Abstract
Mycorrhizal symbiosis between plants and fungi is ubiquitous, and has been played key roles in plant terrestrialization and diversification. Although arbuscular mycorrhizal (AM) symbioses with Glomeromycotina fungi have long been recognized as both ancient and widespread symbionts, recent studies showed that Mucoromycotina fungi were also ancestral symbionts and would thus be expected to co-exist with many land plants. To explore whether Mucoromycotina colonize fern gametophytes, we subjected fungal associations with gametophytes of two distantly related ferns, Angiopteris lygodiifolia (Marattiales) and Osmunda japonica (Osmundales), to molecular analysis. Direct PCR amplification from intracellular hyphal coils was also performed. We detected Mucoromycotina sequences in the gametophytes of A. lygodiifolia and O. japonica at rates of 41% (7/17) and 50% (49/98) of gametophytes, respectively, and assigned them to 10 operational taxonomic units of Endogonales lineages. In addition, we used AM fungal-specific primers and detected Glomeromycotina sequences in all individuals examined. The results suggest that Glomeromycotina and Mucoromycotina colonized fern gametophytes simultaneously. We found that Mucoromycotina were present in fern gametophytes of Marratiales and Osmundales, which implies that a variety of fern taxa have Mucoromycotina associations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mycorrhizal symbiosis between plants and fungi is ubiquitous; it is evident in 85% of land plants (Brundrett and Tedersoo 2018) and has played key roles in plant terrestrialization and diversification during evolution (Pirozynski and Malloch 1975; Strullu-Derrien et al. 2018). This plant–fungal association involves the fungal phyla Mucoromycota, Basidiomycota, and Ascomycota (Spatafora et al. 2016; Wang and Qiu 2006). Arbuscular mycorrhizal (AM) fungi, which are the most common plant-associated fungi of the Mucoromycota subphylum Glomeromycotina, are associated with 74% of land plants, including liverworts, pteridophytes, and gymno- and angiosperms; the Basidiomycota and Ascomycota form mycorrhizae with specific plant lineages such as ectomycorrhiza, ericoid, and orchid mycorrhiza (van der Heijden et al. 2015). The presence of fossil AM in Early Devonian Rhynie Chert plants has long supported the suggestion that Glomeromycotina associations are ancestral mycorrhizal symbiosis (Remy et al. 1994; Taylor et al. 1995).
However, recent studies suggest that symbiotic associations with members of the Mucoromycota subphylum Mucoromycotina are also widespread among land plant lineages (Hoysted et al. 2018). An endophytic Mucoromycotina association was first recognized in the basal liverwort Haplomitriopsida, which associated exclusively with Mucoromycotina fungi (Bidartondo et al. 2011). The fossil evidence of dual colonization by Glomeromycotina and Mucoromycotina in the Rhynie Chert plant Horneophyton indicates that Mucoromycotina were also the ancestral symbionts, and that the associations between ancient plants and symbiotic fungi were more diverse than previously assumed (Field and Pressel 2018; Strullu-Derrien et al. 2014).
Although Mucoromycotina symbiosis was initially largely considered to be restricted to early diverging non-vascular plants, except for one occurrence in the fern Anogramma leptophylla (Bidartondo et al. 2011), it was shown to be widespread in non-Haplomitriopsida liverworts (Field et al. 2016), hornworts (Desirò et al. 2013), and lycophytes (Rimington et al. 2015), which were often simultaneously colonized with AM fungi. More recently, fine root endophytes (FRE) that were previously thought to be Glomeromycotina due to their arbuscule-forming feature were reclassified into Mucoromycotina based on molecular phylogenetic analysis (Orchard et al. 2017a). These fungi are distinguished from AM fungi by small-diameter fungal hyphae and vesicles and detected in many vascular plant families, including ferns and flowering plants (Orchard et al. 2017b).
