genus Oceanobacillus. Int J Syst Evol Microbiol 55:1521–1524
Acknowledgments
We are grateful to Shu-Kun Tang for his help with some of the experiments. This work is supported by the Fundamental Research Funds for the Central University and Sichuan Science and Technology Project No. 2014JY0199.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Long, X., Ye, R., Zhang, S. et al. Oceanobacillus damuensis sp. nov. and Oceanobacillus rekensis sp. nov., isolated from saline alkali soil samples. Antonie van Leeuwenhoek 108, 731–739 (2015). https://doi.org/10.1007/s10482-015-0529-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-015-0529-9