Abstract
Clustering ensemble has been increasingly popular in the recent years by consolidating several base clustering methods into a probably better and more robust one. However, cluster dependability has been ignored in the majority of the presented clustering ensemble methods that exposes them to the risk of the low-quality base clustering methods (and consequently the low-quality base clusters). In spite of some attempts made to evaluate the clustering methods, it seems that they consider each base clustering individually regardless of the diversity. In this study, a new clustering ensemble approach has been proposed using a weighting strategy. The paper has presented a method for performing consensus clustering by exploiting the cluster uncertainty concept. Indeed, each cluster has a contribution weight computed based on its undependability. All of the predicted cluster tags available in the ensemble are used to evaluate a cluster undependability based on an information theoretic measure. The paper has proposed two measures based on cluster undependability or uncertainty to estimate the cluster dependability or certainty. The multiple clusters are reconciled through the cluster uncertainty. A clustering ensemble paradigm has been proposed through the proposed method. The paper has proposed two approaches to achieve this goal: a cluster-wise weighted evidence accumulation and a cluster-wise weighted graph partitioning. The former approach is based on hierarchical agglomerative clustering and co-association matrices, while the latter is based on bi-partite graph formulating and partitioning. In the first step of the former, the cluster-wise weighing co-association matrix is proposed for representing a clustering ensemble. The proposed approaches have been then evaluated on 19 real-life datasets. The experimental evaluation has revealed that the proposed methods have better performances than the competing methods; i.e. through the extensive experiments on the real-world datasets, it has been concluded that the proposed method outperforms the state-of-the-art. The substantial experiments on some benchmark data sets indicate that the proposed methods can effectively capture the implicit relationship among the objects with higher clustering accuracy, stability, and robustness compared to a large number of the state-of-the-art techniques, supported by statistical analysis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
It is a very reasonable task in any field that anybody wants to group a host of data (Wang et al. 2017; Ma et al. 2018), also called dataset. It can be necessary due to lack of any insight on dataset. So, a first grouping in dataset, where there is no primary insight, can provide us with a good initial understanding and insight. Grouping unlabeled data samples into meaningful groups is a challenging unsupervised Machine Learning (ML) (Deng et al. 2018; Yang and Yu 2017) problem with a wide spectrum of applications (Li et al. 2017a, b; Alsaaideh et al. 2017; Song et al. 2017), ranging from image segmentation (Chakraborty et al. 2017) in computer vision to data modeling in computational chemistry (Spyrakis et al. 2015).
Due to its value, clustering has emerged as one of the leading tasks of multivariate analysis. It has been shown that Cluster Analysis (ClA) is among the most versatile concepts for analyzing datasets with many (more than 3) features (Kettenring 2006). The ClA is also an essential tool for capturing the structure of data (Li et al. 2017a, b, 2018a).
Data clustering is a preliminary task in the field of data mining and pattern recognition (Jain 2010). The clustering goal is to detect the underlying structures of a given dataset and divide the dataset into a number of homogeneous subsets, called clusters. Recently, different clustering methods have been suggested by employing various techniques (Xu et al. 1993; Ng et al. 2002; Frey and Dueck 2007; Wang et al. 2011, 2012). Any of these methods has its weak and strong points, and consequently may outperform the others for some specific applications. Clustering samples according to an effective metric and/or vector space representation is a challenging unsupervised learning task. This kind of methods finds application in several fields.
It can be claimed that no clustering method is superior to all other methods for all applications (for example if clusters of a dataset are of linear type, model based clustering methods is weak or if its clusters are extracted from a normal distribution, linkage based clusters may be non-suitable). Different partitions of a given dataset can be attained by applying different clustering methods, or by applying a clustering algorithm with different initializations or different parameters. So, selection of the best clustering algorithm for a given dataset is very challenging; or if we want to use a specific clustering algorithm, selection of the best parameters for it is still not possible. A lot of clustering techniques have been suggested, but the No Free Lunch (NFL) theorem (Wolpert and Macready 1996) recommends that there is no particular, best technique that fits all cluster shapes and structures absolutely.
In some applications of clustering, usage of a robust one is inevitable; for example, if clustering mechanism is employed in a preliminary stage of a classification algorithm (to find and omit outliers) (Frey and Dueck 2007). Another reason to desire a robust clustering is hidden in the strong relationship between the ClA and Robust Statistics (Schynsa et al. 2010). Robust clustering methods are discussed in detailed by García-Escudero et al. (2010). Coretto and Hennig (2010) have shown experimentally that the RIMLE method, their previously proposed method, can be recommended as optimal in some situations, and always acceptable.
The outlier detection problem is a field that is very involved in ClA. It is also very involved in the robust covariance estimation problem. It is expected that if the outliers of a dataset are ignored, the maximum likelihood estimation can be employed to learn dataset with a perfect accuracy. It is believed that outliers affect the maximum likelihood estimation method and the estimated parameters are pushed toward them. So many works have been done to deal with the robust detection of outliers and present methods free from model assumptions. A very fast approach with low computational complexity has been designed by use of a special mathematical programming model to find the optimal weights for all the observations, such that at the optimal solution, outliers are given smaller weights and can be detected (Nguyen and Welsch 2010). In their work, the parameters of data are detected by a maximum likelihood estimation method in such a weighted manner that the participation weight of an outlier is very low. While the algorithm is efficient, its computational complexity is reasonable compared with its competitors.
The transfer distance between partitions (Charon et al. 2006; and Denoeud 2008) is equal to the minimum number of elements that must be moved from one class to another (possibly empty) to turn one partition into another. This distance has studied under the name of “partition-distance”. On the base of the concept of the transfer distance between partitions, a consensus partition has been proposed by Guénoche (2011). Given a host of primary partitions over a dataset, Guénoche (2011) has proposed a consensus partition containing a maximum number of joined or separated pairs in dataset that are also joined or separated in the primary partitions. So, a score function has been defined mapping a partition to score value. In the first step, the problem is transformed to a graph partitioning problem. Then, using some mathematical tricks converts it to an integer linear programming optimization. Finally, a partition that maximizes the score is explored. The score can be maximized by an integer linear programming only in certain cases. It has been shown that in those certain cases that the method converges to a solution the produced consensus partition is very close to the optimal solution. Although the method is a very simple and clear one, it fails in many cases as the paper declared it.
