Abstract
A new geminivirus was isolated from field-grown tomatoes with leaf yellowing and curly top symptoms in Fukushima, Japan in 2009–2010. Sequence comparisons of the full-length genome of the virus and other geminiviruses revealed its close relationship to common bean curly stunt virus (CBCSV), with 86.1% nucleotide sequence identity. The virus encodes six open reading frames (ORFs) that had a very high sequence identity (84.3–92.6%) with those of CBCSV, but it lacks ORF V3. The agroinoculation of the cloned genome induced stunting on the tobacco plants, and the graft inoculation of tobacco scions onto tomato rootstock reproduced curly top symptoms. The name "tomato curly top virus" is proposed for this virus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Geminiviridae viruses present a significant threat to agricultural crop production in tropical and temperate zones (Rojas et al. 2018). Geminiviruses are composed of one (or two in bipartite begomoviruses) circular single-stranded DNA genome and a twinned icosahedral capsid composed of coat proteins (CPs). The family Geminiviridae includes 520 species that are classified into 14 genera based on genome organization, host range, and vector species (Fiallo-Olivé et al. 2021). Begomovirus is its largest genus, including 445 species that are transmitted by the whitefly Bemisia tabaci, and it only infects dicotyledonous plants, including economically important crops such as tomato, cotton, bean, and cassava. Other major dicot-infecting geminiviruses are from the genera Becurtovirus, Curtovirus, and Turncurtovirus, which are transmitted by the leafhoppers Circulifer haematoceps or Circulifer tenellus (Varsani et al. 2014a, b). Among them, becurtoviruses and curtoviruses have three open reading frames (ORFs) in virion-sense strands (V1, V2, and V3), whereas turncurtoviruses only have V1 and V2. Turncurtovirus includes three species, turnip curly top virus, turnip leaf roll virus, and sesame curly top virus, which occur in Iran (Briddon et al. 2010; Hasanvand et al. 2018; Kamali et al. 2016). Common bean curly stunt virus (CBCSV), which is a new geminivirus that is phylogenetically related to turncurtoviruses, was reported from China. It encodes V3, whereas no sequence similarity was found with proteins of any other geminiviruses (Zhang et al. 2020).
In Japan, 10 geminiviruses were found to naturally infect wild plants and agricultural crops, including tomato (Fuji et al. 2022). Tomato crops with geminivirus-like symptoms such as leaf yellowing, leaf curling, and internode shortening were observed during field surveys from 2008 to 2010 in Fukushima Prefecture, Japan (Fukushima Prefectural Plant Protection Office 2011). Polymerase chain reaction (PCR) analysis was done on diseased plant samples using universal primer pairs specific for begomoviruses (Briddon et al. 1994). Products with expected lengths for begomoviruses were obtained. However, their partial sequences putatively encoding parts of ORFs C1 and C2 only showed low similarities to those of known geminiviruses, prompting us to further characterize this virus and name it tomato curly top virus (ToCTV). Sequence comparison and phylogenetic analysis of the full-length genomic DNA and encoded proteins of ToCTV revealed its apparent relationship with CBCSV. The results raised further questions about the evolution and geographic distribution of curtovirus- and turncurtovirus-like viruses in Asia.
Materials and methods
Collection and maintenance of virus samples
Tomato (Solanum lycopersicum cv. Momotaro-Natsumi) plant shoots with leaf yellowing or curling and curly top symptoms were collected from the Fukushima Prefecture in July 2009 and in July 2010. The leaves were stored separately at − 80 °C until further analysis. The virus collected in 2009 was maintained by periodically grafting a symptomatic tomato shoot onto a healthy tomato (S. lycopersicum cv. Rutgers) rootstock.
Virion purification and electron microscopy
Virions were purified from 10 g of symptomatic leaves of graft-inoculated tomato plants (Luisoni et al. 1995); however, instead of Cs2SO4 density gradient centrifugation, sucrose density gradient centrifugation (10–40% gradient) was performed for 90 min. The purified virions were negatively stained with 4% uranyl acetate and observed under a transmission electron microscope (JEOL, Tokyo, Japan). The length and diameter of 20 virions were obtained, and mean values were calculated.
