Abstract
Glycosyltransferases (GTs) regulate many physiological processes and stress responses in plants. However, little is known about the function of GT in rice development. In this study, molecular analyses revealed that the expression of a rice GT gene (Cold-Upregulated Glycosyltransferase Gene 1, CUGT1) is developmentally controlled and stress-induced. OsCUGT1 was knocked out by using the clustered regularly interspaced short palindromic repeats (CRISPR) system to obtain the mutant oscugt1, which showed a severe dwarf and sterility phenotype. Further cytological analyses indicated that the dwarfism seen in the oscugt1 mutant might be caused by fewer and smaller cells. Histological pollen analysis suggests that the spikelet sterility in oscugt1 mutants may be caused by abnormal microsporogenesis. Moreover, multiple transgenic plants with knockdown of OsCUGT1 expression through RNA interference were obtained, which also showed obvious defects in plant height and fertility. RNA sequencing revealed that multiple biological processes associated with phenylpropanoid biosynthesis, cytokinin metabolism and pollen development are affected in the oscugt1 mutant. Overall, these results suggest that rice OsCUGT1 plays an essential role in rice development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The development of stems and inflorescences in crop plants is fundamentally important to their growth and productivity (Wang et al. 2018). In rice (Oryza sativa L.), plant height and spikelet fertility rely on a sophisticated regulatory network in which thousands of genes work in a coordinated and integrated fashion during development (Liu et al. 2018; Wang et al. 2018; Zeng et al. 2017). In the past few decades, genetic and genomic studies in model plant species have uncovered many of the molecular mechanisms underlying the control of plant height, which is primarily determined by several endogenous phytohormones, including gibberellins, brassinosteroids, cytokinins, auxin and strigolactones (Liu et al. 2018). It has been reported that functional alleles in the gibberellin biosynthesis gene semidwarf 1 (sd1) result in dwarfism to different degrees (Spielmeyer et al. 2002; Ueguchi-Tanaka et al. 2005). In addition, cytokinins also exert essential roles in the regulation of plant height because they can control many agriculturally important processes, including cell division, stem elongation and other developmental processes (Heyl and Schmulling 2003). Decapitation of cytokinin catabolism or the disturbance of cytokinin signaling dramatically decreases plant yield and height (Xiao et al. 2020). For example, reduced expression of CYTOKININ OXIDASE/DEHYDROGENASE 2 (OsCKX2), an enzyme that functions in cytokinin oxidative cleavage and homeostasis, enhanced grain yield (Ashikari et al. 2005). Overexpression of OsCKX2 resulted in developmental defects due to cytokinin level reduction (Yan et al. 2020). In addition, it has been reported that overexpression of a B-TYPE CYTOKININ RESPONSE REGULATOR (OsORR2) also reduces rice height (Shi et al. 2020).
Spikelet fertility is also vital for crop production. The genetic basis of floral organ initiation and development has been extensively studied in Arabidopsis (Arabidopsis thaliana) and rice (Wang and Li 2008). Plant reproductive success is influenced by various environmental and developmental factors (Nubankoh et al. 2020). Numerous physical factors, important molecular components, and complex regulatory interactions contribute to spikelet fertility by affecting pollen formation and viability, anther dehiscence, and flower opening (Hu et al. 2021; Jagadish et al. 2007; Qi and Wu 2022; Zhou et al. 2017). Accumulating genetic evidence underscores the importance of multiple transcription factors, lipid metabolism- and sugar metabolism-related enzymes in spikelet fertility. For example, TAPETUM DEGENERATION RETARDATION (OsTDR) encodes a putative basic helix–loop–helix transcription factor that functions in tapetum degradation and anther development (Li et al. 2006). SUCROSE PHOSPHATE SYNTHASE (OsSPS1) is essential in pollen germination by modulating sucrose synthesis (Hirose et al. 2014). Despite extensive efforts to elucidate the molecular regulation of plant height and spikelet fertility in rice, much still remains unknown.
Glycosyltransferases (GTs) are the most diverse group of enzymes present in all eukaryotic and prokaryotic cells (Cao et al. 2008; Hu and Walker 2002). They can transfer glycosyl groups from the activated donor (usually a UDP-glycosyl group) to the hydroxyl group of numerous substrates, such as lipids, proteins, hormones and secondary metabolites (Hou et al. 2004). The glycosylation catalyzed by GTs functions as an important modulator in regulating hormone homeostasis, stem growth, pollen and seed maturation, defense responses and abiotic stress tolerance (Bowles 2002; Gachon et al. 2005; Rehman et al. 2022; Wang et al. 2011, 2022; Yang et al. 2022). For example, OsGT1 genes have been reported to be essential for the production of viable pollen grains in rice (Moon et al. 2013). Moreover, a series of recent studies have shown that GTs can determine phenylpropanoid pathway flux, and therefore affect plant development and abiotic tolerance (Dare et al. 2017; Dong et al. 2020; Peng et al. 2017). In apple plants, dysfunction of a phloretin-specific glycosyltransferase UGT88F1 led to a severely dwarfed phenotype by modulating the phenylpropanoid pathway. In rice, OsUGT707A2 regulates flavone accumulation and UV tolerance (Peng et al. 2017).
Although much evidence has suggested that GTs participate in many physiological processes, growth/development and stress responses in plants, little is known about the molecular function of OsCUGT1 (MSU: LOC_Os01g43380). Previously, OsCUGT1 was isolated as a cold-induced glycosyltransferase based on the cold treatment transcriptome library of Guizhou landrace rice ‘Ping Tang Wild-type’ (PTWT, Oryza sativa ssp. japonica) (Cai et al. 2021). One of OsCUGT1’s homologous gene At5g04480 in Arabidopsis was reported to participate in pollen tube development (Hoedemaekers et al. 2015). However, the biological function of OsCUGT1 in rice is still unknown. In this study, we report that OsCUGT1 plays a prominent role in regulating stem height and spikelet fertility in rice. OsCUGT1 is generally expressed in rice and most highly expressed in sheaths and panicles. oscugt1 mutants obtained by means of clustered regularly interspaced short palindromic repeats (CRISPR)-mediated genome editing had a short plant stature and defects in microspore development. Moreover, the OsCUGT1 RNAi transgenic lines exhibited a weaker but similar morphological phenotype to the oscugt1 mutants. A combination of cytological observations and comparative transcriptome analysis suggested that OsCUGT1 regulates rice growth and development by modulating essential genes involved in phenylpropanoid biosynthesis, cytokinin metabolism and pollen development. These findings may provide a more comprehensive understanding of GT functions in rice.