Different partitions created by different methods (or the same clustering method with different initializations and parameters) may reveal different viewpoints of the data. The mentioned different partitions create a diverse ensemble. Then a partition is extracted out of the ensemble in such a way that it exploits the maximum information among the ensemble. This partition that is similar to all partitions of the ensemble as much as possible is named consensus partition. The function that takes a diverse ensemble as input and produces a consensus partition as output is named consensus function. The framework that first creates a diverse ensemble (by any method mentioned at the start of this paragraph) and then uses a consensus function to aggregate them into a final partition is named clustering ensemble (Strehl and Ghosh 2003; Cristofor and Simovici 2002; Fern and Brodley 2004; Fred and Jain 2005; Topchy et al. 2005; Li et al. 2018b).
Clustering ensemble usually produces a more robust, accurate, novel, and stable partition (it is worthy to be mentioned that a robust clustering method is the one whose performance doesn’t fall very much in the presence of noise; an accurate clustering method is the one whose predicted clusters are equal to real predefined classes in the dataset; a novel clustering method is the one that uncovers some new clusters dissimilar to the clusters detected by the ordinary simple clustering algorithms; a stable clustering method is the one that produces the same results irrespective to initialization) (Wu et al. 2013). One of the most challenging tasks in Cluster Analysis is how to obtain the robustness of a clustering. Hennig (2008) acclaimed that this problem is mainly due to the fact that robustness and stability in Cluster Analysis are not only data dependent, but even cluster dependent.
In clustering ensemble, there are two vital elements that both have effect on the quality of consensus partition: diversity among the base partitions and their quality. Some efforts have been done to give a weight to each base clustering based on its quality, while they have ignored diversity among base clusterings. Some other efforts have been done to select a subset of ensemble to maximize the diversity among the selected ensemble. Indeed, they have defined a metric for measuring diversity of an ensemble at first. After selecting a subset of ensemble, then they have used a mentioned weighting mechanism (Li and Ding 2008; Yu et al. 2014; Huang et al. 2015). By the way, all these efforts have assumed implicitly that all clusters of a base partition have the equal dependability. Although this assumption doesn’t cause bad partitions to be selected, it sometimes causes some clusters to be omitted because they are inside an unsuitable partition while they are suitable clusters and even make our ensemble more diverse. It means that some clusters in a partition may be more trustworthy than other clusters in the same partition. Based on this fact Zhong et al. (2015) have proposed a metric to access the cluster dependability. By the way, our clustering ensemble is inherently different from their method. On contrary to their methods, our method doesn’t need the original features of dataset and it doesn’t assume any distribution in dataset.
This paper also wants to suggest a clustering ensemble based on assessing the clusters’ undependability. It also considers weighting mechanism. The proposed mechanism defines diversity in cluster level. It considers the quality and diversity of clusters during clustering ensemble in order to improve consensus partition.
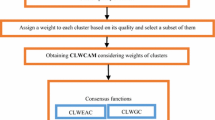
The dependability of any cluster is computed based on its undependability of the cluster. The undependability of any cluster is computed based on the entropy among the labels of its data points throughout all partitions of the ensemble. Through assessing and then emphasizing the more trustworthy clusters (the ones with more dependability) in the ensemble, an innovative cluster-wise weighing co-association matrix has been proposed. Then, a cluster-wise weighting bi-partite graph has been also introduced to represent the ensemble based on its corresponding cluster-wise weighing co-association matrix. Finally, the consensus partition is extracted based on two mechanisms: (a) by considering the cluster-wise weighing co-association matrix as a similarity matrix and then applying a simple hierarchical clustering algorithm, and (b) by partitioning the cluster-wise weighting bi-partite graph into a certain number of parts (or clusters).
To sum up, the novelties of the paper includes:
-
1.
Proposing a mechanism to assess the dependability of a cluster in the ensemble.
-
2.
Proposing a clustering ensemble framework to let clusters in the ensemble contribute in the consensus partition according to their dependabilities.
-
3.
Proposing a cluster-wise weighing co-association matrix considering the cluster dependability.
-
4.
Proposing a cluster-wise weighting bi-partite graph considering the cluster-wise weighing co-association matrix.
We have compared our proposed mechanism with some other state-of-the-art clustering ensemble methods by applying them to 19 real-world datasets. We have evaluated the results using Normalized Mutual Information (NMI), F-Measure (FM), Adjacent Rand Index (ARI), and Accuracy criterion (Acc). We assess the following aspects of the proposed method:
-
1.
Effect of varying φ on undependability assessment quality.
-
2.
Performance of the proposed method versus the base clustering algorithms.
-
3.
Performance of the proposed method versus other state-of-the-art clustering ensemble methods.
-
4.
Effect of varying the number of base partitions on quality of the proposed method and other methods.
-
5.
Effect of noise on the proposed method performance.
-
6.
Execution time.
2 Related works
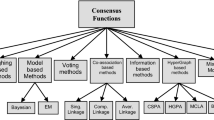
Clustering ensemble integrates several base partitions to attain a more robust, accurate, novel, and stable consensus partition. In a wide categorization, they are divided into two groups based on their base clustering structures: (1) hierarchical clustering ensemble (Mirzaei et al. 2008) and (2) partitional clustering ensemble (Parvin and Minaei-Bidgoli 2015). The consensus function of partitional clustering ensemble is divided into two groups: (1) optimization based methods (Alizadeh et al. 2014) and (2) intermediate space clustering (Parvin and Minaei-Bidgoli 2015). Intermediate space clustering (i.e. the subgroup of partitional clustering ensemble) is divided into three groups: (1) the pair-wise co-association based methods, (2) the graph partitioning based methods (Strehl and Ghosh 2003; Fern and Brodley 2004), and the median partition based methods (Cristofor and Simovici 2002; Topchy et al. 2005; Franek and Jiang 2014).
Some approaches are based on Co-Association (CA) (Fred and Jain 2005; Wang et al. 2009; Iam-On et al. 2011; Wang 2011). They first create a CA matrix whose (i, j) th element indicates how many times ith data point and jth data point belong to a shared cluster through all base partitions of the ensemble. Then by considering the CA matrix as a similarity matrix, a simple clustering technique that performs clustering task based on similarity matrix, like a hierarchical clustering method, can be employed to make the final consensus clustering (Jain 2010). The CA matrix has been introduced first in the name of the Evidence Accumulation Clustering (EAC) (Fred and Jain, 2005). Then, it has been generalized by introducing the probability accumulation method (Wang et al. 2009). Wang (2011) has introduced a CA-tree to enhance the consensus partition. Then, the CA matrix has been modified by taking the joint neighbors among clusters into consideration to enhance the consensus partition (Iam-On et al. 2011).
