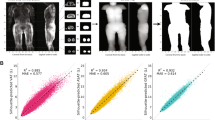
Abstract
Background
There is increasing appreciation of the association of obesity beyond co-morbidities, such as cancers, Type 2 diabetes, hypertension, and stroke to also impact upon the muscle to give rise to sarcopenic obesity. Phenotypic knowledge of obesity is crucial for profiling and management of obesity, as different fat—subcutaneous adipose tissue depots (SAT) and visceral adipose tissue depots (VAT) have various degrees of influence on metabolic syndrome and morbidities. Manual segmentation is time consuming and laborious. Study focuses on the development of a deep learning-based, complete data processing pipeline for MRI-based fat analysis, for large cohort studies which include (1) data augmentation and preprocessing (2) model zoo (3) visualization dashboard, and (4) correction tool, for automated quantification of fat compartments SAT and VAT.
Methods
Our sample comprised 190 healthy community-dwelling older adults from the Geri-LABS study with mean age of 67.85 ± 7.90 years, BMI 23.75 ± 3.65 kg/m2, 132 (69.5%) female, and mainly Chinese ethnicity. 3D-modified Dixon T1-weighted gradient-echo MR images were acquired. Residual global aggregation-based 3D U-Net (RGA-U-Net) and standard 3D U-Net were trained to segment SAT, VAT, superficial and deep subcutaneous adipose tissue depots (SSAT and DSAT). Manual segmentation from 26 subjects was used as ground truth during training. Data augmentations, random bias, noise and ghosting were carried out to increase the number of training datasets to 130. Segmentation accuracy was evaluated using Dice and Hausdorff metrics.
Results
The accuracy of segmentation was SSAT:0.92, DSAT:0.88 and VAT:0.9. Average Hausdorff distance was less than 5 mm. Automated segmentation significantly correlated R2 > 0.99 (p < 0.001) with ground truth for all 3-fat compartments. Predicted volumes were within ± 1.96SD from Bland–Altman analysis.
Conclusions
DL-based, comprehensive SSAT, DSAT, and VAT analysis tool showed high accuracy and reproducibility and provided a comprehensive fat compartment composition analysis and visualization in less than 10 s.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
World Health Organization defines obesity or overweight as abnormal body fat accumulation, which is harmful to health. Improper diet, lack of physical activities, or heredity lead to obesity [1]. Globally, obesity is growing rapidly, about 15% of the world population (adults and children in developing and underdeveloped countries) is obese or overweight [2]. Obesity-related chronic condition management can be expensive, annually US healthcare spends about 21% (~ 190 billion) of its expenditure on obesity-related illnesses. Overweight and obesity are associated with a wide range of co-morbidities, such as cancers, type 2 diabetes, hypertension, stroke, Heart Failure, depression, sleep disturbances, renal failure, asthma, chronic back pain, osteoarthritis, pulmonary embolism, gallbladder disease, and increased risk of disability. Obesity is considered the world’s most preventable leading cause of death, accounting for more than three million deaths worldwide annually [3] and 14% of premature deaths in Europe [4].
In the recent years, there is increasing attention about the geriatric syndrome of sarcopenic obesity, whereby synergistic complications from both sarcopenia and obesity lead to negative health impacts such as loss of independence, disability, reduced quality of life, and increased mortality [5]. With the increasing prevalence of obesity and its tremendous health effects, it is important to invest time and effort into the research of obesity and its associated diseases to develop better care and prevention [6, 8]. Worldwide there are many clinical trials, such as GENYAL (prevention of obesity in childhood); SWITCH (community-based obesity prevention trial); and clinical trials exploring exercise intervention on obese women Similarly, in Singapore, pertinent studies include SAMS (Singapore adult Metabolism Study) [6] where it was identified that different body fat compartments have varied influence in metabolic syndrome as well differential body fat partitioning and abnormalities in muscle insulin signaling associated with higher levels of adiposity [7]; and GUSTO (Growing towards a healthy outcome in Singapore) [8] showed association of early life weight gain with abdominal fat compartments in different sex, ethnicity [9], and Longitudinal Assessment of Biomarkers for characterization of early Sarcopenia and predicting frailty and functional decline in community-dwelling Asian older adults Study” (Geri-LABS) which highlighted the deleterious impact of sarcopenic obesity on muscle performance [5]. Many countries, such as Singapore have declared war on obesity. Understanding the phenotypes of obesity is crucial for the risk profiling and management of the condition.
Advanced cross-sectional imaging like Computed Tomography (CT) and Magnetic resonance (MRI) are now part of large cohort studies, allows in vivo visualization and quantification of fat compartments, helps in monitoring interventional changes, and characterization of inter- and intrasubject differences. Anatomically, abdominal fat depots are broadly classified as Subcutaneous Adipose Tissue (SAT) and Visceral Adipose Tissue (VAT) depots. SAT and VAT depots are the two major anatomic distributions with unique anatomic, metabolic, or endocrine properties. SAT region is defined as the region that is superficial to the abdominal wall musculature, whereas visceral fat is deep to the muscular wall and includes the mesenteric, subperitoneal, and retroperitoneal components.
Abdominal SAT is further subdivided into superficial and deep SAT (SSAT and DSAT) separated by a fascial plane (fascia superficialis). SAT segmentation is easier than VAT as it is a continuous region and is enclosed between the internal and external abdominal boundaries, whereas VAT is distributed around the organs, and is discontinuous (with small and large regions).