Materials and methods
Plant materials and growth conditions
The rice japonica cultivars ‘Zhonghua11 (ZH11)’ and ‘PTWT’, a black glutinous rice landrace in Guizhou, were used in this study. ZH11 and mutant plants were grown in a greenhouse at 30 °C (16-h light) and 22 °C (8-h dark) or in a paddy field at Guizhou University in Guiyang, China, from March to November of each year.
Plasmid construction and rice transformation
High-fidelity DNA polymerase (Genestar) was used for PCR amplification; final plasmids were also verified by Sanger sequencing. OsCUGT1 knockout mutants were obtained via CRISPR/CRISPR-associated protein 9 (Cas9)-mediated genome editing. The guide RNAs targeting the first exon of OsCUGT1 were cloned into the rice CRISPR/Cas9 vector pHUE411-2gR with a U3 promoter (Xing et al. 2014). pTCK303 (Wang et al. 2004) was used as the binary vector for hpRNA-producing construction. A 360 bp fragment of OsCUGT1 was inserted into the sense/antisense orientation of pTCK303. All resulting recombinant plasmids were then introduced into ZH11 by means of a modified Agrobacterium (Agrobacterium tumefaciens EHA105)-mediated transformation method. The coding sequences of OsCUGT1 were PCR amplified from PTWT rice plants and cloned into the binary vector pCAMBIA1300-UBI (Bio-Transduction) to obtain OsCUGT1 overexpression lines.
RNA extraction and quantitative real-time PCR (qRT‒PCR)
Total RNA was isolated from rice plants at the mature stage using an RNA Extraction Kit (Omeaga). qRT‒PCR was performed using a 2 × SYBR Green Power PCR Master Mix kit (Genestar) as previously described (Huang et al. 2022). Values are the means ± standard deviations (SD) of three repeats, with Student’s t test used for statistical analysis (Miura et al. 2007). All primers used in this study are listed in Table S1.
Microscopy observations
Paraffin sections were prepared as previously described (Huang et al. 2022). Internodes and spikelets from ZH11 and oscugt1 plants were collected and fixed in 75% (v/v) or 50% (v/v) formaldehyde-acetic-acid (FAA) ethanol fixative solution. The treated samples were dehydrated, embedded and double-stained with safranin O and fast green or toluidine blue (Service-bio), then finally observed, imaged and measured with the Case-Viewer digital microscopy application (3DHISTECH). Fresh pollen grains were examined using scanning electron microscopy (SEM) (Huang et al. 2022). Pollen grains were collected, fixed and washed with 2.5% (w/v) glutaraldehyde (Ser-vice-bio), 0.1 M sodium phosphate buffer (pH 7.4) and 1% (w/v) osmic acid sequentially. Then, the treated samples were dehydrated using a graded series of ethanol, dried and coated with gold films before SEM observation. (HITACHI, SU8100).
Transcription analysis
Total RNA was isolated from the leaves and spikelets of ZH11 and OsCUGT1 rice plants at the mature stage. Both sets of experiments were performed by the Metware, Jiaxing. mRNA libraries for transcriptome deep sequencing (RNA-seq) were sequenced on the Illumina platform. Sequencing data were collected from three independent experiments for each sample. Differentially expressed genes (DEGs) between OsCUGT1 and WT plants were determined with the R package DEseq2 (Love et al. 2014). The Benjamini–Hochberg multiple comparison adjustment (p < 0.05) was used to control false discovery rate (FDR) (Benjamini et al. 2001). Genes with FDR lower than 0.05 and |log2Fold Change|> = 1 were identified as DEGs.
Results
Molecular expression patterns of OsCUGT1
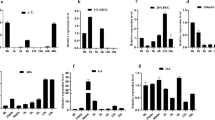
Previously, OsCUGT1 was isolated as a cold-induced glycosyltransferase based on the cold treatment transcriptome library of Guizhou landrace rice PTWT (Cai et al. 2021). According to the CAZy (Carbohydrate-Active EnZymes) database (http://www.cazy.org/GlycosylTransferases.html), OsCUGT1 belongs to the glycosyl transferase family 1 which is known to be involved in the modification of certain secondary metabolites and hormones (Cao et al. 2008). Glycosyl transferase family 1 consisting of 35 members in rice, which can be further clustered into six groups (Fig. S1). OsCUGT1 and its rice homolog LOC_Os10g39900 belongs to the Group II of family 1 (Fig. S1). Multiple sequence alignment of OsCUGT1, LOC_Os10g39900 and their Arabidopsis homolog (At4g01210 and At5g04480) revealed that these proteins all contain glycosyl transferase group 1 domains (PF00534) and display relatively high-level sequence conservation in the C-terminal domains (Fig. 1a). BLAST analysis showed that At4g01210 and At5g04480 in Arabidopsis and LOC_Os10g39900 in rice share approximately 39%, 28% and 26% amino acid sequence identity with OsCUGT1, respectively, which suggests functional similarity among these proteins. These results indicated that OsCUGT1 may act on plant development like At5g04480. In addition, divergence in the spatial expression patterns of OsCUGT1 was examined to explore the possible biological function. qRT‒PCR analyses indicated that OsCUGT1 is ubiquitously expressed in rice, with relatively low expression in roots, leaves, seedlings and mature panicles, and the highest expression in sheaths and young panicles (Fig. 1b). This finding suggested that OsCUGT1 may function mainly in rice sheaths and panicles. In addition, promoter analysis showed that OsCUGT1 had several cis-acting elements, including ABA-responsive elements (ABRE), drought-responsive elements (DRE) and anoxic-responsive elements (ARE). qRT‒PCR confirmed that OsCUGT1 was indeed upregulated by multiple external stress stimuli. For example, the expression of OsCUGT1 was upregulated nearly 48-fold and sixfold by NaCl and mannitol treatment, respectively. Moreover, OsCUGT1 was also upregulated by many hormone treatments, especially by cytokinin, which increased the expression of OsCUGT1 22-fold (Fig. 1c). These results indicate that OsCUGT1 may play roles in regulating plant growth and their adaptation to environmental conditions.