Ferns are of great interest to those studying symbiotic mycorrhizal evolution in vascular plants. Their life cycle typically features two independent generations, the gametophyte and sporophyte. Gametophytes grow independently of sporophytes and must acquire nutrients in the absence of a root system, indicating that fungal associations may play important roles in nutrient and water uptake. Although both fern gametophytes and sporophytes have long been recognized as AM plants (Boullard 1957; Gemma et al. 1992; Kessler et al. 2010), the sporophyte of several ferns in the Equisetales (Hodson et al. 2009) and Polypodiales (Cooper 1976; Hall 1977; Turnau et al. 1999) often had FRE in their roots. Molecular evidence of a Mucoromycotina association was demonstrated in Anogramma leptophylla (Polypodiales) (Rimington et al. 2015), while no Mucoromycotina were detected in fern sporophytes of another 17 species using molecular and microscopic methods (Rimington et al. 2015). In gametophytes, FRE was successfully colonized in Pellaea viridis (Polypodiales) under cultivated condition (Turnau et al. 2005), but little is understood of the symbiotic relationships between fern gametophytes and Mucoromycotina fungi (Pressel et al. 2016).
Here, we performed a molecular study of fungal associations within fern gametophytes to test whether fern gametophytes are associated with Mucoromycotina. We studied gametophytes of Angiopteris lygodiifolia Rosenst. (Marattiales) and Osmunda japonica Thunb. (Osmundales), both of which we previously showed to have AM associations (Ogura-Tsujita et al. 2013, 2016).
Materials and methods
Plant materials
The DNA samples of Angiopteris lygodiifolia gametophytes that we used previously to detect AM fungi (Ogura-Tsujita et al. 2013) were employed for the molecular identification of Mucoromycotina (Table 1). Gametophytes of Osmunda japonica were newly collected at four sites in Japan (Table 1). Gametophytes were growing on exposed soil surfaces along forest road side. Collected gametophytes were washed in water; the rhizoids were removed using tweezers under a stereomicroscope to avoid contamination by surface-inhabiting fungi.
DNA analysis
Osmunda gametophytes were crushed in a ball mill and DNA was extracted using a DNeasy Plant Mini Kit (Qiagen, Valencia, California, USA). Fungal 18S ribosomal DNA was amplified using the Mucoromycotina-specific primers EndAD1f and EndAD2r (Desirò et al. 2013), Ampdirect Plus (Shimadzu, Kyoto, Japan), and TaKaRa Ex Taq DNA polymerase (Takara-Bio, Shiga, Japan) following the manufacturers’ protocols. PCR initially proceeded at 94 °C for 3 min, followed by 40 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 90 s, and a final extension at 72 °C for 7 min. For samples that had insufficient PCR amplification, a second round of PCR was performed with 20 cycles under the same conditions using 1 μL of first round PCR product. PCR amplicons were purified using the ExoProStar system (GE Healthcare, Buckinghamshire, UK) and sequenced on an Applied Biosystems 3500×L genetic analyzer using a BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA, USA). The samples whose PCR products were difficult to sequence directly were amplified again from extracted DNA by using PrimeStar GXL DNA Polymerase (Takara-Bio) and cloned into the pGEM-T vector (Promega, Madison, WI, USA). Between 4 and 26 colonies were sequenced for each clone.
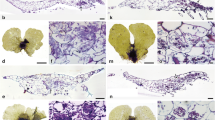
As shown by Ogura-Tsujita et al. (2013) mycorrhizal fungi form intracellular hyphal coils in the inner tissues of the thick midribs of Angiopteris and Osmunda gametophytes. Amplification of total plant DNA often yields amplicons from non-symbiotic surface-inhabiting fungi. To avoid this, the intracellular hyphal coils from 51 Osmunda gametophytes obtained at site OJ4 were PCR-amplified. The gametophytes were cut into pieces, and hyphal coils were released into TE buffer. Single coils were collected using a micropipette and 0.5 μL TE buffer under a stereomicroscope, transferred to 0.2-mL tubes containing PCR mix, and PCR-amplified as detailed above. Residual gametophyte tissues were subjected to DNA extraction using a DNeasy Plant Mini Kit as described above, followed by routine PCR amplification.
The sequences were compared to GenBank data using BLAST (http://www.ncbi.nlm.nih.gov/BLAST) to discover taxonomic affinities and then clustered into operational taxonomic units (OTUs) at the 97% sequence similarity level. The Mucoromycotina sequences determined were deposited in the DNA Data Bank of Japan (DDBJ) under accession numbers LC429228 to LC429294. For phylogenetic analysis, these sequences and GenBank reference sequences were aligned using the MUSCLE program (Edgar 2004) of MEGA ver. 6.06 software (Tamura et al. 2013), followed by manual adjustment. Alignment gaps were treated as missing data. Maximum likelihood analysis was performed with raxmlGUI ver. 1.31 software (Silvestro and Michalak 2012) using the GTR +G model. Support values were estimated via 1,000 bootstrap replicates using the rapid bootstrap option. Mortierella chlamydospora and Basidiobolus ranarum served as the outgroup taxa.