Literature indicates that each fat compartment has different risk profiles for obesity-related comorbidities [10]. To understand the influence of each fat compartment on the body, accurate quantification of abdominal fat regions, such as VAT, DSAT, and SSAT becomes essential. Large cohort studies generate a large number of imaging datasets, and the time needed for quantitative analysis data increases accordingly. Labor-intensive manual segmentation and intra-/interobserver variability are other common pain points. Manual quantification by an expert will be accurate and; therefore, ideal, but with large datasets, this becomes impractical and expensive. There is a need for an accurate, precise, robust, automated, or at least, a semi-automated framework that performs segmentation and quantification in a timely and consistent fashion. Various methods listed in the literature for abdominal fat compartments (like SAT and VAT) segmentation are based on the fuzzy clustering [11, 12], morphology [13], registration [14, 15], deformable model [16], and graph cuts [17] have limited scalability options. Advancements in deep learning-based methods [18] have brought feasibility for quantification of fat compartments either using 2D/3D CT or MR images. Estrada and colleagues [19], proposed FATSegNet using 2D-competitive dense fully convolutional networks (CDFNet), segmenting the images in axial, coronal, and sagittal planes and reported an accuracy of 97% for SAT and 82% for VAT in 641 subjects from Rhineland Study, where only 38 datasets were annotated by the experts. Most literature reports a new segmentation technique for SAT and VAT, but not for all the 3 fat compartments (SSAT, DSAT, and VAT). In addition, a comprehensive tool that (1) accurately segments SAT (SSAT and DSAT) and VAT, rapidly with high reproducibility; (2) visualizes data and statistics; and (3) corrects for errors, is lacking.
Study proposition
In this paper, we propose a Residual Global Aggregation-based 3D U-Net (RGA-U-Net) for segmentation of fat compartments (SSAT, DSAT, and VAT) and evaluate its performance and suitability for automated analysis in future large cohort studies.
In addition, we built a Comprehensive Abdominal Fat (Analysis) Tool (CAFT) around the proposed deep learning algorithm that includes:
-
Deep learning model zoo: (1) a standard U-Net (2) RGA-U-Net;
-
Dashboard for the data presentation and analytics, which allows automatic lumbar-based quantification and analysis—3D visualization, percentage analysis of the whole abdomen (volumetric) or lumbar level-based (cross-sectional area);
-
Interactive correction tool that allows manual editing of contours (between background and outer abdominal boundary, between SSAT and DSAT—correction of Fascia Superficialis, and inner abdominal boundary which separates SAT and VAT) for correction of segmentation errors.
which facilitates expanding the repository of models, allows correction of the results, and presents a complete visualization of fat depots quantification.
Materials and methods
MR data acquisition
The “Longitudinal Assessment of Biomarkers for characterization of early Sarcopenia and predicting frailty and functional decline in community-dwelling Asian older adults Study” (Geri-LABS) is a prospective cohort study involving cognitively intact and functionally independent adults aged 50 years and older residing within the community [20]. We acquired 190 abdominal MRI scans from these subjects using a 3 T MRI scanner (Siemens Magnetom Trio, Germany) and a 6-channel body matrix external phased-array coil. Written consent was obtained by all subjects and the study was reviewed and approved by the National Healthcare Group institutional review board. The cohort's mean age was 67.85 ± 7.90 years, BMI 23.75 ± 3.65 kg/m2, and predominantly female (69.5%) and Chinese (91.6%) in ethnicity. Common comorbidities, include hypertension (46.8%), hyperlipidemia (65.8%), and Type II diabetic mellitus (21.1%).
3D-modified Dixon T1-weighted gradient-echo images (dual-echo VIBE with T2* correction) images were acquired for each patient. Axial HASTE was also acquired as a routine structural scan in the study, but the images from this sequence were not used for any computations. Scans of the abdomen and pelvis spanning the diaphragm to the perineum were acquired. Each pulse sequence was completed in a single breath hold of 20 s, with subjects in supine position and arms placed at the sides. Selected images of the abdomen between L1-L5 and T10-L3 levels of the lumbar spine were extracted for analysis based on which regions were imaged.
In each patient, 60–80 axial slices in the abdominal region, 20–30 in thoracic–abdominal cavity of 5 mm slice thickness, with no interslice gaps, and 1.56 × 1.56 mm in-plane resolution was acquired. For the T1-weighted sequence, settings were: TR 6.62 ms, TE 1.225 ms, FA 10o, and acquisition bandwidth 849 Hz/pixel respectively. Water-only and fat-only images were generated by a linear combination of in-phase and out-phase images. Fat–water swap distortions in the acquired images were corrected during the reconstruction processes. The dataset had a mix of different age groups, fat-mass, thoracic, and lumbar spine regions, and variations in dimensions. Out of the 190 datasets, only 26 datasets had manually drawn ground truth which was considered for model training. The data augmentation was performed using 26 original datasets to increase the total number to 130 datasets, as described in the data augmentation section.
Data augmentation
MR data acquisition relevant data augmentation was performed on the fly using the Torchio library [21]. Four different data augmentations performed were (1) RandomBiasfield artifacts—generated by randomly changing the intensity of very low frequency across the whole image (2) Blur artifacts- using random-sized Gaussian filter and varying standard deviations (3) Random Ghosting artifacts—along the phase-encode direction, modeled by choosing the number of ghosts in the image (4) Random Noise—by adding Gaussian noise under normal distribution with random mean and standard derivation. Data augmentation provided a total number of 130 datasets = 26 (original) + 4 (augmentations) × 26. The datasets were randomly (blinded) divided into training (~ 80%) and testing (~ 20%) datasets, i.e., 104 for training/validation and 26 for testing. Further 104 datasets were divided for training and validation using a ratio of 80: 20 [22]. In our study, data augmentation was done only once before the training, whereas the framework provides option for augmentation on the fly in each epoch of training. Care was taken to ensure that datasets used for testing were not included in the training. From each subject about 7000 patches were generated and in total, we had about 700 K patches from the thoracic and lumbar regions for training the model.