Characterization of the OsCUGT1 sequence and expression pattern. a Multiple sequence alignment of OsCUGT1 and its homologs from rice and Arabidopsis. These proteins all contain glycosyl transferase group 1 domains (PF00534). b The spatial expression patterns of OsCUGT1 were determined by qRT‒PCR. Total RNA was extracted from various rice tissues. Abbreviations: R, root; S, seedling; YL, young leaves; SH, sheaths; FL, flag leave; C, culm; MP, mature panicle; YP, young panicle. c The expression level of OsCUGT1 was upregulated by multiple external stress stimuli and hormone treatments. Values are presented as means ± SD (n = 3); **P < 0.01 by Student’s t-test. d Analysis of cis-acting elements in the promoter of OsCUGT1. TCA-element, salicylic acid responsive elements; ABRE ABA-responsive elements, DRE drought-responsive elements, LRE light-responsive elements, ARE anoxic-responsive elements
oscugt1 mutants exhibit a dwarf phenotype
To investigate the biological function of OsCUGT1, we used a genetic approach to understand the role of OsCUGT1 in the rice cultivar ZH11 via CRISPR/Cas9-mediated genome editing. Nineteen (90.5%) plants were successfully edited from 21 T0 transgenic plants. Many independent lines carrying different types of homozygous mutations in the OsCUGT1 gene were obtained (Fig. 2a). For example, the oscugt1-1 allele harbored a 1 bp A insertion, while oscugt1-2 carried a 1 bp T deletion 317 bp downstream of the ATG in OsCUGT1 (Fig. 2a). All homozygous mutants exhibited a dwarf phenotype compared to their WT (Fig. 2b). Statistically, the plant height of oscugt1 mutants was approximately three-fifths of that of WT at the maturity stage (Fig. 2c). The rice internodes from the first to the fifth main tiller were dramatically shorter in oscugt1 mutants (Fig. 2d). These results suggested that OsCUGT1 plays an important role in rice height.
Isolation and characterization of mutants in OsCUGT1. a Alignment of genomic sequences from the wild type (WT) in several independent oscugt1 mutants. b Dwarf phenotype of the oscugt1 mutant during the mature period. Bar = 10 cm. c Plant height of WT and five independent oscugt1 mutant lines. d Internode length comparison of WT and oscugt1. Values are expressed as the mean ± SD (n = 3); **P < 0.01 by Student’s t-test
Cell division and expansion of the stem are affected in the oscugt1 mutant
We characterized the morphology of epidermal cells in the first internode in the WT and the oscugt1 mutants through microscopic observations (Fig. 3). Longitudinal sections of the first internode in oscugt1-1 and oscugt1-2 showed shorter cell lengths compared to WT (Fig. 3a). We obtained support for this observation by quantifying the epidermal cell length in oscugt1 mutants with the imaging software Case-Viewer, which revealed a dramatic reduction of nearly 40% in cell length compared to WT (Fig. 3b). We also estimated the cell number of the WT and mutants by measuring their first internode length and dividing it by the average cell length. Analysis showed that the length of the first internode in oscugt1-1 and oscugt1-2 was approximately 50% and 42% of that in the WT, respectively (Fig. 3c). Therefore, the cell number in oscugt1-1 and oscugt1-2 decreased by approximately 20 and 24%, respectively, compared to that in the WT (Fig. 3d). These results indicated that the dwarf stature of oscugt1 is caused by the reduction in both cell number and cell size. Moreover, the expression of two genes that are mainly related to cell elongation (OsXTH8, Xyloglucan endotransglucosylases/hydrolases 5) (Fig. 3e) and cell division (OsRAN2, Ras-related nuclear protein GTPase 2) (Fig. 3f) downregulated in the oscugt1 mutant, suggesting that both cell expansion and cell division might be affected in the oscugt1 mutant.
Histological characterization of stems and roots from wild-type (WT) and the oscugt1 mutant. a Comparison of the first internode’s longitudinal section between WT and oscugt1. b Comparison of the first internode’s mean cell length between WT and oscugt1. Values are presented as means ± SD (n = 100 cells). c Comparison of the first internode’s mean length between WT and oscugt1. Values are presented as means ± SD (n = 3). d Comparison of the first internode’s mean cell number between WT and oscugt1. The cell number were calculated by dividing the length of the first internode by the average cell length. Values are presented as means ± SD (n = 3); *P < 0.05, **P < 0.01 by Student’s t-test. e Relative expression levels of OsXTH8 as determined by qRT‒PCR. f Relative expression levels of OsRAN2 as determined by qRT‒PCR. Both of OsXTH8 and OsRAN2 with lower expression in oscugt1 compared to WT, **P < 0.01 by Student’s t-test
The oscugt1 mutant is defective in microspore development
We noticed a shorter panicle and abnormal seed production in the oscugt1 mutant (Fig. 4a). Panicle seed-setting rates were 0% in the oscugt1 mutant, in sharp contrast to 94% in the WT plants (Fig. 4b). To investigate the possible causes of sterility in the oscugt1 mutant, we performed crosses between homozygotes (oscugt1-5) and the wild type. When oscugt1-5 was used as a pollen receiver (female), their seed setting rates reached 41%, while the seed settings rates of crosses between WT reached about 60%. However, oscugt1-5 cannot serve as a pollen donor due to the lack of mature pollen. Therefore, the above results indicated that dysfunction of OsCUGT1 causes defects in the male gamete. Then, we conducted detailed histological and cytological observations of male reproductive tissues. Compared to the WT, the length of anthers in the oscugt1 mutant was approximately two-thirds that of the WT (Fig. 4c). The whole stamens from the oscugt1 mutant showed clear morphological abnormalities, as they were much smaller and a lighter shade of yellow compared to WT (Fig. 4d-g). We determined from pollen grains with Lugol solution (KI-I2) staining that the pollen number and viability in oscugt1 were markedly decreased (Fig. 4h, i). In addition, SEM confirmed that the aborted pollen grains in oscugt1 still had an exine layer and aperture (Fig. 4j, k), although they exhibited an abnormal ornamentation with larger bacula structures (Fig. 4i-m).