Cloning and sequencing of the ToCTV genome
DNA was extracted from symptomatic tomato leaves using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Using the DNA as a template, rolling circle amplification (RCA) was performed using Ø29 DNA polymerase (illustra TempliPhi Amplification Kit; Cytiva, Marlborough, MA, USA) (Inoue-Nagata et al. 2004). The RCA products were digested through a series of restriction enzymes and separated by agarose gel electrophoresis. A 3.0-kb single band obtained from PstI restriction enzyme digestion was purified from the agarose gel and cloned into an appropriately digested pUC19 vector. The cloned fragments were sequenced using a BigDye Terminator v1.1 Cycle Sequencing Ready Reaction Kit and ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) with primer walking strategy. The sequences around the PstI site were confirmed by direct sequencing of the PCR products with appropriately designed primers. Then, the complete genome sequence was assembled using a CodonCode Aligner software (CodonCode Corporation, Dedham, MA, USA).
Sequence comparison, phylogenetic, and recombination analyses
ORFs located within the ToCTV genome and deduced amino acid (aa) sequences encoded by those were predicted using DNADynamo sequence analysis software (Blue Tractor Software Ltd., UK). Corresponding sequences of other geminiviruses were retrieved from GenBank. Sequence identities for the pairwise sequence alignments of full-length genome and aa sequences of CP and replication-associated protein (Rep) were calculated using an SDT v1.2 program (Muhire et al. 2013) using the MAFFT option. For the phylogenetic tree analysis, nucleotide (nt) sequences of full-length genome or aa sequences of CP and Rep were aligned using MAFFT program Ver. 7 (Katoh and Standley 2013). Phylogenetic trees were constructed using MEGA X (Stecher et al. 2020), applying the neighbor-joining method. The reliability of each tree branch was evaluated by bootstrap testing, with 1,000 pseudoreplicates. For the recombination analysis, all full-length genome sequences of CBCSV, becurtoviruses, curtoviruses, and turncurtoviruses available in GenBank were collected and aligned by MAFFT. Then, the alignment was analyzed by RDP5 using default settings (Martin et al. 2020).
ToCTV agroinoculation
A pBI121 (Takara Bio, Kusatsu, Japan)-derived binary vector carrying a tandem repeat of ToCTV clone was constructed. The full-length ToCTV-K4 sequence was amplified by PCR from the cloned RCA product using primers GemKFc/HindIII (5′-ACCTCAAGCTTCAGCATCATTAGACGTCTGTTG-3′) and GemKRc/PstI (5′-GCTGCTGCAGAAGCCTTAAACGCAGG-3′), which harbor restriction sites (underlined) and correspond to nt 2,414 to 2,435 and nt 2,413 to 2,392, respectively. The PCR product was digested with HindIII and PstI and inserted between the corresponding sites of pUC19. Similarly, another full-length fragment was amplified using primers GemKFc2/PstI (5′-GCTTCTGCAGCAGCATCATTAGACGTCTG-3′) and GemKRc2/SmaI (5′-CTACCCGGGAAGCCTTAAACGCAGGTAACG-3′), which correspond to nt 2,408 to 2,435 and nt 2,407 to 2,388, respectively, and was inserted into the corresponding sites of pUC19. The sequences of the cloned DNA fragments were verified, and HindIII–PstI and PstI–SmaI fragments from the corresponding plasmids were tandemly inserted into the HindIII–SacI-blunted site of the binary vector pBI121 to yield pBI-ToCTV-K4.
The Rhizobium radiobacter strain C58C1 (pGV2260) (hereafter, agrobacterium) harboring pBI-ToCTV-K4 or pBI121 was generated by electroporation. Agrobacterium-mediated inoculation (agroinoculation) was performed, as described previously (Erickson et al. 1999). An overnight culture of agrobacterium in an LB medium supplemented with appropriate antibiotics was centrifuged and washed with agroinfiltration medium [10 mM 2-[N-morpholino] ethanesulfonic acid-HCl (pH 5.85), 10 mM MgCl2, and 150 µM acetosyringone]. Then, the cells were suspended in the same medium to achieve an OD600 of 0.6 and incubated for 3 h at room temperature. The suspension was infiltrated into the fourth true leaf of tobacco (Nicotiana tabacum cv. Xanthi nc) or to several true leaves and petioles of tomato (S. lycopersicum cv. Rutgers). Infiltrated plants were incubated in a growth chamber at 22 °C for 2 d and subsequently grown in a glasshouse at 26 °C/22 °C.