Data segmentation
Fat compartments, segmentation was performed in 3 stages: Preprocessing—removal of imaged arms and other nonabdominal/thoracic regions; and data augmentation; Segmentation—using 3D U-Net or RGA-U-Net for SAT/VAT (two class), and SSAT/DSAT/VAT (three class) region segmentation, and Postprocessing—for spine positions based quantification of fat compartments.
Preprocessing
Quality assurance was performed to make sure that every scan is free of artifacts like fat–water swaps, and motion artifacts from heavy breathing or patient movements. The number of slices in each training dataset was matched with the marked ground-truth slices and the extra slices were removed from the original datasets. Arm regions-based artifacts were automatically removed using the projection-based method, with morphological and connected-component analysis (supplementary notes and figures).
Network models
As a proof-of-concept, we have used (1) 3D U-Net and (2) modified U-Net with residual global aggregation for global information fusion. These modifications overcome the limitations of U-Net and avoid an increase of depth which is computationally expensive with delayed convergence [23].
U-Net
A popular semantic segmentation network has convolutional layers similar to FCN [24] and SegNet [25]. U-Net [26] has symmetric architecture where the encoder extracts spatial image features and decoder reconstructs output segmentation map. The encoder uses a convolutional network, i.e., a sequence of MxM convolutions, followed by max-pooling operation with stride parameters. A convolutional sequence is generally repeated four times, with filter size doubled after each down-sampling, and the output of the encoding section’s fully connected layer connected to the input of the decoder at the same level. The decoding section up samples the feature map using transposed convolution [27]. In the last layer, a 1 × 1 convolution operation is performed to generate the final semantic segmentation map. At each convolutional layer ReLU (rectified linear unit) is used as the activation function [28], except at the final one where the sigmoid activation function is employed. U-Net uses skip connections in all the levels that allow the network to retrieve the spatial information lost due to pooling operations. In our study, the standard 3D-UNet available in Tensorflow was considered as the first model to test its efficacy for the segmentation of different abdominal fat depots. Recently, a PyTorch implementation based on the state-of-the-art NN-UNet has been proposed and tested on diversified biomedical opensource datasets [29] (Fig. 1).
Residual global aggregation (RGA-U-Net)
We employed a modification of 3D U-Net with residual global aggregation that allows attention on targeted fat compartments, fascial plane, and its varying shapes and sizes to improve semantic segmentation. Short-range residual connections (skip connections with summation) were introduced in encoding as in ResNet [30] which facilitates better performance. Attention modules act like an up-sampling residual connection ensuring relevant spatial information to be brought across in the skip connection and reduced the number of redundant filters in the network. Residual Global Aggregation U-Net (RGA-U-Net) architecture used in the study is illustrated in Fig. 2, consists of a regular residual block with two consecutive 3 × 3 × 3 convolutions with a stride of 1, along with batch normalization [31] and Rectified Linear Units (ReLU) [32]. The residual block functions as an input block, followed by down-sampling blocks with increased filter size to extract spatial information at each convolutional layer. The latent space at the end of the encoder network contains fully connected features which are transferred to the decoder network. The decoder network uses three up-sampling blocks [33] followed by an output 1 × 1 × 1 convolution block with a stride of 1, output dropout rate [34] of 0.5 before the final 1 × 1 × 1 convolution, and a weight decay [35] of \(2{\text{e}}^{ - 6}\). Each up-sampling decoder block (Fig. 3) contains enriched feature maps with prominent input features, due to the global aggregation of information. Gating signal, \({\varvec{g}}\), is created using feature maps, which are used as a reference while pruning irrelevant feature responses. The low-level feature map from the decoder path \({{\varvec{x}}}^{{\varvec{l}}},\) undergoes a stride-based convolution to match the dimensions of \({\varvec{g}}\). The two signals are summed elementwise, where relevant weights found in both signals become accentuated. The output is passed through a RELU layer and undergoes a 1 × 1 × 1 convolution to have the same number of feature maps as \({{\varvec{x}}}^{{\varvec{l}}}.\) The output is then passed through a sigmoid layer to generate attention coefficients \({\alpha }_{i}\in \left[0, 1\right]\), where relevant weights in filters contain larger attention coefficients. These coefficients are up-sampled through trilinear interpolation and multiplied element-wise with the original signal \({{\varvec{x}}}^{{\varvec{l}}}\) to scale \({{\varvec{x}}}^{{\varvec{l}}}\) and retain only relevant feature maps. This technique is known as additive attention. Since SSAT and DSAT have broken fascia superficialis in some slices with no clear boundary, incorporating self-attention prevents the network from creating too many false positives in the separation of SSAT, DSAT, and VAT.
Network training
Fat-only image (single contrast) was used as input, which was randomly cropped into 16 × 16 × 16 patches to have sufficient training data. 1500 epochs, batch size of 16, and ADAM optimizer [36] with a learning rate of 1e−3 was used for the gradient-descent algorithm with cross-entropy loss function [37]. To avoid overfitting, weighted decay of 2e−6 and a dropout rate of 0.5 were employed to train the model with patience, i.e., the number of epochs to wait before early stop if no progress on the validation was 5 epochs. The neural network was trained on NVIDIA GPU Titan X 24 GB with 128 GB RAM in Ubuntu-18.04 LTS, using python-3.6 and Tensorflow-2.2 [38] with the on-premises computing device. The patch size was based on the average number of slices available in the datasets. Most subjects had about 60 slices whereas some of the subjects had about 30 slices in their thoracic–lumbar region scans. Hence, we decided to consider a 16 × 16 × 16 patch size with an overlap of 8. In addition, we considered balancing between discontinuous smaller VAT regions and continuous SAT regions representations in patch size. Different batch sizes (16, 32, 64) were evaluated over 250 epochs for their computation time, accuracy, and entropy loss before putting the model for full training. We found the batch size of 16 was most efficient in terms of accuracy, time, and entropy loss and hence selected it for full model training.