In some approaches, the clustering ensemble problem is first transformed into a graph (Strehl and Ghosh 2003; Fern and Brodley 2004). They have extracted the consensus partition through partitioning the mentioned graph into a predefined number of disjoint parts. The first set of these approaches includes Cluster-based Similarity Partitioning Algorithm (CSPA), Hyper Graph Partitioning Algorithm (HGPA), and Meta-CLustering Algorithm (MCLA). A Bi-Partite Graph Partitioning Algorithm then has been introduced to speed-up these approaches (Fern and Brodley 2004).
In another direction of research, some researchers have transformed the clustering ensemble problem into an optimization problem, and then they have tried to find a consensus partition that its distance to all base partitions is minimum (Cristofor and Simovici 2002; Topchy et al. 2005; Franek and Jiang 2014). The found consensus partition is named median partition. This approach is named median partition based clustering ensemble. Finding the median partition is an NP-hard problem and its state space is extremely large; so finding the best partition as median partition through a brute-force search is impossible for datasets even with moderate sizes. So some researchers have proposed a mechanism to reach an approximate solution by heuristic optimization methods like genetic algorithm (Cristofor and Simovici 2002). In another work, the Expectation Maximization (EM) has been employed to find an approximate solution to the median partition problem (Topchy et al. 2005). In another recent research, the mentioned problem has been first transformed into a new Euclidean median problem. This has been possible by considering each partition as a vector. Then the median vector can be found by the method proposed by Weiszfeld and Plastria (2009). Finally the median vector is retransformed into a partition (i.e. the median partition).
All of the mentioned approaches have tried to find a solution to the clustering ensemble problem using different methodologies. Nonetheless, there is a shared limitation to almost all of those approaches. Their limitation is due to their inability to treating different clusters according to their qualities. Indeed, they have considered all clusters (or even base clusterings) equally irrespective to their quality. This make them exposed to low-quality clusters (or low-quality base clusterings). For managing this inadequacy, some recent works have been done (Li and Ding 2008; Yu et al. 2014; Huang et al. 2015). Li and Ding (2008) transform the clustering ensemble problem into an optimization problem. They have also suggested assigning a weight to each clustering. The Attribute Selection systems have been recently extended to be employed to select (or to weight) the base clusterings (Yu et al. 2014). Indeed, the clustering ensemble weighing problem can be considered as a binary weighting system by Yu et al. (2014), where a weight one (or a weight zero) for a base clustering shows that clustering is selected for (or correspondingly removed from) the ensemble. In their work, the clustering ensemble weighing problem indeed is the clustering ensemble selection problem. In another more recent research, a more mature work has been proposed (Huang et al. 2015). The authors have proposed a clustering ensemble weighing scheme where each base clustering is assigned to a weight according to its Normalized Crowd Agreement Index. All the mentioned works employ a weighting in the level of the base clusterings. So, all of them suffer from ignoring the cluster dependability.
Alizadeh et al. (2014) have introduced a cluster ensemble selection approach. Their approach is done through ranking of all clusters according to their stabilities (i.e. their averaged NMI values) and then selecting a portion of the best clusters. In another work, Alizadeh et al. (2015) have introduced a cluster ensemble problem with cluster-wise weighing. They have employed a binary weighting system, where a weight one (or a weight zero) for a base cluster shows that cluster is selected for (or correspondingly removed from) the ensemble. In their work, the cluster ensemble weighing problem is indeed the cluster ensemble selection problem.
Zheng et al. (2015) have proposed an instance-wise weighted nonnegative matrix factorization for aggregating partitions with locally reliable clusters. Zhong et al. (2015) uses spatial data to approximate the cluster dependability. While these works seem to be cluster-wise weighting schemes, after producing the ensemble they still need original features of dataset.
3 Definitions
Definition 1
A dataset denoted by D is a set of \( \left\{ {d_{1} ,d_{2} , \ldots , d_{n} } \right\} \) where di is ith data point and n stands for the size of dataset. dij stands as jth feature in ith data point. F stands as the number of features.
Definition 2
A sub-sampling of dataset D is a random subset of dataset D and is denoted by DS where S is the size of subset and is a positive integer.
Definition 3
A partition or clustering is a vector π = [π1, π2, …, πn]T where πi is a positive integer indicating to which cluster ith data point belongs. Note that it is assumed that no cluster should be empty.
Definition 4
A clustering pool or ensemble is a set of B partitions denoted by Π and defined by Eq. (1).
where πi is ith partition and has ci cluster, i.e. π i j ∊ {1, 2, …, ci}. As it can be inferred from Eq. (1), clustering pool Π is a matrix with the size of n × B whose each column is a partition. An exemplary ensemble of three partitions is presented in Table 1. In the first and third partitions of this ensemble, there are 3 clusters, but in the second partition there are only 2 clusters. Any of partitions inside the ensemble has been done on a dataset with 15 data points.
Definition 5
A cluster representation (CR) of a partition is defined as Eq. (2).
where \( \tau_{j}^{i} \) is jth cluster inside ith partition and is a vector defined as Eq. (3).
Definition 6
A cluster representation of a clustering pool Π is defined as Eq. (4).
where T is a matrix with the size of n × b and b = ∑ B i=1 ci. Cluster representation of the exemplary ensemble of Table 1 is depicted in Table 2. Ti stands for ith column of matrix T. Tij is equal to 1 if jth data point belongs to ith cluster.
Definition 7
For a discrete random variable Y, the entropy denoted by H(Y) is defined as Eq. (5).
where Y is the set of values that y can take, and p(y) is the probability mass function of y.
Lemma 1
The H(Y) will be maximum when\( p\left( y \right) = \frac{1}{\left| Y \right|} \)where |Y| is the number of different possible values in discrete random variable Y, or is the size of state space of the discrete random variable Y. The maximum H(Y) will be log 2|Y| as computed in Eq. (6).
Definition 8
The joint entropy of a pair of discrete random variables Y and X denoted by H(Y, X) is described as Eq. (7).
where p(y, x) is the joint probability of (Y, X). It should be noted that H(Y, X) = H(Y) + H(X) if Y and X are independent. In general, if a number of discrete random variables Yi are independent form each other we can have Eq. (8).