ToCTV graft inoculation
The apical part of tobacco plants (approximately 5-cm long, with leaves of > 1 cm removed), which were agroinoculated for 10–20 d prior to grafting, was employed as an inoculum. Healthy tomato (S. lycopersicum cv. Rutgers) plants with 5–8 true leaves were obtained, and a 3 cm-deep cleft was made at the cut surface. The apex of the tobacco scion was beveled and tightly fastened to the cleft of tomato rootstock using a parafilm. Then, the plants were covered with plastic bags for several days to avoid dehydration. The lateral shoots of the tomato rootstock that developed thereafter were investigated for viral infection symptoms. To test whether ToCTV was graft-transmissible from tomato to tomato, the apices of the lateral shoots were used as an inoculum instead of the tobacco apex.
PCR analysis for ToCTV detection
The total DNA was extracted from the upper non-inoculated leaves of agroinoculated tobacco plants or lateral shoots of graft-inoculated tomato plants (Dellaporta et al. 1983). PCR was performed using the extracted DNA as a template, Ex Taq DNA Polymerase (Takara Bio), and ToCTV-specific primers GemFK4-v1 (5′-CCGTTTGGTGACCCAGCGT-3′) and GemFK4-c1 (5′-TGGACTCTACAAAATTCAAAAATCA-3′), corresponding to nt 483 to 501 and nt 1,829 to 1,805, respectively. The PCR conditions included an initial denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 90 s. PCR products were analyzed by 1% agarose gel electrophoresis.
Results and discussion
ToCTV was isolated from afflicted tomato plants grown under plastic rain shelters lacking insect-proof nets in Fukushima Prefecture, Japan. ToCTV infection resulted in leaf yellowing and curling and curly top symptoms (Fig. 1a and b). The disease was graft-transmissible between tomato plants and produced similar symptoms. Electron microscopic images of virus particles purified from graft-inoculated tomato plants revealed geminivirus-like geminate particles of approximately 32.9 nm in length and 20.2 nm in diameter (Fig. 1c).
Diseased tomato crops in Fukushima, Japan, naturally infected with tomato curly top virus (ToCTV) showing a leaf yellowing and curling or b severe curly top symptoms. c Electron microscopy of ToCTV virions, indicated by arrows, purified from graft-inoculated tomato plants. Scale bar, 100 nm. d Schematic representation of the genome architecture and open reading frame (ORF) composition of ToCTV-K4. The ORFs encoded on virion (V)-sense and complementary (C)-sense strands are shown by blue and red arrows, respectively
A full-length ToCTV genome was amplified using RCA for molecular characterization. Of the seven tomato leaf samples collected in the field in 2009, four did not yield RCA products and were negative for geminivirus infection, while three yielded 3.0-kb PstI restriction fragments; these restriction fragments were cloned and sequenced to yield the complete genome sequences of the isolates. The cloned sequences shared features with the genome of geminiviruses, and the three isolates were designated as ToCTV-[JP-Fuk-K1-09] (ToCTV-K1), ToCTV-[JP-Fuk-K2-09] (ToCTV-K2), and ToCTV-[JP-Fuk-K4-09] (ToCTV-K4). In a field survey done in 2010, ToCTV-[JP-Fuk-K5-10] (ToCTV-K5) and ToCTV-[JP-Fuk-K6-10] (ToCTV-K6) were cloned. The nt sequences determined in this study were deposited in the DDBJ/ENA/GenBank database, with accession numbers AB935396–AB935398, LC160267, and LC160268.