Ground-truth generation
Ground truth is important, especially for supervised learning. However, the generation of ground truth with enough abdominal/thoracic scans becomes laborious. Hence, we combined a semi-supervised method for ground-truth generation. We selected datasets from 50 subjects out of datasets the total cohort of 190 subjects based on BMI (low-, medium-, and high-fat subjects) and visual inspection of different fat compartments. From the 3 different groups, we further selected 26 of them with almost equal representations from low-fat, medium-fat, and high-fat groups. Ground truth was established by manually segmenting the boundaries of the fat compartments by a trained technician (C.W.X), and reviewed by an experienced abdominal radiologist (C.H.T.). For VAT, we included omental, mesenteric, and retroperitoneal fat. Small depots of intermuscular fat within the psoas and abdominal wall musculature were disregarded. Using the ground truth, we calculated the total fat volume (TFV) and average SSAT, DSAT, and VAT per slice to distribute them into different groups, i.e., SSAT + DSAT + VAT and classified as low if TFV < 3000 cc; medium 3000 ≥ TFV < 6000; and high if TFV ≥ 6000 cc, respectively. The in-house tool allowed clinicians (with experience greater than 10 years) to correct and draw the fat compartments that were saved as ground truths [39]. The training set had the following characteristics:
-
Age matched to the study data: 69.42 ± 6.82
-
Gender matched: M: 8 and F: 18
-
BMI—23.92 ± 5.78
-
Anatomy: Thoracic and abdominal regions and their mix.
-
Low BMI data sets—6; Medium: 9 and High: 11 proportional to the total cohort.
-
Good mix of clean and artifact images of various kind—bias field inhomogeneities, motion artifacts, skin folding etc. (Fig. 6A–D).
Postprocessing: SPINE disc-based fat compartment analysis
Discs were segmented using the sagittal plane image by thresholding, morphological operations, and connected component analysis (Fig. 3) to automatically localize and associate the fat regions to disc-based regions (refer to supplementary notes for the algorithm).
Evaluation metrics
Multiclass Dice ratio (DR) and 3D-Hausdorff distance (3D-HD) and averaged Hausdorff distance (AVD) [40] were used for evaluating segmentation/classification results at two levels—(1) Total fat region and (2) individual subregions – C1:SSAT, C2:DSAT, and C3:VAT. Binary masks of the Total-fat region and class-based subregions were generated for evaluation metric computations. Fat sub-region volumetric analysis \(Vr\) was computed using Eq. 1,
where \({\mathrm{TP}}_{\mathrm{sat}}\) represent predicted voxel count of C1, \({\mathrm{TP}}_{\mathrm{dsat}}\) for C2, \({\mathrm{TP}}_{\mathrm{vat}}\) for C3, and \({V}_{\mathrm{xyz}}\) represent voxel resolution of each subject as shown in Table 1.
Percentage subregion volumes \(\% V\mathrm{c}\) was calculated using Eq. 2, where \(\mathrm{TPv}\) is the true positive volume of a class and \(\sum \mathrm{TPi}\) is the total volume of the fat region.
Dashboard and visualization
Clinical management and large cohort studies need a dashboard for data analysis and visualization. The proposed dashboard (Fig. 4) populates the whole abdomen, and lumbar position-based fat distribution information required for a clinician, or a researcher which is useful in investigating fat depots, analyze their possible effects, understand the effects of interventions on fat compartments, etc.
Fat segmentation and analysis tool dashboard with its features (a) Plots of slice-based volume analysis and fat percentage calculation. b Slice-wise SSAT, DSAT, and VAT depot volume analysis. c Spine disc and inter disc-based volume analysis of SSAT, DSAT, VAT, and Total fat depots. d Fat percentage-based analysis for Spine disc- and inter disc-based SSAT, DSAT, VAT, and Total fat depots. e 2D and 3D Visualization of SSAT, DSAT and VAT depots
The tool developed using python can be deployed on any platform, has the following features (Fig. 4B).
-
Visualization of different fat regions—SSAT, DSAT and VAT—2D slice-wise and 3D volume
-
Total fat Analysis—Profile, Percentage and Volume
-
Subregion fat analysis—slice-based distribution and profile stats
-
Spine position-based fat volume and percentage analysis.
-
Calculations of BMI and Waist to hip ratio (WHR).
Correction tool
The correction tool enables interactive correction, of the segmentation results, by the user. The correction window allows the user to select the boundary to edit from the set of detected SAT-VAT/SSAT-DSAT/Background—outer abdominal wall boundaries. The edges of each compartment are converted as editable contours by placing 40 points evenly on the edges as shown in Fig. 5. These points can be manipulated (drag, add, delete) to correct the inaccurate segmentation regions. This feature is only enabled for those slices that are already segmented. In the case of VAT depot correction, the inner abdominal wall contour is used as a mask to exclude the SAT depots. A painting tool is created to paint the discontinuous regions of the VAT depot.
Results
Multiclass fat compartment quantification plays an important role in the evaluation of different fat depots and their influence on various conditions like metabolic syndrome, obesity, cardiovascular risks, etc. Both 3D U-Net and RGA-U-Net performed accurate segmentation and quantification of total abdominal fat and individual fat compartments, i.e., Superficial-SAT (C1), Deep-SAT (C2), and Visceral fat (C3). Training and testing dice indices and Hausdorff distance metrics (Mean ± SD) for two-class (SAT and VAT) and three class (SSAT, DSAT, and VAT) are described in Table 1.