Definition 9
The undependability of ith cluster of an ensemble Π (i.e. cluster Ti) with regard to a reference set (usually the reference set is the same ensemble Π) is denoted by \( U\left( {T_{i} ,\varPi } \right) \) obtained according to Eq. (9).
where \( u\left( {T_{i} ,\pi^{j} } \right) \) is calculated based on Eq. (10).
where n is the number of data points and \( T_{i} \cdot \tau_{k}^{j} \) is the inner product of two vectors Ti and τ j k .
When all data points of Ti belong to a same cluster in πj, u(Ti, πj) will hit its minimum at zero and when all data points of Ti equally belong to a all clusters of πj, it will reach its maximum value. We assume that all partitions πj in the ensemble Π are independent and so we averaged the undependability of ith cluster with regard to all partitions πj in Eq. (9).
The dependability of a cluster is in contradiction of its undependability. Its dependability should have the highest value when its undependability has the minimum value (i.e. 0). Its dependability should also have the lowest value when its undependability has the maximum value (i.e. + ∞). As we want dependability values (denoted by R) to be in a certain range, we use an exponential transform.
Definition 10
Given an ensemble Π with B base partitions, the exponential dependability (ED) for a cluster Ti with regards to the ensemble Π is defined according to Eq. (11).
where φ is a positive parameter that controls the effect of the cluster undependability.
Definition 10 describes ED formally. As U(Ti, Π) is a non-negative real value, so ED(Ti, Π) is always a real number in the range (0, 1]. As cluster undependability increases ED value decreases. For all clusters in ensemble of Table 1, the ED values are calculated, and then their ED values are represented in Table 3. The parameter φ has been set to 0.4.
When the undependability of a cluster Ti is its lowest possible value, i.e. U(Ti, Π) = 0, its ED will thereby rises to its maximum, i.e., ED(Ti, Π) = 1. The ED of a cluster \( T_{i} \) tends to its lowest possible value, i.e. \( ED\left( {T_{i} ,\varPi } \right) = 0 \), as its undependability tends to infinity.
By trial and error it has been understood that for very small values of φ, i.e. φ < 0.2, the ED decreases considerably as the cluster undependability increases. It has been also understood that if we use very large values for φ, i.e. φ > 2, the cluster undependability and ED value will be linearly correlated. Empirically, it is suggested that the parameter φ should be set in the interval of [0.2, 1]. We will empirically show latter that the best φ is 0.4.
Definition 11
The normalized undependability (NU) of ith cluster of an ensemble Π (i.e. cluster Ti) with regard to a reference set (usually the reference set is the same ensemble Π) is denoted by \( NU\left( {T_{i} ,\varPi } \right) \) obtained according to Eq. (12).
where cj is the number of clusters in partition πj. Based on Lemma 1, it is straightforward to prove that NU(Ti, Π) is in the interval of [0, 1].
Definition 12
Given an ensemble Π with B base partitions, the normalized dependability (ND) for a cluster Ti with regards to the ensemble \( \varPi \) is defined according to Eq. (14).
Definition 12 describes ND formally. As \( NU\left( {T_{i} ,\varPi } \right) \) is a real number in the interval \( \left[ {0,1} \right] \), so \( ND\left( {T_{i} ,\varPi } \right) \) is always a real number in the same range. As NU value of a cluster increases, its ND value decreases. For all clusters in ensemble of Table 1, the nu values are calculated, then their NU values are calculated, and finally their ND values are represented in Table 4.
Fred and Jain (2005) have introduced the co-association matrix. Its (i, j)th entry shows how many partitions of the ensemble Π put the ith data point and the jth data point in the same cluster.
Definition 13
The co-association matrix of an ensemble Π is denoted by CA and is calculated based on Eq. (15)
where \( \theta_{ij}^{k} \) is defined based on Eq. (16).
There are many efforts in the clustering ensemble literature that have used co-association matrix (Fred and Jain, 2005; Wang, 2011; Yu et al. 2015; Mimaroglu and Erdil, 2011). Although co-association based clustering ensemble efforts are numerous, they almost always suffer from the inability to consider cluster quality or dependability. The NCAI index has recently been used to weigh the base partitions. Then they have made a weighted co-association matrix (Huang et al. 2015). By the way, they have also ignored cluster-wise weighing. In this paper, cluster-wise weighing co-association (CWWCA) matrix is proposed.
Definition 14
Given an ensemble \( \varPi \), the cluster-wise weighing co-association matrix is computed based on Eq. (17).
where \( W_{ij}^{k} \) is defined based on Eq. (18).
where \( \tau_{{\pi_{i}^{k} }}^{k} \) is \( \pi_{i}^{k} \) th cluster of kth partition and \( Dep\left( {\tau_{{\pi_{i}^{k} }}^{k} ,\varPi } \right) \) can be either \( ED\left( {\tau_{{\pi_{i}^{k} }}^{k} ,\varPi } \right) \) or \( ND\left( {\tau_{{\pi_{i}^{k} }}^{k} ,\varPi } \right) \).
We now have a weighting mechanism that sets a weight to each cluster in order to consider the cluster dependability. Through this mechanism a data point that usually falls into a dependable cluster is highly likely to be placed into a correct cluster. Also by the cluster-wise weighing co-association matrix we take into consideration the dependability of clusters. For more details, Table 5 provides CWWCA matrix for the ensemble presented in Table 1. We have used ED(Ti, Π) as cluster dependability during computation of Table 5.
We compute Table 6 like Table 5. We employ ND(Ti, Π) instead of ED(Ti, Π) as cluster dependability during computation of Table 6.
Definition 15
The cluster-wise weighting bi-partite graph (\( CWWBG \)) is defined based on Eq. (19).
where \( {\mathbb{N}}\left( {\varPi ,D} \right) = \left\{ {1,2, \ldots ,n,n + 1, \ldots ,n + b} \right\} \) is the set of nodes and \( {\mathbb{E}}\left( {\varPi ,D} \right) \) is the set of edges and \( b = \sum\nolimits_{i = 1}^{B} {c^{i} } \). The edge weight between two nodes x and y is defined based on Eq. (20).