Floret phenotypes in wild-type (WT) and the oscugt1 mutant. a Representative panicles of WT and oscugt1 mutants. Bar = 4 cm. b Average seed-setting rates of WT and oscugt1 mutants. c Average anther length of WT and oscugt1 mutants. Values are presented as means ± SD (n = 6). Data were analyzed using Student’s t-test; **P < 0.01. Floret phenotypes of WT (d) and oscugt1 mutants (e–g). Bar = 2 mm. Pollen stained with Lugol solution in WT (h) and oscugt1 mutants (i). Normal pollen appears dark. Bar = 50 μm. j–m Scanning electron micrographs of pollen grains. General view of WT pollen (j) and oscugt1 pollen (k) with exine and aperture ornamentation. Enlarged view of WT exine ornamentation with normal bacula structures (l) and oscugt1 exine ornamentation with larger bacula structures (m). Bar = 1 μm (j and k), Bar = 0.1 μm (l and m)
To further investigate the exact role of OsCUGT1 in microsporogenesis, we characterized the progression of anther development in WT and oscugt1-1 over eight developmental stages (Fig. 5). We detected no obvious differences in either the early pre-meiosis stage or the young microspore stage between WT and oscugt1-1, as both genotypes went through normal meiosis and released free microspores from their tetrads (Fig. 5a–d, i–l). Micro-spores from WT were plump and spherical, as expected for healthy microspores (Fig. 5e), whereas oscugt1-1 microspores were adhesive, shriveled and deformed (Fig. 5m). The differences between the two genotypes became obvious at the vacuolated pollen stage, during which WT pollen transformed into large vacuolated microspores with degraded tapetal layer cells (Fig. 5f, g). However, most of the central vacuole in oscugt1-1 appeared deformed and shrunken (Fig. 5n, o). At the mature pollen stage, each anther lobe in WT was full with viable pollen grains that accumulated plentiful starch granules and nutrients on their surface (Fig. 5h). In contrast, the structures of the four anther lobes were highly abnormal in the oscugt1-1 mutant, as nearly all lobes failed to produce viable and plump pollen grains, instead presenting very shrunken and deformed pollen grains (Fig. 5p). Collectively, these data indicated that the male sterility phenotype observed in the oscugt1 mutant might be due to defective microspore development.
Histological analysis of wild-type (WT) and oscugt1 mutant microspores at eight developmental stages. Transverse sections of wild-type (WT) and oscugt1 single anther locules at the meiosis stage (a–c, i–k), the tetrad stage (d and l), the young microspore stage (e and m), the vacuolated pollen stage (f and m) and the pollen mitosis stage (g and o). Cross-sections of WT and oscugt1 mutant whole anthers with four locules at the mature pollen stage (h and p). BP, bicellular; DP, degraded microspore; M, microspore; MP, mature pollen, VP, vacuolated pollen. Bars, 20 µm (a–g, j–o); 40 µm (h, p)
Knockdown of OsCUGT1 by RNAi results in a reduction in rice height and fertility
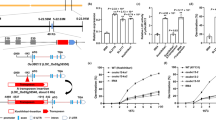
To further elucidate OsCUGT1’s functions in the regulation of plant height and spikelet fertility, the OsCUGT1-RNAi vector was constructed (Fig. 6a) and used to transform the rice ZH11. Based on hygromycin-resistance selection, twenty-five T0 RNAi plants were obtained. Semiquantitative PCR revealed that OsCUGT1 expression was knocked down in several RNAi lines (Fig. 6b). The height and seed setting rate of these plants decreased to varying degrees compared with those of the WT (Fig. 6c-f). In addition, two RNAi lines from the twenty-five T0 plants exhibited complete sterility, similar to oscugt1 (Fig. 6f). These results suggested that the RNAi lines exhibited a weaker morphological phenotype than the knockout oscugt1 mutants and demonstrated that OsCUGT1 plays a crucial role in the regulation of plant development.
The generation of OsCUGT1 RNAi transgenic plants through the RNA interference system. a pTCK303 was used as the binary vector for hpRNA-producing construction. A 360 bp fragment of OsCUGT1 was inserted into the sense/antisense orientation of pTCK303. b Relative expression levels of OsCUGT1 in the RNAi transgenic plants, as determined by semi-quantitative PCR. c Dwarf phenotype of the RNAi transgenic plants during the mature period. d Representative panicles of WT and RNAi transgenic plants. e Plant height of WT and nine independent RNAi transgenic lines. f Seed setting rate of WT and nine independent RNAi transgenic lines
RNA-seq analysis of the oscugt1 mutant
To unravel the underlying reason for the shorter stature and abnormal pollen grains in the oscugt1 mutant, we performed RNA-seq using the leaves (L) and spikelets (SP) of WT and oscugt1 mutant plants. From two comparison groups (WT-L vs. GT-L; WT-SP vs. GT-SP), 2108 and 4224 differentially expressed genes (DEGs) were identified, respectively (Fig. 7a). Only 375 genes were shared between groups “L” and “SP” (Fig. 7b), indicating that gene expression patterns and the regulatory pathway in leaves vs. spikelets are quite different. Pearson correlation analysis revealed that there was little variation among experimental replicates (Fig. 7c). Therefore, these RNA-seq data have high reliability and could be used for further analysis.