The genomes of the five isolates contained a canonical nonanucleotide sequence (ATATATT/AC) in their 420-nt long putative intergenic region (IR). The length of the complete genome of ToCTV-K1 was 2,970 nt, whereas the other four had a length of 2,969 nt, as a result of 1-nt deletion in the putative IR. The absence of RCA products derived from the DNA-B, alpha-satellite, or beta-satellite sequences that accompany certain begomoviruses suggests the lack of association of such components with ToCTV. Pairwise nt sequence alignments of the complete genome of the isolates revealed > 99.8% sequence identity. Thus, ToCTV-K4 was employed for further analyses. Compared to the complete genome of ToCTV-K4, ToCTV shared the highest identity to CBCSV (86.1%), followed by turncurtoviruses (TCTV, SCTV, and TLRV), sesame yellow mosaic virus (a putative turncurtovirus), and beet curly top virus (a curtovirus) (62.6–60.5%). The other geminiviruses showed < 59.0% nt sequence identity (Table 1).
ToCTV-K4 encodes two ORFs on virion-sense strand [V1 (nt 506–1270) and V2 (nt 265–576) encoding 29.0-kDa and 11.8-kDa proteins, respectively] and four ORFs on complementary-sense strand [C1 (nt 2816–1713), C2 (nt 1945–1427), C3 (nt 1704–1306), and C4 (nt 2656–2399) encoding 41.8-kDa, 19.5-kDa, 15.7-kDa, and 9.5-kDa proteins, respectively] (Fig. 1d). Blastp search indicated that V1 contains a domain conserved in CPs of geminiviruses. On the v-sense strand of ToCTV-K4, there was another small ORF (nt 744–980, completely overlapping within the V1 ORF), which putatively encodes a protein composed of 78 amino acids with no significant sequence similarity to any other known proteins. The V2 protein showed a sequence similarity to that of CBCSV, TLRV, and SYMV, while proteins C2, C3, and C4 shared a sequence similarity to those of diverse geminiviruses (data not shown). The proteins V1 and C1 of ToCTV-K4 showed the highest aa sequence identities of 84.3% and 92.6% with those of CBCSV, respectively, whereas it was only < 34.3% for V1 (TLRV) and < 72.4% for C1 (SeYMV) (Table 1). The aa sequence identities between the ToCTV-K4 and CBCSV of proteins V2, C2, C3, and C4 were 85.4%, 82.7%, 92.4%, and 88.2%, respectively.
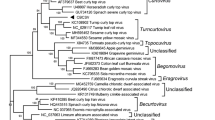
In a phylogenetic tree based on the complete nt sequences of ToCTV and representative geminiviruses, ToCTV was subclustered together with CBCSV, and they formed a clade with turncurtoviruses, curtoviruses, and becurtoviruses (Fig. 2a). Similarly, a tree for the aa sequences of V1 indicated the closest relationships between ToCTV and CBCSV and with turncurto-, curto-, and becurtoviruses (Fig. 2b). In a phylogenetic tree for C1, ToCTV and CBCSV formed a clade with tunrcurtoviruses, becurtoviruses, and horseradish curly top virus (Fig. 2c). No recombination events were detected in the ToCTV-K4 genome (data not shown).
Phylogenetic relationships of K4 isolates of the tomato curly top virus (ToCTV-K4) and selected viruses from the family Geminiviridae. Analyses were performed using a complete nucleotide sequence alignment or b amino acid sequence alignments for coat protein (V1) and c replication-associated protein (C1). The numbers at each node represent the percentage of the bootstrap values, with those < 60% omitted. The scale bars represent the number of residue substitutions per site. The GenBank accession numbers of the proteins used for phylogenetic analyses are reported. Viruses from each genus are indicated by colored dots. ToCTV sequences are shown in bold