After creation of cluster-wise weighting bi-partite graph based on Definition 15, we partition the graph using the hyper-graph partitioning algorithm (Li et al. 2012). The nodes {1, 2, …, n} are the representatives of data points {1, 2, …, n}. So, each of them are clustered according to their clusters in the results of the hyper-graph partitioning algorithm.
Cluster-wise weighting bi-partite graph is presented in Table 7 where ND(Ti, Π) is employed as cluster dependability for the ensemble presented in Table 1.
4 Proposed method
The proposed method is a clustering ensemble framework that is based on integration of undependability concept (definition 9 and definition 11) and cluster weighting. The pseudo code of the proposed method is presented in Fig. 1. The inputs of the algorithm are as follows: a real number of cluster denoted by c, a base clustering algorithm alg, a number of base clustering B; a dataset D; ground truth labels or the target partition P; the sampling rate r; the exponential transfer parameter φ. The outputs of the algorithm are as follows: the first output is the consensus partition denoted by πALED where it is obtained by the average linkage as the consensus function and the exponential dependability as the cluster evaluator metric; the second output is the consensus partition denoted by πALND where it is obtained by the average linkage as the consensus function and the normalized dependability as the cluster evaluator metric; the third output is the consensus partition denoted by πGED where it is obtained by applying Tcut algorithm as the consensus function and the exponential dependability as the cluster evaluator metric; the fourth output is the consensus partition denoted by πGND where it is obtained by applying Tcut algorithm as the consensus function and the exponential dependability as the cluster evaluator metric; and NMI values of πALED, πALND, πGED, and πGND are respectively returned in nmiOutALED, nmiOutALND, nmiOutGED, and nmiOutGND.
In line 1, we first initialize SampleSize and size of dataset. In lines 2-8 an ensemble is created. In this ensemble, each base partition is consisted of vc clusters where \( 2 \le vc \le \sqrt {SampleSize} \) and SampleSize is size of each sampling. In each iteration of the for statement, first a sub-sample with the size SampleSize is produced. Then we partition this sub-sample into vc clusters through a simple base clustering algorithm denoted by alg. In the following step the output of the base clustering alg is generalized by applied it on the total dataset resulting in a partition denoted by πi (line 6). Finally in the line 7, the cluster representation of partition πi is constructed according to definition 5. After the loop, the ensemble is created by concatenating B partitions, π1, π2, …, πB. In the next line, i.e. line 10, the cluster representation of ensemble Π is created according to definition 6. Then according to definition 10 and definition 12, ED(Ti, Π) and ND(Ti, Π) are respectively computed for all i ∊ {1, 2, …, b}. Lines 13 and 14 respectively compute CWWCA with ED(Ti, Π) and ND(Ti, Π) as dependability measure. Lines 15 and 16 respectively compute CWWBG with ED(Ti, Π) and ND(Ti, Π) as dependability measure. By applying hierarchical average linkage clustering algorithm (Jain, 2010) on CWWCAED and CWWCAND, we compute consensus partitions πALED and πALND in the lines 17 and 18. In the lines 19 and 20, we obtain consensus partitions πGED and πGND by partitioning the bi-partite graphs CWWBGED and CWWBGND using the Tcut algorithm. In the last four lines of the pseudo code of the proposed approach, we calculate respectively Normalized Mutual Information (NMI), F-Measure (FM), Accuracy (Acc), and Adjacent Rand (AR) of πALED, πALND, πGED, and πGND.
Hierarchical agglomerative clustering as a well-known basic clustering algorithm (Jain, 2010) usually takes a similarity matrix as input and produces a partition as output. If the CWWCAED matrix is employed as the input similarity matrix, partition πALED can be produced as output. If the CWWCAND matrix is employed as the input similarity matrix, partition πALND can be produced as output.
Consensus partitions πALED, πALND, and πGND are presented in Table 8 for the exemplary ensemble presented in Table 8 when φ is 0.4.
5 Experimental study
In this section, the proposed methods have been evaluated versus the state-of-the-art clustering ensemble methods on a diverse real-world collection of datasets. All experiments have been performed in Matlab R2015a 64-bit on a workstation.
In the current section, the proposed methods will be compared to some of the state-of-the-art methods. Some of the methods that are compared to our methods include Hybrid Bi-Partite Graph Formulation (HB-PGF) (Fern and Brodley 2004), Sim-Rank Similarity (S-RS) (Iam-On et al. 2008), Weighted Connected Triple (W-CT) (Iam-On et al. 2011), Cluster Selection Evidence Accumulation Clustering (CS-EAC) (Alizadeh et al. 2014), Weighted Evidence Accumulation Clustering (W-EAC) (Huang et al. 2015), Wisdom of Crowds Ensemble (ECE) (Alizadeh et al. 2015), Graph Partitioning with Multi-Granularity Link Analysis (GPM-GLA) (Huang et al. 2015), and Two_level co-association Matrix Ensemble (TME) (Zhong et al. 2015), Elite Cluster Selection Evidence Accumulation Clustering (ECS-EAC) (Parvin and Minaei-Bidgoli 2015).
5.1 Benchmarks
In our experiments, ninety real-world datasets have been used. Dataset names and their details have been presented in Table 9. All datasets are from UCI machine learning repository (Bache and Lichman 2013) except datasets with the number 5 and 9. Prior doing any experimentation, all datasets have been first normalized so as to any feature in any dataset of the paper mapped into range [0, 1]. It means that before doing any step, a preprocessing step is taken just like Eq. (21).
where \( \dot{d}_{ij} \) is the jth normalized feature in the ith data point.
5.2 Evaluation metrics
We have employed the Normalized Mutual Information (NMI) (Parvin and Minaei-Bidgoli 2015), F-Measure (FM) (Parvin and Minaei-Bidgoli 2015), Accuracy ratio (Acc) (Parvin and Minaei-Bidgoli 2015), and Adjacent Rand Index (ARI) (Azimi and Fern 2009) to evaluate the quality of the partitions obtained whether by the proposed method or by the state-of-the-art methods. A better partition has a larger value of NMI. It also has a larger value of other mentioned metrics. The range of all mentioned metrics belongs to [0,1], i.e. they don’t exceed one and aren’t negative. The larger the NMI, FM, Acc, or ARI, the better the clustering result. To be more precise in our conclusions, any result, reported in Tables or depicted in Figures, is averaged over 30 independent runs.