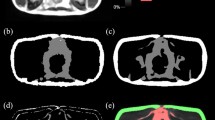
Figure 6A illustrates the variability in datasets (low- to high-fat, multiple fasciae, skin folding, bias field variations, discontinuous fascia, etc.) used in the study and its predicted results along with the ground truth. Figure 6B, shows a comparison of predicted results and ground truth from a few sample datasets (low-t, medium-, and high-fat subjects). Figure 6C, illustrates the learning process of standard 3D U-Net and RGA-U-Net during the training of the models. During the initial iterations, RGA-U-Net functions like a 2-class classifier segmenting the SAT and VAT and later builds on segmenting SAT into SSAT and DSAT, whereas the standard U-Net functions as a 3-class classification from the initial iterations. Figure 6D, showcases the misclassification and under segmentation examples in low-, medium-, and high fat subjects.
A Illustrates the dataset variability of the training and testing cohort used along with the comparison of predicted results against the ground truth. a Bias field artifact, b low-fat subject having discontinuous fascia, c skin folding, and movement artifact, d an example of multiple fasciae, e low contrast discontinuous fascia. B Shows comparison of predicted results and ground truth from a few selected sample datasets of low-, medium-, and high-fat subjects. C Illustrates the comparison of U-Net and RGA-U-Net’s training phase at different epochs. The figure demonstrates the differences in the learning process of different fat compartments like SSAT, DSAT, and VAT regions. RGA-U-Net excludes spine region and inter-disc regions whereas U-Net seems to classify some regions of the spine as VAT. D. Showcases the examples of misclassification, and under segmentation of DSAT and SSAT. The top two rows correspond to low-fat subjects, the third and fourth rows correspond to medium-fat subjects while the last two rows correspond to high-fat subjects respectively
Box plots (Supplementary figure S2) indicate an accuracy of segmentation in original and augmented datasets by both network models, which reinforces that the networks were good at generalization and efficiently handles data variability. Further, it emphasizes that the proposed RGA-U-Net network had better accuracy across subject categories and for SSAT, DSAT, and VAT than U-Net. We observed (Fig. 7) varied distribution of fat compartments (SSAT, DSAT, and VAT) in low-, medium- and high-fat subjects.
Agreement and responsiveness of the method with ground truth were evaluated using concordance correlation analysis, correlation coefficient [41], and Bland–Altman analysis (Fig. 8A). Correlation studies illustrate the relationship between segmentation and ground truth, but not their differences, whereas Bland–Altman analysis, based on the evaluation of agreement between two quantitative measurements using the mean difference and limits of agreement helps to understand the differences.
A Correlation analysis of segmentation result with ground truth for U-Net and RGA-U-Net predicted segmentation volumes. Graphs indicate a good correlation for all the fat compartments though there is under segmentation of VAT by U-Net. B Bland–Altman plots analyzing the agreement/mismatch between the ground truth and segmentation for the training datasets. It is evident from the graph that U-Net had under-segmentation for all the fat compartments whereas RGA-U-Net shows better accuracies
This technique is useful to evaluate bias of the network; estimate agreement interval; and identity possible over-and under-estimation (Fig. 8B). Our analysis shows DSAT is underestimated in U-Net whereas RGA-U-Net performs well. Both network models under-segmented the SSAT region, pointing to the error in the identification of the right facial plane from multiple fasciae. In VAT segmentation RGA-U-Net outperforms U-Net which produces under-segmentation especially in smaller regions near the pelvic area and near the spine. The correlation coefficients for U-Net were 0.9933, 0.9908, 0.9780, and 0.9875 for SSAT, DSAT, VAT, and Total fat, respectively. Similarly, RGA-U-Net had 0.9933, 0.9963, 0.9972, 0.990 for SSAT, DSAT, VAT, and Total fat, respectively indicating that RGA-U-Net had a better correlation with GT for all the fat compartments as compared to U-Net. RGA-U-Net performed well especially in VAT segmentation as compared to U-Net. Correlation plots indicate RGA-U-Net was good at generalization and adaption to data variability. Bland–Altman plots indicate the performance of both networks was consistent across low to high-fat volumes even though some low-fat volumes had higher errors in SSAT and DSAT separation.
Discussion
General obesity is caused due to excess accumulation of body fat, and abdominal obesity is known to have strong influences on metabolic syndrome and other morbidities. Accurate segmentation of different fat depots; subcutaneous (SAT) and visceral adipose tissue depots (VAT), and superficial (SSAT) and deep (DSAT) subcutaneous adipose tissue depots from cross-sectional imaging is essential to understand the clinical impact in patients. Single slice-based analysis of the fat compartments at a specific lumbar position (e.g. L2–L3 or L3–L4) is practical and has been suggested by prior studies [42,43,44] to correlate with associated clinical conditions. However, due to heterogeneity of patient body habitus, the optimal level for analysis could theoretically vary. Thomas et al. [45] indicated that uncertainty of prediction or correlation increases with a reduction in the number of slices used to quantify adipose tissue depots. We believe that volumetric analysis of a large segment of the abdomen could achieve a better correlation. However, manual quantification on a large scale would be nearly impossible without automated or at least, a semi-automated technique. In this study, we have proposed a Residual global aggregation-based 3D U-Net (RGA-U-Net) for segmentation and validated CAFT as a comprehensive tool that deploys a deep learning RGA-U-Net algorithm that reliably segments and quantifies fat compartments (SSAT, DSAT, and VAT) using abdominal MR images. Our algorithm takes less than 10 s for simultaneous quantification of all the 3 fat compartments in the volume data, making it feasible for use in large clinical trials, and foreseeably, clinical routine.
Segmentation
The proposed framework of Comprehensive Fat Analysis tool (CAFT) is built with 5 components—(1) Preprocessing (2) Data Augmentation (3) Neural Network model zoo containing a standard 3D U-Net and Residual Global Aggregation U-Net (RGA-U-Net)-based segmentation models which can be extended to any number of models (4) Dashboard and visualization for data presentation and analysis, and (5) Editing tool to correct the contours of segmentation.