To confirm co-symbiosis with AM and Mucoromycotina, PCR amplification using the AM-specific primer pair NS31/AML2 (Lee et al. 2008; Simon et al. 1992) was also performed on DNA from eight Osmunda gametophytes containing Mucoromycotina sequences (Table S1). All PCR products were cloned and sequenced as described above. Between 2 and 17 colonies of each clone were sequenced, and the sequences were assigned to virtual taxa (VTX) defined by ≥ 97% sequence similarities using the MaarjAM database (Öpik et al. 2010) that includes published AM fungal sequences and creates virtual taxa at roughly the level of species.
All collected gametophytes were identified by reference to their chloroplast rbcL sequences; gametophytes are difficult to distinguish morphologically. PCR amplification was performed as described by Ebihara et al. (2008) using the primer pair rbcL1-1 (Hasebe et al. 1995) and rbcLHIR1 (Ebihara et al. 2002). The sequences were compared to those from sporophytes of A. lygodiifolia and O. japonica.
Results
We detected Mucoromycotina sequences in the gametophytes of A. lygodiifolia and O. japonica at rates of 41% (7/17) and 50% (49/98), respectively (Tables 1, S1). Direct PCR amplification from fungal coils also detected Mucoromycotina sequences in Osmunda gametophytes from 15 of 51 individuals sampled at site OJ4. The Mucoromycotina sequences were assigned to 10 OTUs using the 97% sequence similarity criterion (Fig. 1). OTU6 was the most common (22%; 15 sequences), and OTU10 was widely distributed in Osmunda gametophytes from all collection sites. Fungal infection of a single gametophyte by two or three OTUs was observed in nine individuals, either via cloning or combined analysis of normal and fungal coil PCR-amplified products (Table S1).
Phylogenetic analysis indicated that the 10 OTUs belonged to several Endogonales lineages, including the Endogonaceae and Densosporaceae (Fig. 2). OTU1 was distantly related to these two families and clustered with Mucoromycotina group I of Desirò et al. (2013). OTU8 clustered with Mucoromycotina sequences from Lycopodiella inundata (Rimington et al. 2015) (98–99% sequence identity). The sequence from Anogramma leptophylla (KJ952232) was the sister group of OTU7 (98% sequence similarity). OTU1, OTU6, and OTU9 included Mucoromycotina from liverworts (Bidartondo et al. 2011) and hornworts (Desirò et al. 2013) (98–99% sequence similarity).
A maximum likelihood tree constructed using the 18S ribosomal DNA sequences of Mucoromycotina fungi derived from fern gametophytes and GenBank. Bootstrap values ≥ 50% are shown on the branches, and those ≥ 70% are indicated by thick lines. Mortierella chlamydospora and Basidiobolus ranarum served as the outgroup taxa. The fungal sequences obtained in this study are shown by filled squares (Angiopteris lygodiifolia) or filled circles (Osmunda japonica). Open squares denote symbionts of liverworts or hornworts; open circles those of lycophytes; triangles those of ferns; and diamonds ectomycorrhiza. Country and GenBank accession number are shown in parentheses. AS Ascension Island, AU Australia, CA Canada, CL Chile, CN China, EN England, FR France, IT Italy, JP Japan, MX Mexico, MY Malaysia, NZ New Zealand, SA South Africa, SC Scotland, US United States
Arbuscular mycorrhizal colonization was confirmed in both fern species using an AM-specific primer set. Previously, we showed that seven Angiopteris gametophytes containing Mucoromycotina sequences (Tables 1, S1) host AM fungi (Ogura-Tsujita et al. 2013). Of the Osmunda gametophytes, all eight individuals that harbored Mucoromycotina sequences yielded AM sequences that were grouped into 12 VTXs based on the MaarjAM database (Öpik et al. 2010); all were members of the orders in Glomeromycotina, the Glomerales, Archaeosporales, and Paraglomerales (Table S1).