5.3 Parameter setting
Parameter alg is always simple k-means clustering algorithm. The initialization approach used for k-means is the deterministic Kaufman’s initialization (which was shown to be better than random initializations (Peña et al. 1999), and is expected to be adopted unless otherwise justified). So it will produce the same exact clustering results for any given k value for a given dataset. As we choose k values randomly from 2 to the square root of the number of objects, which will be for the largest dataset, L.R., 141, for the second largest dataset, U., 104, and for the rest of the datasets less than 100. So if instead of choosing random k values, brute-force approach was adopted and all k values were chosen, we will not get 100 different clustering results for any dataset other than the two aforementioned ones. Therefore, one may suggest two mechanisms: (1) adopting different clustering methods in the consensus as that (i) allows for the datasets to be viewed by different methods with different implicit assumptions, which enriches the ensemble and covers the different downfalls of any of the methods, and (ii) increases the number of different clustering results in the pool of results which we were trying to produce, and (2) using resampling methods. We use the latter as we want our method to be as simple and general as possible.
Parameter B is chosen among the set {10, 20, 30, 40, 50, 60, 70, 80, 90, 100}. But its default value is 40. Parameter r is chosen among the set {0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1.0}. The default value for r is 0.9. Parameter φ is chosen among the set {0.1, 0.2, 0.4, 0.6, 0.8, 1, 2, 4, 8}. But its default value is 0.4.
To compare all methods fairly and make a suitable and valid conclusion, we evaluate each method over 30 independent runs. In each run, the ensemble Π is the same for all methods. We finally have presented their averaged performances.
5.4 Parameter φ
The parameter φ has a big influence on the relation of dependability-undependability. When it is small, even a little increment in undependability value results in a considerably reduction in dependability value. Effect of the parameter φ on nmiOutALER and nmiOutGER have been studied respectively in Figs. 2 and 3. As it can be observed from Figs. 2 and 3, setting a value around 0.3 or 0.7 for parameter φ should be recommended; indeed the results in these figures suggest that a value around 0.3 or 0.7 is the best. So we, from here to the end of the paper, use value 0.4 for parameter φ.
5.5 Sampling ratio
The sampling ratio has a big influence on efficacy of the proposed approach. From Fig. 4, it can be inferred that the best value for sampling ratio is a value in the range [0.85, 0.95], that we set 0.9 as the default value for this parameter in the rest of the paper.
5.6 Ensemble size
To examine the best value for parameter B, an experimentation has been conducted and Fig. 5 has been presented. Effect of the parameter B on nmiOutGER has been studied in Fig. 5. As it can be observed from Fig. 5, setting a value around 40 for parameter B should be recommended. So we, from here to the end of the paper, use value 40 for parameter B.
Additionally, we have conducted another experiment to assess the performances of our methods and the state-of-the-art methods in terms of NMI. The final results of this experimentation are reported in Fig. 6. Indeed, we first evaluate a method in terms of NMI in 30 different independent runs on a dataset (it is worthy to be mentioned that in each run the ensemble size is fix and is β), and then the performance averaged over the 30 runs is considered as the performance of the method on that dataset for ensemble size β. After computing performances of that method on all of 19 datasets for ensemble size β, the performance averaged over all of 19 datasets is considered as the performance of that method for ensemble size β. After computing performances for all methods in all different ensemble sizes, we plot the Fig. 6. Indeed, Fig. 6 summarizes the performance of the different methods over the datasets by averaging their NMI values. Figure 7 also summarizes the performance of the different methods over the datasets by averaging their FM values. Indeed, previous experiment has been done for FM index instead of NMI index in Fig. 7.
NMI is not independent of the datasets and therefore is not comparable across them. In other words, averaging NMI across datasets is not very informative. For instance, its values are around 0.2 in the F.C.T. dataset, 0.6 in S. dataset, and 0.8 in O.D.R. dataset. So, an NMI value of 0.5 would not be known if it is good or bad unless it is compared with other methods over the same dataset. So, we should use some normalized form of NMI which would be meaningful while comparing a single method over the datasets. Our approach is to use the methods’ ranks. The method has the rank of 1 at a given dataset if it shows the highest NMI value at that dataset compared with the other methods, etc. Averaging ranks can be meaningful as an averaged rank of 2.5 for example means that, on average, this method scores as the second or the third best method across the datasets. Figure 8 depicts the methods’ ranks on all of 19 datasets for different values of ensemble size. The Figs. 6 and 7 prove that the proposed methods outperform the state-of-the-art in any ensemble size. Figure 8 also approves the results of Figs. 6 and 7. Figure 8 states among the proposed methods, CWWBG based on Definition 15 and using ND(Ti, Π) as dependability measure is the best. So, usage of the normalized cluster dependability as cluster weight can be better than exponential cluster dependability.
5.7 Experimental results
Different numbers of clusters have been employed during obtaining the consensus partition of any different method; and accordingly NMI, FM, Acc, and ARI values are evaluated. Any reported performance is a performance that is averaged over 30 different runs. The averaged performances of the different methods over 30 different independent runs in terms of NMI, are reported in Table 10. It is worthy to be mentioned that for abstracting the results, the two best dominant methods among the four proposed methods are reported here. This two methods are selected based on previous results and are πALND and πGND. Based on the results of Table 10, our proposed methods outperform all of the state-of-the-art methods in almost all datasets in terms of NMI.
The same results of Table 10, have been reproduced in terms of FM, ARI, and Acc respectively in Tables 11, 12, and 13. The averaged performances of the different methods over 30 different independent runs in terms of FM, ARI, and Acc are reported respectively in Tables 11, 12 and 13. Based on the results of Table 11, our proposed methods outperform all of the state-of-the-art methods in almost all datasets in terms of NMI.
According to the results reported in Tables 10, 11, 12 and 13 the proposed method is superior to the state-of-the-art ensemble methods in terms of NMI, FM, ARI, and Acc.
According to results reported in Table 13, our proposed method Cluster-Wise Weighting Bi-Partite Graph with ND as dependability measure outperforms almost all datasets in terms of Acc. In Table 13, the sign “ + ” (or “- ”) indicates that the result of the πGND is meaningfully better (or worse) than the competent method validated by t test with the confidence level 0.95. The sign “ ∼ ” indicates that the result of the πGND is meaninglessly better (or worse) than the competent method validated by t-test with the confidence level 0.95. The last row of the table summarizes the results of the table. The results reported in Table 13 indicate the proposed method completely outperforms the state-of the-art methods. Those reported results have been validated by paired t-test.