Pre-processing improved the accuracy of segmentation since the arms contain SAT, and being similar in contrast to the abdominal fat, would have interfered with automated segmentation. Furthermore, an error would occur if the arms were imaged to be abutting the abdomen. Concurrently, our postprocessing method localized the spine and individual lumbar discs to build correspondence between data slices to anatomy. By identifying the correspondences, we were able to aggregate the lumbar-based segmentation stats for the visualization. This automatic processing eliminated the need for manual aggregation and computation of fat volumes, contributing to improved efficiency and accuracy. Importantly, we found high accuracy of our technique, in comparison to the ground truth (manual segmentation by our human readers).
The multilayer attention and global aggregation module at each level of U-Net architecture help in the consolidation and merging of attention features at each level. Attention modules captured important features (fascia boundary, smaller VAT components around spine without including spine or its discs, Fig. 6C) at different resolutions and Residual connection blocks facilitated the improved separation of SSAT and DSAT, which is physically separated by a thin fascia superficialis that is not visible on some slices. Localization of right fascial separation was the most difficult aspect in the segmentation of SSAT and DSAT where RGA-U-Net excels. RGA-U-Net starts with a 2-class model of SAT and VAT and during the later iterations divides the SAT into SSAT and DSAT, whereas standard U-Net starts with 3 class-based classifications from the initial iterations itself (Fig. 6C). We observed a higher initial error in RGA-U-Net than U-Net as it starts with a 2-class classification in the initial stages. The error decreases exponentially in the later phase of training and achieves faster convergence. Further, RGA-U-Net is fast and can be deployed in a low-end computation system as the inference is patch-based which reduces the computational time.
Our results show reasonably accurate quantification in both 2-class and 3-class-based segmentations. The mean dice coefficient was about 95% for total fat (sum of VAT and SAT) and greater than 90% for SAT and VAT (Table 1). For segmenting between SSAT and DSAT, the accuracies as compared to our ground truth were around 91% and 89% respectively. RGA-U-Net had greater than 94% accuracy in distinguishing between fat and nonfat tissues and was accurate in differentiating between bone and fat especially in the spine and pelvic regions, where the bone contours can be complex. Average Hausdorff distance for RGA-U-Net was marginally better for 2- class (SAT and VAT) segmentation whereas it was significant for 3-class (SSAT, DSAT and VAT) when compared to standard U-Net. RGA-U-Net proved its worth in VAT segmentation (Table 1) where the spine and its disc regions were not segmented as shown in Fig. 6C. In lean or low-fat subjects, a fascial plane could not be observed between the SSAT and DSAT near L1 and L2 regions. In such patients, RGA-U-Net was superior to U-Net, which tends to “under-segment” all fat compartments. Nevertheless, both network models had comparably high accuracies for original and augmented dataset segmentation (Fig. 7), bearing testament to their abilities to generalize and handle data variability across patients with diverse body habitus. Correlation analysis and Bland–Altman plots analysis (Fig. 8A, B) show a high agreement between the ground truth and segmentation for the training datasets. Some intermuscular fat regions that are closer to pelvic bone is being segmented as VAT by both models. These false positives did not significantly contribute to the DICE statistics (Figs. 5, 6D). In some cases, we observed part of pelvic bone considered as VAT due to the presence of bone fat and intermuscular fat (Fig. 6D). In some cases, we observed some false positives (DSAT classified as VAT) especially in the pelvic cavity and in cases where the intermuscular fat is closer to the inner abdominal boundary.
Our study data was retrospectively derived from the GeriLabs cohort study and only one acquisition per pulse sequence was performed in each MRI study. Hence, image technical reproducibility could not be evaluated. We addressed reproducibility by augmenting each subject’s data with MR acquisitional variations to simulate variations in practice. The dice scores of the augmented subject data exhibited good consistency (Supplementary Figure S2).
Visualization
Different patterns in fat compartment distribution were observed across low-, medium- and high-fat subjects. In the low-fat subjects, the volume difference between SSAT and DSAT is significantly high, whereas there is no significant volume difference between SSAT and VAT (Fig. 7). The SSAT and DSAT volume difference start reducing with increasing fat accumulation (medium- and high-fat subjects). VAT volume increases linearly as obesity increases. DSAT volume seems to be increasing more than SSAT volume across different groups. Such insights will be useful to monitor the progress of nutritional or exercise intervention programs that target obese older adults [46].
We observed that the SAT accumulation profile generally changed as we move from the thoracic to lumbar regions for every patient. In some regions (like L1 and L2) we noticed an equal quantity of SSAT and DSAT, often with a prominent fascia superficialis, whereas progressing more caudally towards L5, there were multiple fascial lines whereby some appeared discontinuous. This was more pronounced in our older adult cohort dataset due to skin folds and loosely bound fat compartments. Identifying a single boundary between SSAT and DSAT at the lower lumbar levels can be challenging even for radiologists, raising susceptibility to errors in delineating and consequently, quantifying the fat compartments.
Assumptions and limitations
In the study, we assumed that the MR scans are acquired in a standardized manner with proper placement of field-off view; no swap of fat and water pixels (in Dixon sequence); arms are at a distance from the trunk; low field inhomogeneities, etc. We also consider that the ground truths are drawn by the clinician as a gold standard for training the model. Care was taken to include datasets with variability in fat quantity, fat profiles, and body types (low-, medium-, and high fat), data from young adults and elderly, male, and female, data from different anatomical locations (lumbar and thorax), slice thickness, data dimensions, MR acquisitional variations like bias field, ghosting, blur, and random noise to avoid any possible biases, improve the segmentation performance and generalization.