Transcriptome profiling of wild-type (WT) and oscugt1 plants. Transcriptome profiling of wild-type (WT) and oscugt1 plants. a Overview of differentially expressed genes (DEGs) in the leaves (L) and spikelets (SP) of WT and oscugt1 mutant plants (GT). b The DEGs were picked out from two comparison groups (WT-L vs. GT-L; WT-SP vs.GT-SP) to draw the Venn diagram. c Pearson correlation coefficients of RNA-seq data from all samples. d DEGs in phenylpropanoid pathway. The color in each cell indicates the value of the gene expression levels as fragments per kilobase of transcript per million mapped reads (FPKM). (e) Differential expression of OsPAL1 and Os4CL5 was analyzed based on the transcriptome data. f DEGs in cytokinin biosynthesis pathway. g DEGs in cytokinin degradation/inactivation pathway. The color in each cell indicates the value of FPKM. h Differential expression of OsCKX2 was analyzed based on the transcriptome data. i DEGs in sugar metabolism during pollen development. j Differential expression of OsUGP2 and OsINV4 was analyzed based on the transcriptome data. Values are presented as means ± SD (n = 3). Data were analyzed using Student’s t-test; *P < 0.05, **P < 0.01
Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway classification of the DEGs from the two comparison groups showed that a wide spectrum of biological processes and metabolic pathways were enriched (Fig. S2-S5). Further analysis revealed that many DEGs in the comparison group “WT-L vs. GT-L” are related to the biosynthesis of secondary metabolites (Fig. S3). Since phenylpropanoids are termed ‘secondary metabolites’, we noticed that a large number of phenylpropanoid pathway genes, such as several PHENYLALANINE AMMONIA LYASE (PAL) genes, including OsPAL1, OsPAL2, OsPAL3, OsPAL4, OsPAL6, OsPAL7 and CINNAMYL ALCOHOL DEHYDROGENASE (OsCAD2), OsCAD3, CAFFEIC ACID 3-O-METHYLTRANSFERASE (OsCOMT), 4-COUMARATE:COENZYME A LIGASE 1 (Os4CL1), Os4CL2, Os4CL3, Os4CL5, and FLAVANONE 3'-HYDROXYLASE (OsF3'H), which are involved in phenylpropanoid compound biosynthesis in rice, were all downregulated in the oscugt1 mutant relative to WT (Fig. 7d, e). These results suggested that the loss of OsCUGT1 function disturbs the transcript levels of genes encoding phenylpropanoid biosynthetic enzymes, thereby impairing the accumulation of related metabolites that are important modulators of plant growth and development.
Additionally, we found that many DEGs from the “WT-L vs. GT-L” group, which participate in cytokinin metabolism, were affected in the oscugt1 mutant. Genes involved in cytokinin biosynthesis, such as LONELY GUY OsLOG1 and OsLOG10, which encode cytokinin-activating enzymes (Kurakawa et al. 2007), were both repressed in oscugt1 (Fig. 7f). By contrast, genes functioning in cytokinin degradation/inactivation, such as cytokinin-O-glucosyltransferases (CGTs) and cytokinin oxidase/dehydrogenase (CKX) family genes, were highly expressed in the oscugt1 mutant compared to the WT (Fig. 7g, h). These results suggested that the loss of OsCUGT1 may affect the cytokinin levels in plant leaf cells.
Furthermore, DEG analysis was conducted in the comparison groups “WT-SP vs. GT-SP”. KEGG analysis revealed that the top three enriched pathways were metabolic pathways, biosynthesis of secondary metabolites and starch and sucrose metabolism (Fig. S5). Defective sugar metabolism during anther and pollen development is known to contribute to male sterility (Liu et al. 2021). Consistently, we found that a large number of genes involved in sugar and starch synthesis during microsporogenesis were significantly downregulated in the oscugt1 mutants (Fig. 7i, j). For example, SUCROSE PHOSPHATE SYNTHASE (OsSPS1), UDP-GLUCOSE PYROPHOSPHORYLASE GENE (OsUGP2), ANTHER-SPECIFIC CELL WALL INVERTASE GENE (OsINV4), and LARGE SUBUNIT OF ADP-GLUCOSE PYROPHOSPHORYLASE (OsAGPL4) were all significantly less expressed in oscugt1. In addition, the transcripts of several anther development-specific genes, including MALE STERILITY 1 (OsMS1), OsMS2, MONOSACCHARIDE TRANSPORTER GENE (OsMST5), RICE ANTHER-SPECIFIC GENE (OsRTS), and TAPETUM DEGENERATION RETARDATION (OsTDR), were all nearly abolished in the oscugt1 mutant (Fig. S6).
Together, these results indicated that loss of OsCUGT1 affects the expression of genes related to multiple biological processes, especially in the phenylpropanoid biosynthetic pathway, cytokinin homeostasis and pollen development.
Discussion
GTs are encoded by large multigene families, which sometimes comprise hundreds of genes in plants (Cao et al. 2008). Many identified GTs participate in essential processes, including pathogen defense (Ke et al. 2019; Zhang et al. 2022), stress adaptation (Li et al. 2017; Wang et al. 2022) and plant growth and development (Moon et al. 2013; Zhang et al. 2014, 2022). Although a large number of GTs have been functionally characterized in rice, the biological roles of OsCUGT1 remain unknown. In this study, we characterized OsCUGT1 and revealed its vital roles in rice height and spikelet fertility. Our results confirm the conserved function of OsCUGT1 in maintaining the balance of cellular phenylpropanoid metabolite levels and highlight its potential functions in cytokinin homeostasis and pollen development, especially in pollen starch and sugar metabolism.