All the results presented in Tables 10, 11, 12, and 13 indicate the two following statements:
-
(a)
Usage of our proposed cluster dependability as the cluster weight can improve clustering performance; because not only one consensus function, but two different consensus functions have been employed during results and both almost outperforms the state-of-the-art methods in all benchmarks. It means that enabling our cluster weighting has been effective.
-
(b)
Usage of Tcut should be preferred to the average linkage method.
5.8 Robustness analysis
To study the robustness of all the methods against noise, a number of noisy datasets by adding some synthetic data points generated uniformly distributed in feature space are produced. The number of the added synthetic data points is ϱ percent of the data size (i.e. ϱ * n). It is worthy to be mentioned that ϱ ∊ {0.00, 0.05, 0.10, 0.15, 0.20, 0.25, 0.30, 0.35, 0.40, 0.45}. The NMI results on the mentioned datasets with different noise levels are depicted in Fig. 9. The results depicted in the Fig. 9 are the ones averaged on all 19 datasets, where each result for any dataset is also an averaged over 30 independent runs. According to Fig. 9, our proposed method, Cluster-Wise Weighting Bi-Partite Graph with ND as dependability measure, outperforms is the method with the most robustness against noise.
To sum up, as the experimental results on various datasets illustrated, the proposed methods outperform the state-of-the-art clustering ensemble approaches. It is worthy to be mentioned that our method, Cluster-Wise Weighting Bi-Partite Graph with ND as dependability measure, not only outperform the other methods in terms of NMI, FM, ARI, and Acc, but also it always consumes less time to produce consensus partition. It means in all datasets, the proposed method, i.e. Cluster-Wise Weighting Bi-Partite Graph with ND as dependability measure, has produced consensus partition in much less time. As the number of used datasets is large, and they have different numbers of clusters, it is a fair conclusion that our method is faster than the state-of-the-art clustering ensemble approaches.
6 Conclusion
The paper presents a method for performing consensus clustering by exploiting the estimated uncertainty of clusters. It proposes a clustering ensemble method through a combination of several steps: First, the dependability of each cluster is estimated via an entropy measure and an exponential transformation, which reflects the spread of the cluster among different clusters of a clustering solution. The proposed method builds upon the clustering ensemble paradigm. The multiple clusters are reconciled through the cluster uncertainty. The paper proposes two approaches to achieve this goal: locally weighted evidence accumulation and locally weighted graph partitioning. The former approach is based on hierarchical agglomerative clustering and co-association matrices, while the latter is based on bi-partite graph formulating and partitioning. The proposed approached are then evaluated on 19 real-life datasets. The experimental evaluation reveals that the proposed method has better performance than the competing methods; indeed, wide experimentations on real-world datasets show that our framework outperforms the state-of-the-art.
The main novelty of the paper is the incorporation of the uncertainty concept into the consensus functions. The contributions stated in the paper have been formulated to better reflect them in the paper. The proposed methods are indeed adaptations of some existing concepts for consensus clustering. Furthermore, dependability measure is a derivative of the uncertainty measure. Based on the definitions and intuitively, the proposed uncertainty measure is highly sensitive to the cluster size: It punishes clusters with larger sizes. This is the only drawbacks of the proposed method. So to solve this drawback, we have proposed normalized dependability measure. This measure is less sensitive to the cluster size.
References
Alizadeh H, Minaei-Bidgoli B, Parvin H (2014) To improve the quality of cluster ensembles by selecting a subset of base clusters. J Exp Theor Artif Intell 26(1):127–150
Alizadeh H, Yousefnezhad M, Minaei-Bidgoli B (2015) Wisdom of crowds cluster ensemble. Intell Data Anal 19(3):485–503
Alsaaideh B, Tateishi R, Phong DX, Hoan NT, Al-Hanbali A, Xiulian B (2017) New urban map of Eurasia using MODIS and multi-source geospatial data. Geo-Spat Information Science 20(1):29–38
Azimi J, Fern X (2009) Adaptive cluster ensemble selection. In: Proceedings of IJCAI, pp 992–997
Bache K, Lichman M (2013) UCI machine learning repository [Online]. http://archive.ics.uci.edu/ml
Chakraborty D, Singh S, Dutta D (2017) Segmentation and classification of high spatial resolution images based on Hölder exponents and variance. Geo-spatial Inf Sci 20(1):39–45
Charon I, Denoeud L, Guénoche A, Hudry O (2006) Maximum transfer distance between partitions. J Classif 23(1):103–121
Coretto P, Hennig Ch (2010) A simulation study to compare robust clustering methods based on mixtures. Adv Data Anal Classif 4:111–135
Cristofor D, Simovici D (2002) Finding median partitions using information-theoretical-based genetic algorithms. J Univers Comput Sci 8(2):153–172
Deng Q, Wu S, Wen J, Xu Y (2018) Multi-level image representation for large-scale image-based instance retrieval. CAAI Trans Intell Technol 3(1):33–39
Denoeud L (2008) Transfer distance between partitions. Adv Data Anal Classif 2:279–294
Dueck D (2009) Affinity propagation: clustering data by passing messages, Ph.D. dissertation, University of Toronto
Fern XZ, Brodley CE (2004) Solving cluster ensemble problems by bi-partite graph partitioning. In: Proceedings of international conference on machine learning (ICML)
Franek L, Jiang X (2014) Ensemble clustering by means of clustering embedding in vector spaces. Pattern Recogn 47(2):833–842
Fred ALN, Jain AK (2005) Combining multiple clusterings using evidence accumulation. IEEE Trans Pattern Anal Mach Intell 27(6):835–850
Frey BJ, Dueck D (2007) Clustering by passing messages between data points. Science 315:972–976
García-Escudero LA, Gordaliza A, Matrán C, Mayo-Iscar A (2010) A review of robust clustering methods. Adv Data Anal Classif 4:89–109