We aim to further expand our datasets in subsequent studies to include more variability in terms of subjects and data acquisition, such as usage of multiple contrasts (in-phase, out-phase, etc.), extending to other MR sequences, and training on pediatric datasets. The cause for the over-estimation of VAT was due to imaging errors (errors in reconstruction due to some fat–water pixel swaps). Over-estimation of DSAT and under-estimation of SSAT were due to multiple boundaries created by different fasciae like fascia superficialis, deep fascia, skin ligaments, and fascia of the obturator internus. While drawing the ground truth, the clinicians use their knowledge and experience to draw a contiguous boundary, whereas in applications like patch-based deep learning architectures, since the whole anatomy information is lost (to use causality conditions), the learning methods instead rely on locally available information for segmentation which could be erroneous (Fig. 6D).
Conclusion
In this study, we propose a comprehensive deep-learning RGA-U-Net-based tool (complete processing pipeline) along with other features like data augmentation on the fly, pre-processing, automatic whole abdomen (volumetric) or lumbar level-based (cross-sectional area) fat quantification, automatic spine segmentation, 2D and 3D visualizations, and correction tool which are essential for large cohort studies. Our framework for abdominal fat compartments segmentation (SAT-SSAT and DSAT, VAT, Total Fat), demonstrated that the deep learning model is highly accurate and takes just about 10 s (using standard computational hardware) to segment data containing about 80 slices. The editing module allows easy navigation and manipulation of the contours across the data and corrects the errors in segmentation to aid in continuous learning. The model trained with a large number of patches and with high variability data (low-, medium-, and high-fat volume subjects, from different regions, from young to elderly subjects, etc.) is scalable, deployable, and useful for large cohort studies. The proposed framework alleviates laborious manual segmentation and saves precious time of clinicians and money.
Availability of data and material
Data cannot be shared due to National policy.
Code availability
Can be made available based on the approval from the funding institute.
Change history
04 September 2021
A Correction to this paper has been published: https://doi.org/10.1007/s10334-021-00956-7
References
World Health Organization (2016) World Health Organization. http://www.who.int/mediacentre/factsheets/fs311/en/
Murray CJL (2021) Institute for health metrics and evaluation. http://www.healthdata.org/news-release/nearly-one-third-world%E2%80%99s-population-obese-or-overweight-new-data-show
Djalalinia S, Qorbani M, Peykari N, Kelishadi R (2015) Health impacts of obesity. Pakistan J Med Sci 31:239–242
U.S National Library—The World’s Largest Medical Library (2016) https://www.ncbi.nlm.nih.gov/pubmedhealth/behindtheheadlines/news/2016-07-14-obesity-now-a-leading-cause-of-death-especially-in-men/. Accessed 14 Jul 2016
Khor EQE, Lim JP, Tay L, Yeo A, Yew S, Ding YY, Lim WS (2020) Obesity definitions in sarcopenic obesity: Differences in prevalence, agreement and association with muscle function. J Frailty Aging 9:37–43
Article on SAMS. http://medicine.nus.edu.sg/medi/doc/education/combating_diabetes.pdf
Khoo CM, Leow MK, Sadananthan SA, Lim R, Venkataraman K, Khoo EY, Velan SS, Ong YT, Kambadur R, McFarlane C, Gluckman PD, Lee YS, Chong YS, Tai ES (2014) Body fat partitioning does not explain the interethnic variation in insulin sensitivity among Asian ethnicity: the Singapore adults metabolism study. Diabetes 63(3):1093–1102. https://doi.org/10.2337/db13-1483
Sadananthan SA, Tint MT, Michael N, Aris IM, Loy SL, Lee KJ, Shek LP, Yap FKP, Tan KH, Godfrey KM, Leow MK, Lee YS, Kramer MS, Gluckman PD, Chong YS, Karnani N, Henry CJ, Fortier MV, Velan SS (2019) Association between early life weight gain and abdominal fat partitioning at 4.5 years is sex, ethnicity, and age dependent. Obesity (Silver Spring) 27(3):470–478. https://doi.org/10.1002/oby.22408
NCD-RisC (2014) Risk factor collaboration. http://www.ncdrisc.org/obesity-prevalence-map.html
Positano V, Gastaldelli A, Sironi A, Santarelli M, Lombardi M, Landini L (2004) An accurate and robust method for unsupervised assessment of abdominal fat by MRI. J Magn Reson Imaging 20(4):684–689
Sussman D, Yao J, Summers R (2010) Automated measurement and segmentation of abdominal adipose tissue in MRI. In: IEEE international symposium on biomedical imaging: from nano to macro, pp 936–939
Kullberg J, Ahlström H, Johansson L, Frimmel H (2007) Automated and reproducible segmentation of visceral and subcutaneous adipose tissue from abdominal MRI. Int J Obes 31:1806–1817
Joshi AA, Hu H, Leahy R, Goran M, Nayak K (2013) Automatic intra-subject registration-based segmentation of abdominal fat from water-fat MRI. J Magn Reson Imaging 37(2):423–430
Leinhard OD, Johansson A, Rydell J, Smedby Ö, Nyström F, Lundberg P, Borga M (2008) Quantitative abdominal fat estimation using MRI. In: 2008 19th international conference on pattern recognition, pp 1–4
Mosbech TH, Pilgaard K, Vaag A, Larsen R (2011) Automatic segmentation of abdominal adipose tissue in MRI. In: Heyden A, Kahl F (eds) Image analysis. SCIA 2011. Lecture notes in computer science, vol 6688. Springer, Berlin
Sadananthan SA, Prakash B, Leow MK, Khoo C, Chou H, Venkataraman K, Khoo E, Lee Y, Gluckman P, Tai E, Velan S (2015) Automated segmentation of visceral and subcutaneous (deep and superficial) adipose tissues in normal and overweight men. J Magn Reson Imaging 41(4):924–934