It has been reported that many GTs exhibit stress induction and are involved in plant stress tolerance via different pathways (Li et al. 2017, 2020; Wang et al. 2022). Previously, OsCUGT1 was isolated as a cold-induced glycosyltransferase based on the cold treatment transcriptome library of Guizhou landrace rice PTWT. The analysis of transcript expression patterns in this study indicated that OsCUGT1 was indeed induced by multiple external stimuli (Fig. 1c), pointing to a possible role in environmental adaptation. qRT‒PCR analyses suggested that OsCUGT1 may function mainly in rice sheaths and panicles (Fig. 1b). In agreement, the oscugt1 knockout mutant exhibited distinct abnormalities, including dwarfism and sterility. The height of the main tillers in the oscugt1 mutant reached only three-fifths that of the WT (Fig. 2b). RNA-seq revealed that a large number of genes contributing to secondary metabolite biosynthesis, especially the phenylpropanoid pathway, showed significant changes in the leaves and spikelets of the oscugt1 mutant (Fig. 7). In the comparison group “WT-L vs. GT-L”, the transcript of OsPAL1, a key enzyme that acts as an entry point to the phenylpropanoid pathway (Howles et al. 1996), was downregulated in the leaves of oscugt1. The significant reductions in gene expression for several important genes encoding phenylpropanoid biosynthetic enzymes, including OsPALs, Os4CL, OsCOMT and OsF3’H in oscugt1, would therefore lead to a decreased supply of precursors for the phenylpropanoid pathway and may reduce the accumulation of downstream metabolic compounds and in turn influence rice development. It is not surprising to suggest such a hypothesis, as many reports have shown that glycosylation mediated by GTs exerts an important role in regulating phenylpropanoid metabolic flux redirection and governing shoot morphology, grain size and abiotic stress tolerance (Dong et al. 2020; Li et al. 2017; Yin et al. 2014).
It is noteworthy that inside the stem of oscugt1, both cell number and cell length were reduced (Fig. 3). qRT‒PCR confirmed that two important genes that function in cell division and expansion were downregulated, suggesting that both of them were affected in the oscugt1 mutant. Hormones, especially auxin and cytokinin, can regulate many aspects of growth and development, including cell elongation and division (Di Mambro et al. 2017; Street et al. 2016). A number of studies have also demonstrated the effects of phenylpropanoid metabolic compounds on auxin and CK metabolism in Arabidopsis and rice, resulting in growth pattern changes in the plant (Brown et al. 2001; Dong et al. 2020; Kurepa et al. 2018; Wang et al. 2011; Yin et al. 2014). For example, disrupted activity of two Arabidopsis UGTs (UGT78D1 and UGT78D2) would contribute to the accumulation of high levels of a kaempferol glycoside and lead to dwarfed plants by perturbing polar auxin transport (Yin et al. 2014). In addition, nucleotide variations in GSA1 reduce grain size by disturbing cell proliferation and expansion, which is the result of flavonoid-mediated imbalance of auxin homeostasis (Dong et al. 2020). Logically, the loss of OsCUGT1 would be expected to affect auxin-related gene expression; however, RNA-seq results showed that the transcripts of some representative genes that function in auxin polar transport, synthesis and response were insignificantly changed in the oscugt1 mutant (Fig. S7). These results indicated that OsCUGT1 regulated rice height by modulating cell division and expansion, which may not be achieved by affecting the auxin regulatory pathway.
Intriguingly, RNA-seq revealed that the relative expression levels of a number of cytokinin metabolism-related genes were notably affected in the oscugt1 mutant (Fig. 7). Several cytokinin biosynthesis-related genes, such as OsLOG1, OsLOG10 and LOC_Os04g44354, which function in the final step of bioactive cytokinin synthesis in rice (Kurakawa et al. 2007), were repressed in oscugt1. In contrast, genes that negatively regulate cytokinin accumulation and signaling (Ashikari et al. 2005), such as the OsCKX2, OsCKX11, OsCGT genes and cytokinin response regulator OsORR2, were all highly expressed in the oscugt1 mutant compared to the WT. OsCKX2 can modulate rice tillering, spikelet number and organ size by regulating cytokinin metabolism (Yeh et al. 2015). Reduced OsCKX2 expression increases tiller number and yield in transgenic rice. CGT genes, such as Arabidopsis UGT76C2, the endogenous cytokinin level and plant growth and development are affected in its overexpression lines (Wang et al. 2011). OsORR2 gene, a B-type cytokinin response regulator, the cytokinin signaling and rice height are affected in its overexpression lines (Shi et al. 2020). The higher expression levels of these putative cytokinin-inactivating genes could impair cytokinin homeostasis and contribute to cytokinin signaling disturbance, which would partially account for the dwarf phenotype and abnormal inflorescences of the oscugt1 mutant. Many studies have reported that the glycosylation modifications of cytokinins are mostly catalyzed by family 1 glycosyltransferases (Hou et al. 2004; Li et al. 2020). The update of the CAZy database suggested that OsCUGT1 belongs to glycosyl transferase family 1 (http://www.cazy.org/GlycosylTransferases.html). In addition, OsCUGT1 was upregulated by certain hormone treatments, especially by cytokinin (Fig. 1c). These results raised the question of whether OsCUGT1 is a cytokinin glycosyltransferase from rice. We attempted to examine the glycosyltransferase activity of OsCUGT1 by purifying a glutathione S-transferase (GST)-OsCUGT1 fusion protein after expression in Escherichia coli. Unfortunately, we failed to obtain the labeling protein for further enzymatic activity assays. To date, few cytokinin glycosyltransferase genes in rice have been reported (Li et al. 2020), possibly because of the difficulty of establishing prokaryotic protein expression and performing enzyme activity assays. Further attempts may help to solve this problem and elucidate the underlying mechanisms of how OsCUGT1 modulates the phenylpropanoid pathway and cytokinin metabolism.