To investigate whether ToCTV is the causative agent of curly top symptoms in tomato plants, agroinoculation experiments on tomato and tobacco plants using a ToCTV clone, pBI-ToCTV-K4, were conducted. Despite several trials, the agroinoculated tomato plants failed to show any geminivirus infection symptoms. Although we did not pursue the reason of the failure, it has been reported that the C58C1 strain of R. radiobacter is applicable to gene expression in tomato (Wroblewski et al. 2005), and conditions of agroinoculation can be important for successful infection of geminiviruses (Koeda et al. 2018). The agroinoculation of tobacco plants with pBI-ToCTV-K4 resulted in leaf curling at approximately 10 d after infiltration, with subsequent stunting and severe leaf crinkle on the upper non-infiltrated leaves (Fig. 3a and b). As shown in Table 2, the symptoms appeared on eight out of nine infiltrated tobacco plants, and ToCTV infection in all symptomatic plants was confirmed by PCR using ToCTV-specific primers 11 d after infiltration. Meanwhile, no symptom development nor ToCTV infection was observed in the tobacco plants agroinoculated with the negative control, pBI121. Furthermore, to test whether ToCTV infection reproduces curly top symptoms, tomato plants were graft-inoculated with symptomatic tobacco apices previously agroinoculated with ToCTV. After 3–4 weeks, the tomato plants grafted with pBI121-agroinoculated tobacco remained healthy and uninfected (Fig. 3c), while the grafted tomato plants exhibited typical leaf curling and yellowing on the lateral shoots that expanded subsequent to grafting (Fig. 3d). Curly top symptoms were observed in the later stages (data not shown). The virus was also confirmed to be graft-transmissible between tomato plants by grafting healthy tomato rootstocks with symptomatic tomato shoots. This resulted in the development of curly top symptoms on the lateral shoots of the inoculated rootstock (Fig. 3e), which were nearly identical to the symptoms of infected crops in the fields of Fukushima Prefecture (Fig. 1b). ToCTV was detected by PCR in all symptomatic tomato plants (Table 3). Thus, we concluded that ToCTV was responsible for the observed disease in tomato plants.
Symptoms of tomato curly top virus (ToCTV) infection on tobacco and tomato plants. a Tobacco plants agroinoculated with either pBI121 (left) or pBI-ToCTV-K4 (right). ToCTV infection of tobacco resulted in severe stunting. A photograph was taken at 33 d after agroinoculation. b A close-up view of the apical part of ToCTV-infected tobacco, showing leaf crinkle. c and d Symptoms on the lateral shoots of tomato graft-inoculated with agroinoculated tobacco at 21 d after grafting. c A tomato grafted with tobacco that was agroinoculated with pBI121 as a negative control. d Tomato grafted with symptomatic tobacco shoots that were previously agroinoculated with pBI-ToCTV-K4. Leaf yellowing and curling were observed. e Curly top symptoms were reproduced on tomato by graft-inoculating ToCTV-infected tomato shoot (d). Offshoots that developed after graft inoculation were photographed at 33 d post-grafting
In conclusion, the curly top symptoms found in the tomato plants in Fukushima Prefecture were caused by ToCTV. Although sequence comparison and phylogenetic analysis suggest that ToCTV is very closely related to CBCSV, an ORF corresponding to V3, which CBCSV harbors but its homolog is not found in any other geminiviruses (Zhang et al. 2020), is absent in the ToCTV genome. Thus, it remains elusive whether ToCTV and CBCSV are the same or different species. Further biological characterization of the two viruses, including their host range and vector species, would be necessary.
References
Briddon RW, Markham PG (1994) Universal primers for the PCR amplification of dicot-infecting geminiviruses. Mol Biotechnol 1:202–205
Briddon RW, Heydarnejad J, Khosrowfar F, Massumi H, Martin DP, Varsani A (2010) Turnip curly top virus, a highly divergent geminivirus infecting turnip in Iran. Virus Res 152:169–175
Dellaporta SL, Wood J, Hicks JR (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21
Erickson FL, Holzberg S, Calderon-Urrea A, Handley V, Axtell M, Corr C, Baker B (1999) The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J 18:67–75