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. arXiv:1505.04597
Estrada S, Lu R, Conjeti S, Orozco-Ruiz X, Panos-Willuhn J, Breteler MM, Reuter M (2020) FatSegNet: a fully automated deep learning pipeline for adipose tissue segmentation on abdominal dixon MRI. Magn Reson Med 83(4):1471–1483
Chew J, Tay L, Lim JP, Yeo A, Yew S, Tan CN, Ding YY, Lim WS (2019) Serum myostatin and IGF-1 as gender-specific biomarkers of frailty and low muscle mass in community-dwelling older adults. J Nutr Health Aging 23(10):979–986
Pérez-García F, Sparks R, Ourselin S (2020) TorchIO: a Python library for efficient loading, preprocessing, augmentation, and patch-based sampling of medical images in deep learning. arXiv:2003.04696
Uçar MK, Nour M, Sindi H, Polat K (2020) The effect of training and testing process on machine learning in biomedical datasets. Math Prob Eng 2020:2836236
Le Ba T, Khanh D-P, Ho N-H, Yang H-J, Baek E-T, Lee G, Kim S-H, Yoo SB (2020) Enhancing U-Net with spatial-channel attention gate for abnormal tissue segmentation in medical imaging. Appl Sci 10(17):5729
Shelhamer E, Long J, Darrell T (2017) Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell 39(4):640–651
Badrinarayanan V, Kendall A, Cipolla R (2017) SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 39:2481–2495
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. arXiv:1606.06650
Zeiler MD, Krishnan D, Taylor GW, Fergus R (2010) Deconvolutional networks. IEEE CVPR 2010(2528–2535):5539957
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444
Isensee F, Jaeger PF, Kohl SAA et al (2021) nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18:203–211. https://doi.org/10.1038/s41592-020-01008-z
He F, Liu T, Tao D (2020) Why ResNet works? Residuals generalize. IEEE Trans Neural Netw Learn Syst 31:5349–5362
Ba J, Kiros JR, Hinton GE (2016) Layer normalization. arXiv:1607.06450
Agarap AF (2018) Deep learning using rectified linear units (ReLU). arXiv:1803.08375
Zhou Z, Siddiquee MM, Tajbakhsh N, Liang J (2018) U-Net++: a nested u-net architecture for medical image segmentation. DLMIA/ML-CDS@MICCAI, arXiv:1807.10165
Srivastava N, Hinton GE, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15:1929–1958
Wan R, Zhu Z, Zhang X, Sun J (2020) Spherical motion dynamics of deep neural networks with batch normalization and weight decay. arXiv:2006.08419
Kingma DP, Ba J (2015) Adam: a method for stochastic optimization. arXiv:1412.6980
Boer PD, Kroese DP, Mannor S, Rubinstein RY (2005) A tutorial on the cross-entropy method. Ann Oper Res 134:19–67. https://doi.org/10.1007/s10479-005-5724-z
Braiek HB, Khomh F (2019) TFCheck: a tensorflow library for detecting training issues in neural network programs. In: 2019 IEEE 19th international conference on software quality, reliability and security (QRS), pp 426–433
Sadananthan SA, Prakash B, Leow MK, Khoo CM, Chou H, Venkataraman K, Khoo E, Lee YS, Gluckman P, Tai ES, Velan SS (2015) Automated segmentation of visceral and subcutaneous (deep and superficial) adipose tissues in normal and overweight men. J Magn Reson Imaging JMRI 41(4):924–934
Zhao Z, Kuang X, Zhu Y, Liang Y, Xuan Y (2020) Combined kernel for fast GPU computation of Zernike moments. J Real Time Image Proc 1–14
Shamir RR, Duchin Y, Kim J, Sapiro G, Harel NY (2019) Continuous dice coefficient: a method for evaluating probabilistic segmentations. BioRxiv
Demerath E, Shen W, Lee M, Choh A, Czerwinski S, Siervogel R, Towne B (2007) Approximation of total visceral adipose tissue with a single magnetic resonance image. Am J Clin Nutr 85(2):362–368
Shen W, Punyanitya M, Chen J, Gallagher D, Albu J, Pi-Sunyer X, Lewis C, Grunfeld C, Heymsfield S, Heshka S (2007) Visceral adipose tissue: relationships between single-slice areas at different locations and obesity-related health risks. Int J Obesity 31(5):763–769
Ng A, Wai D, Tai E, Ng K, Chan L (2012) Visceral adipose tissue, but not waist circumference is a better measure of metabolic risk in Singaporean Chinese and Indian men. Nutr Diabetes 2(8):e38
Thomas EL, Saeed N, Hajnal J, Brynes A, Goldstone A, Frost G, Bell JD (1998) Magnetic resonance imaging of total body fat. J Appl Physiol 85(5):1778–1785
Villareal DT, Aguirre L, Gurney AB, Waters DL, Sinacore DR, Colombo E, Armamento-Villareal R, Qualls C (2017) Aerobic or resistance exercise, or both, in dieting obese older adults. N Engl J Med 376(20):1943–1955. https://doi.org/10.1056/NEJMoa1616338.s
Acknowledgements
Our sincere thanks to Singapore Bioimaging Consortium, A*STAR, and Tan Tock Seng Hospital for providing funds and data for conducting this study. We would also like to thank the support received from TechSource Systems Pte Ltd, especially Application Engineer Kevin Chng Jun Yan for his technical guidance and help rendered when needed during the development of the correction tool.
Author information
Authors and Affiliations
Contributions
KNB, CS worked on deep learning-based framework development, data processing, concept development, correction tool, and manuscript. LY developed a correction tool and assisted in the manuscript. CHT, WSL, and WC collected data (image acquisition), generated ground truth, and worked on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest.
Ethics approval
Institutional Review Boards have approved the study and written consent was taken from the subjects.
Consent for publication
Authors consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bhanu, P.K., Arvind, C.S., Yeow, L.Y. et al. CAFT: a deep learning-based comprehensive abdominal fat analysis tool for large cohort studies. Magn Reson Mater Phy 35, 205–220 (2022). https://doi.org/10.1007/s10334-021-00946-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10334-021-00946-9