In addition to dwarfism, we also noticed that oscugt1 was completely sterile. It shows defects at a late stage of pollen development, in which pollen initiates starch accumulation. Sugar metabolism is crucial for pollen maturation and viability (Liu et al. 2021). Many studies have shown that defects in sugar metabolism during pollen development often result in male sterility. For example, two pollen-preferential “late genes”, OsUGP2 and OsINV4, whose knockdown transgenic rice failed to accumulate sucrose and starch in pollen and thus contributed to sterility were observed (Mu et al. 2009; Oliver et al. 2005). RNA-seq analysis revealed that both OsUGP2 and OsINV4 and other important sugar metabolism-related genes were significantly repressed in oscugt1. Therefore, we concluded that the defective microspore phenotype in oscugt1 may arise because of abnormal sugar metabolism. These results also demonstrated that OsCUGT1 is necessary for pollen maturation, especially for starch and sugar accumulation in pollen grains.
In conclusion, the combination of phenotypic analysis, microscopic observations and transcriptome analyses in this study revealed that OsCUGT1 regulates rice height and spikelet fertility by modulating essential genes involved in the phenylpropanoid pathway, cytokinin homeostasis and pollen development.
References
Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309:741–745
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125:279–284
Bowles D (2002) A multigene family of glycosyltransferases in a model plant, Arabidopsis thaliana. Biochem Soc Trans 30:301–306
Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126:524–535
Cai ML, Chen R, Huang XZ, Zhao DG (2021) Cloning and mutant construction of cold-upregulated glycosyltransferase-like gene (OsCUGT1) from Guizhou landrace rice “Pingtang Heinuo” (Oryza sativa ssp. japonica). J Agri Biot 29:12–22
Cao PJ, Bartley LE, Jung KH, Ronald PC (2008) Construction of a rice glycosyltransferase phylogenomic database and identification of rice-diverged glycosyltransferases. Mol Plant 1:858–877
Dare AP, Yauk YK, Tomes S, McGhie TK, Rebstock RS, Cooney JM, Atkinson RG (2017) Silencing a phloretin-specific glycosyltransferase perturbs both general phenylpropanoid biosynthesis and plant development. Plant J 91:237–250
Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey PN, Busch W, Novak O, Ljung K, Di Paola L, Maree AFM, Costantino P, Grieneisen VA, Sabatini S (2017) Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc Natl Acad Sci U S A 114:E7641–E7649
Dong NQ, Sun Y, Guo T, Shi CL, Zhang YM, Kan Y, Xiang YH, Zhang H, Yang YB, Li YC, Zhao HY, Yu HX, Lu ZQ, Wang Y, Ye WW, Shan JX, Lin HX (2020) UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat Commun 11:2629
Gachon CM, Langlois-Meurinne M, Saindrenan P (2005) Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci 10:542–549
Heyl A, Schmulling T (2003) Cytokinin signal perception and transduction. Curr Opin Plant Biol 6:480–488
Hirose T, Hashida Y, Aoki N, Okamura M, Yonekura M, Ohto C, Terao T, Ohsugi R (2014) Analysis of gene-disruption mutants of a sucrose phosphate synthase gene in rice, OsSPS1, shows the importance of sucrose synthesis in pollen germination. Plant Sci 225:102–106
Hoedemaekers K, Derksen J, Hoogstrate SW, Wolters-Arts M, Oh SA, Twell D, Mariani C, Rieu I (2015) BURSTING POLLEN is required to organize the pollen germination plaque and pollen tube tip in Arabidopsis thaliana. New Phytol 206:255–267
Hou B, Lim EK, Higgins GS, Bowles DJ (2004) N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J Biol Chem 279:47822–47832
Howles PA, Sewalt VJ, Paiva NL, Elkind Y, Bate NJ, Lamb C, Dixon RA (1996) Overexpression of L-phenylalanine ammonia-lyase in transgenic tobacco plants reveals control points for flux into phenylpropanoid biosynthesis. Plant Physiol 112:1617–1624
Hu Y, Walker S (2002) Remarkable structural similarities between diverse glycosyltransferases. Chem Biol 9:1287–1296
Hu Q, Wang W, Lu Q, Huang J, Peng S, Cui K (2021) Abnormal anther development leads to lower spikelet fertility in rice (Oryza sativa L.) under high temperature during the panicle initiation stage. BMC Plant Biol 21:428
Huang X, Zeng X, Cai M, Zhao D (2022) The MSI1 member OsRBAP1 gene, identified by a modified MutMap method, is required for rice height and spikelet fertility. Plant Sci 320:111201
Jagadish SV, Craufurd PQ, Wheeler TR (2007) High temperature stress and spikelet fertility in rice (Oryza sativa L.). J Exp Bot 58:1627–1635
Ke S, Liu S, Luan X, Xie XM, Hsieh TF, Zhang XQ (2019) Mutation in a putative glycosyltransferase-like gene causes programmed cell death and early leaf senescence in rice. Rice (n y) 12:7
Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445:652–655
Kurepa J, Shull TE, Karunadasa SS, Smalle JA (2018) Modulation of auxin and cytokinin responses by early steps of the phenylpropanoid pathway. BMC Plant Biol 18:278
Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, Wen TQ, Huang H, Luo D, Ma H, Zhang DB (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18:2999–3014
Li P, Li YJ, Zhang FJ, Zhang GZ, Jiang XY, Yu HM, Hou BK (2017) The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J 89:85–103
Li Y, Liu F, Li P, Wang T, Zheng C, Hou B (2020) An Arabidopsis cytokinin-modifying glycosyltransferase UGT76C2 improves drought and dalt tolerance in rice. Front Plant Sci 11:560696
Liu F, Wang P, Zhang X, Li X, Yan X, Fu D, Wu G (2018) The genetic and molecular basis of crop height based on a rice model. Planta 247:1–26