Fiallo-Olivé E, Lett JM, Martin DP, Roumagnac P, Varsani A, Zerbini FM, Navas-Castillo J, ICTV Report Consortium (2021) ICTV virus taxonomy profile: Geminiviridae. J Gen Virol 102:001696
Fuji S, Mochizuki T, Okuda M, Tsuda S, Kagiwada S, Sekine K-T, Ugaki M, Natsuaki KT, Isogai M, Maoka T, Takeshita M, Yoshikawa N, Mise K, Sasaya T, Kondo H, Kubota K, Yamaji Y, Iwanami T, Ohshima K, Kobayashi K, Hataya T, Sano T, Suzuki N (2022) Plant viruses and viroids in Japan. J Gen Plant Pathol 88:105–127
Fukushima Prefectural Plant Protection Office (2011) Special report on forecast of pest occurrence. No. 1. Fukushima Pref. Available via https://www.pref.fukushima.lg.jp/uploaded/attachment/91312.pdf. Accessed 28 Oct 2022
Hasanvand V, Kamali M, Heydarnejad J, Massumi H, Kvarnheden A, Varsani A (2018) Identification of a new turncurtovirus in the leafhopper Circulifer haematoceps and the host plant species Sesamum indicum. Virus Genes 54:840–845
Inoue-Nagata AK, Albuquerque LC, Rocha WB, Nagata T (2004) A simple method for cloning the complete begomovirus genome using the bacteriophage Ø29 DNA polymerase. J Virol Methods 116:209–211
Kamali M, Heydarnejad J, Massumi H, Kvarnheden A, Kraberger S, Varsani A (2016) Molecular diversity of turncurtoviruses in Iran. Arch Virol 161:551–561
Katoh K, Standley DM (2013) MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol Biol Evol 30:772–780
Koeda S, Homma K, Tanaka Y, Onizaki D, Kesumawati E, Zakaria S, Kanzaki S (2018) Inoculation of capsicums with Pepper yellow leaf curl Indonesia virus by combining agroinoculation and grafting. Hort J 87:364–371
Luisoni E, Milne RG, Vecchiati M (1995) Purification of tomato yellow leaf curl geminivirus. New Microbiol 18:253–260
Martin DP, Varsani A, Roumagnac P, Botha G, Maslamoney S, Schwab T, Kelz Z, Kumar V, Murrell B (2020) RDP5: a computer program for analyzing recombination in, and removing signals from, nucleotide sequence datasets. Virus Evol 7:veaa087
Muhire B, Martin DP, Brown JK, Navas-Castillo J, Moriones E, Zerbini FM, Rivera-Bustamante R, Malathi VG, Briddon RW, Varsani A (2013) A genome-wide pairwise-identity-based proposal for the classification of viruses in the genus Mastrevirus (family Geminiviridae). Arch Virol 158:1411–1424
Rojas MR, Macedo MA, Maliano MR, Soto-Aguilar M, Souza JO, Briddon RW, Kenyon L, Rivera Bustamante RF, Zerbini MF, Adkins ST, Legg JP, Kvarnheden A, Wintermantel WM, Sudarshana MR, Peterschmitt M, Lapidot M, Martin DP, Moriones E, Inoue-Nagata AK, Gilbertson RL (2018) World management of geminiviruses. Ann Rev Phytopathol 56:637–677
Stecher G, Tamura K, Kumar S (2020) Molecular evolutionary genetics analysis (MEGA) for macOS. Mol Biol Evol 37:1237–1239
Varsani A, Martin DP, Navas-Castillo J, Moriones E, Hernández-Zepeda C, Idris A, Zerbini FM, Brown JK (2014a) Revisiting the classification of curtoviruses based on genome-wide pairwise identity. Arch Virol 159:1873–1882
Varsani A, Navas-Castillo J, Moriones E, Hernández-Zepeda C, Idris A, Brown JK, Zerbini FM, Martin DP (2014b) Establishment of three new genera in the family Geminiviridae: Becurtovirus, Eragrovirus and Turncurtovirus. Arch Virol 159:2193–2203
Wroblewski T, Tomczak A, Michelmore R (2005) Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol J 3:259–273
Zhang R, Wu X, Jiang X, Wu X, Luan X, Cheng X (2020) Molecular characterization of common bean curly stunt virus: a novel recombinant geminivirus in China. Arch Virol 165:257–260
Acknowledgements
We are grateful to Y. Matsushita for the grafting techniques. We also thank Y. Matsumura, Y. Narita, and J. Sato for their assistance in preparing the biological samples and laboratory work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kubota, K., Tomitaka, Y., Usugi, T. et al. Molecular characterization of a new geminivirus isolated from tomato with curly top symptoms and development of its infectious clone. J Gen Plant Pathol 89, 100–108 (2023). https://doi.org/10.1007/s10327-022-01112-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10327-022-01112-2