Guénoche A (2011) Consensus of partitions: a constructive approach. Adv Data Anal Classif 5:215–229
Hennig B (2008) Dissolution point and isolation robustness: robustness criteria for general cluster analysis methods. J Multivar Anal 99:1154–1176
Huang D, Lai JH, Wang CD (2015) Combining multiple clusterings via crowd agreement estimation and multi-granularity link analysis. Neurocomputing 170:240–250
Iam-On N, Boongoen T, Garrett S (2008) Refining pairwise similarity matrix for cluster ensemble problem with cluster relations. In: Proceedings of international conference on discovery science (ICDS), pp 222–233
Iam-On N, Boongoen T, Garrett S, Price C (2011) A link-based approach to the cluster ensemble problem. IEEE Trans Pattern Anal Mach Intell 33(12):2396–2409
Jain AK (2010) Data clustering: 50 years beyond k-means. Pattern Recogn Lett 31(8):651–666
Kettenring JR (2006) The practice of cluster analysis. J Classif 23:3–30
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2324
Li T, Ding C (2008) Weighted consensus clustering. In: Proceedings of SIAM international conference on data mining (SDM)
Li Z, Wu XM, Chang SF (2012) Segmentation using superpixels: a bi-partite graph partitioning approach. In: Proceedings of IEEE conference on computer vision and pattern recognition (CVPR)
Li C, Zhang Y, Tu W et al (2017a) Soft measurement of wood defects based on LDA feature fusion and compressed sensor images. J For Res 28(6):1285–1292
Li X, Cui G, Dong Y (2017b) Graph regularized non-negative low-rank matrix factorization for image clustering. IEEE Trans Cybern 47(11):3840–3853
Li X, Cui G, Dong Y (2018a) Discriminative and orthogonal subspace constraints-based nonnegative matrix factorization. ACM TIST 9(6):65:1–65:24
Li X, Lu Q, Dong Y, Tao D (2018b) SCE: a manifold regularized set-covering method for data partitioning. IEEE Trans Neural Netw Learn Syst 29(5):1760–1773
Ma J, Jiang X, Gong M (2018) Two-phase clustering algorithm with density exploring distance measure. CAAI Trans Intell Technol 3(1):59–64
Mimaroglu S, Erdil E (2011) Combining multiple clusterings using similarity graph. Pattern Recogn 44(3):694–703
Mirzaei A, Rahmati M, Ahmadi M (2008) A new method for hierarchical clustering combination. Intell Data Anal 12(6):549–571
Ng AY, Jordan MI, Weiss Y (2002) On spectral clustering: Analysis and an algorithm. In: Advances in neural information processing systems (NIPS), pp 849–856
Nguyen TD, Welsch RE (2010) Outlier detection and robust covariance estimation using mathematical programming. Adv Data Anal Classif 4:301–334
Parvin H, Minaei-Bidgoli B (2015) A clustering ensemble framework based on selection of fuzzy weighted clusters in a locally adaptive clustering algorithm. Pattern Anal Appl 18(1):87–112
Peña JM, Lozano JA, Larrañaga P (1999) An empirical comparison of four initialization methods for the K-Means algorithm. Pattern Recogn Lett 20(10):1027–1040
Schynsa M, Haesbroeck G, Critchley F (2010) RelaxMCD: smooth optimisation for the minimum covariance determinant estimator. Comput Stat Data Anal 54:843–857
Song XP, Huang C, Townshend JR (2017) Improving global land cover characterization through data fusion. Geo-Spat Inf Sci 20(2):141–150
Spyrakis F, Benedetti P, Decherchi S, Rocchia W, Cavalli A, Alcaro S, Ortuso F, Baroni M, Cruciani G (2015) A pipeline to enhance ligand virtual screening: integrating molecular dynamics and fingerprints for ligand and proteins. J Chem Inform Model 55(10):2256–2274
Strehl A, Ghosh J (2003) Cluster ensembles: a knowledge reuse framework for combining multiple partitions. J Mach Learn Res 3:583–617
Topchy A, Jain AK, Punch W (2005) Clustering ensembles: models of consensus and weak partitions. IEEE Trans Pattern Anal Mach Intell 27(12):1866–1881
Wang T (2011) CA-Tree: a hierarchical structure for efficient and scalable coassociation-based cluster ensembles. IEEE Trans Syst Man Cybern B Cybern 41(3):686–698
Wang X, Yang C, Zhou J (2009) Clustering aggregation by probability accumulation. Pattern Recogn 42(5):668–675
Wang L, Leckie C, Kotagiri R, Bezdek J (2011) Approximate pairwise clustering for large data sets via sampling plus extension. Pattern Recogn 44(2):222–235
Wang CD, Lai JH, Zhu JY (2012) Graph-based multiprototype competitive learning and its applications. IEEE Trans Syst Man Cybern Part C Appl Rev 42(6):934–946
Wang B, Zhang J, Liu Y, Zou Y (2017) Density peaks clustering based integrate framework for multi-document summarization. CAAI Trans Intell Technol 2(1):26–30
Weiszfeld E, Plastria F (2009) On the point for which the sum of the distances to n given points is minimum. Ann Oper Res 167(1):7–41
Wolpert DH, Macready WG (1996) No free lunch theorems for search. Technical Report. SFI-TR-95-02-010. Citeseer
Wu J, Liu H, Xiong H, Cao J (2013) A theoretic framework of k-means based consensus clustering. In: proceedings of international joint conference on artificial intelligence
Xu L, Krzyzak A, Oja E (1993) Rival penalized competitive learning for clustering analysis, RBF net, and curve detection. IEEE Trans Neural Netw 4(4):636–649
Yu Z, Li L, Gao Y, You J, Liu J, Wong HS, Han G (2014) Hybrid clustering solution selection strategy. Pattern Recogn 47(10):3362–3375
Yu Z, Li L, Liu J, Zhang J, Han G (2015) Adaptive noise immune cluster ensemble using affinity propagation. IEEE Trans Knowl Data Eng 27(12):3176–3189
Zheng X, Zhu S, Gao J, Mamitsuka H (2015) Instance-wise weighted nonnegative matrix factorization for aggregating partitions with locally reliable clusters. In: Proceedings of IJCAI 2015, pp 4091–4097
Zhong C, Yue X, Zhang Z, Lei J (2015) A clustering ensemble: two-level-refined co-association matrix with path-based transformation. Pattern Recogn 48(8):2699–2709
Yang H, Yu L (2017) Feature extraction of wood-hole defects using wavelet-based ultrasonic testing. J For Res 28(2):395–402
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rashidi, F., Nejatian, S., Parvin, H. et al. Diversity based cluster weighting in cluster ensemble: an information theory approach. Artif Intell Rev 52, 1341–1368 (2019). https://doi.org/10.1007/s10462-019-09701-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10462-019-09701-y