Liu S, Li Z, Wu S, Wan X (2021) The essential roles of sugar metabolism for pollen development and male fertility in plants. Crop J 9:1223–1236
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19:1403–1414
Moon S, Kim SR, Zhao G, Yi J, Yoo Y, Jin P, Lee SW, Jung KH, Zhang D, An G (2013) Rice glycosyltransferase1 encodes a glycosyltransferase essential for pollen wall formation. Plant Physiol 161:663–675
Mu H, Ke J, Liu W, Zhuang C, Yip W (2009) UDP-glucose pyrophosphorylase2 (OsUgp2), a pollen-preferential gene in rice, plays a critical role in starch accumulation during pollen maturation. Chin Sci Bull 54:234–243
Nubankoh P, Wanchana S, Saensuk C, Ruanjaichon V, Cheabu S, Vanavichit A, Toojinda T, Malumpong C, Arikit S (2020) QTL-seq reveals genomic regions associated with spikelet fertility in response to a high temperature in rice (Oryza sativa L.). Plant Cell Rep 39:149–162
Oliver SN, Van Dongen JT, Alfred SC, Mamun EA, Zhao X, Saini HS, Fernandes SF, Blanchard CL, Sutton BG, Geigenberger P (2005) Cold-induced repression of the rice anther-specific cell wall invertase gene OsINV4 is correlated with sucrose accumulation and pollen sterility. Plant Cell Environ 28:1534–1551
Peng M, Shahzad R, Gul A, Subthain H, Shen S, Lei L, Zheng Z, Zhou J, Lu D, Wang S, Nishawy E, Liu X, Tohge T, Fernie AR, Luo J (2017) Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat Commun 8:1975
Qi B, Wu C (2022) Potential roles of stigma exsertion on spikelet fertility in rice (Oryza sativa L.) under heat stress. Front Plant Sci 13:983070
Rehman HM, Khan UM, Nawaz S, Saleem F, Ahmed N, Rana IA, Atif RM, Shaheen N, Seo H (2022) Genome wide analysis of family-1 UDP glycosyltransferases in populus trichocarpa specifies abiotic stress responsive glycosylation mechanisms. Genes (Basel) 13:1640
Shi F, Wang M, An Y (2020) Overexpression of a B-type cytokinin response regulator (OsORR2) reduces plant height in rice. Plant Signal Behav 15:1780405
Spielmeyer W, Ellis MH, Chandler PM (2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci U S A 99:9043–9048
Street IH, Mathews DE, Yamburkenko MV, Sorooshzadeh A, John RT, Swarup R, Bennett MJ, Kieber JJ, Schaller GE (2016) Cytokinin acts through the auxin influx carrier AUX1 to regulate cell elongation in the root. Development 143:3982–3993
Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, Matsuoka M (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437:693–698
Wang Y, Li J (2008) Molecular basis of plant architecture. Annu Rev Plant Biol 59:253–279
Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22:409–417
Wang J, Ma XM, Kojima M, Sakakibara H, Hou BK (2011) N-glucosyltransferase UGT76C2 is involved in cytokinin homeostasis and cytokinin response in Arabidopsis thaliana. Plant Cell Physiol 52:2200–2213
Wang B, Smith SM, Li J (2018) Genetic regulation of shoot architecture. Annu Rev Plant Biol 69:437–468
Wang T, Li XK, Liu X, Yang XQ, Li YJ, Hou BK (2022) Rice glycosyltransferase gene UGT2 functions in salt stress tolerance under the regulation of bZIP23 transcription factor. Plant Cell Rep 38:3883
Xiao Y, Zhang J, Yu G, Lu X, Mei W, Deng H, Zhang G, Chen G, Chu C, Tong H, Tang W (2020) Endoplasmic reticulum-localized PURINE PERMEASE1 regulates plant height and grain weight by modulating cytokinin distribution in rice. Front Plant Sci 11:618560
Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327
Yan H, Sun H, Jia X, Lv C, Li J, Zhao Q (2020) Phenotypic, transcriptomic, and metabolomic signatures of root-specifically overexpressed OsCKX2 in rice. Front Plant Sci 11:575304
Yang H, Liu L, Wu K, Liu S, Liu X, Tian Y, Wang Y, Duan E, Lei J, Bao X (2022) FLOURY AND SHRUNKEN ENDOSPERM6 encodes a glycosyltransferase and is essential for the development of rice endosperm. J Plant Biol 65:187–198
Yeh SY, Chen HW, Ng CY, Lin CY, Tseng TH, Li WH, Ku MS (2015) Down-regulation of cytokinin oxidase 2 expression increases tiller number and improves rice yield. Rice (n y) 8:36
Yin R, Han K, Heller W, Albert A, Dobrev PI, Zazimalova E, Schaffner AR (2014) Kaempferol 3-O-rhamnoside-7-O-rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytol 201:466–475
Zeng D, Tian Z, Rao Y, Dong G, Yang Y, Huang L, Leng Y, Xu J, Sun C, Zhang G, Hu J, Zhu L, Gao Z, Hu X, Guo L, Xiong G, Wang Y, Li J, Qian Q (2017) Rational design of high-yield and superior-quality rice. Nat Plants 3:17031
Zhang B, Zhao T, Yu W, Kuang B, Yao Y, Liu T, Chen X, Zhang W, Wu AM (2014) Functional conservation of the glycosyltransferase gene GT47A in the monocot rice. J Plant Res 127:423–432
Zhang D, Wang Z, Yamamoto N, Wang M, Yi X, Li P, Lin R, Nasimi Z, Okada K, Mochida K, Noutoshi Y, Zheng A (2022) Secreted glycosyltransferase RsIA_GT of Rhizoctonia solani AG-1 IA inhibits defense responses in Nicotiana benthamiana. Pathogens 11:1026
Zhou XD, Zhou J, Zhang XS, Jiang XD, Yang LX, Wang YL (2017) Progress in decoding the impact of abiotic stress on spikelet fertility in rice. Ying Yong Sheng Tai Xue Bao 28:4127–4133
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 31500219 and 31960615).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest in the authorship and publication of this document.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, L., Liu, H., Peng, K. et al. Cold-upregulated glycosyltransferase gene 1 (OsCUGT1) plays important roles in rice height and spikelet fertility. J Plant Res 136, 383–396 (2023). https://doi.org/10.1007/s10265-023-01455-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-023-01455-7