Discussion
Our results clearly demonstrated that Mucoromycotina fungi form associations with fern gametophytes. Mucoromycotina sequences were detected in 41–50% of the gametophytes of two fern species (Table 1), and direct PCR amplification from intracellular fungal coils also detected the fungal sequences (Table S1). All examined gametophytes of A. lygodiifolia and O. japonica hosting Mucoromycotina also yielded AM sequences (Table S1), which indicates that Mucoromycotina are mostly associated with gametophytes in combination with Glomeromycotina.
A comprehensive survey of Mucoromycotina in 199 hornwort samples (more than 20 species in 10 genera from six continents) revealed that 40% of the samples harbored Mucoromycotina sequences, and 25% harbored both Glomeromycotina and Mucoromycotina (Desirò et al. 2013). Molecular and cytological analyses of 20 lycophyte and 18 fern species showed that Mucoromycotina were present in 13% and 3% of sporophyte samples, respectively, but only a single fern sample (1 of 17 roots of Anogramma leptophylla) contained both fungal sequences (Rimington et al. 2015). Our detection rates were considerably higher for both Angiopteris and Osmunda gametophytes. We previously showed that 92% (48/52) of Angiopteris and 92% (35/38) of Osmunda gametophytes contained AM sequences (Ogura-Tsujita et al. 2013); 41–50% of gametophytes generated Mucoromycotina sequences in the present study. These results imply that gametophytes of A. lygodiifolia and O. japonica are associated principally with Glomeromycotina, but nearly half are also associated with Mucoromycotina, often co-existing with Glomeromycotina. In a pot-culture experiment using the fern Pellaea viridis, when the gametophytes and sporophytes were inoculated with both Glomeromycotina (Rhizophagus intraradices) and Mucoromycotina (Planticonsortium tenue; i.e., FRE; Walker et al. 2018), almost 90% of the gametophytes were colonized by Mucoromycotina, but the sporophyte roots mainly hosted Glomeromycotina in up to 90% of the entire root length (Turnau et al. 2005). Considering the high Mucoromycotina detection rates of the present study, it is possible that fern gametophytes prefer Mucoromycotina symbionts to a greater extent than do sporophytes. Indeed, all fern sporophytes analyzed by Rimington et al. (2015) were exclusively colonized by Glomeromycotina fungi, with one exception of Anogramma. The frequent association with Mucoromycotina in gametophytes may be attributed to the difference in the plant organ that hosts the mycorrhizal fungi. While the root is the host organ in sporophytes, mycorrhizal fungi mainly colonize a multilayered portion called the cushion in cordate gametophytes (Ogura-Tsujita et al. 2013, 2016); this is similar to mycorrhizal hornworts and liverworts in that fungal hyphae are mainly observed in the central parts of thalli (Desirò et al. 2013; Field et al. 2015). However, a difference in environment or stress conditions may also affect the frequency of Mucoromycotina associations. FRE are often dominant under extreme environmental conditions, such as low temperature and waterlogging (Orchard et al. 2017b). The fern sporophyte of Dryopteris carthusiana, liverworts, and Oxalis growing on the banks of a stream ravine were highly colonized by FRE (Turnau et al. 1999). Small fern gametophytes are frequently exposed to strong physical stresses, which may result in high Mucoromycotina colonization in gametophytes. Further research comparing gametophytes and sporophytes is required.
We detected a range of Mucoromycotina fungi (of 10 OTUs) within two families of the Endogonales and Glomeromycotina (of 12 VTXs) across three fungal orders, implying that Angiopteris and Osmunda gametophytes are associated with a phylogenetically diverse group of fungi. The presence of several Mucoromycotina and Glomeromycotina taxa within single gametophytes implies that individual gametophytes host phylogenetically distant fungi of the Mucoromycotina and Glomeromycotina. The Mucoromycotina sequences detected in this study were closely related to those of the liverworts, hornworts, and lycophytes of Asia to Oceania, Africa, and Europe (Fig. 2). This implies that plant-symbiotic Mucoromycotina are distributed worldwide and are minimally specific for host plants. Plant-symbiotic Mucoromycotina belong to the Endogonales, which consists of two families (Endogonaceae and Densosporaceae) (Desirò et al. 2017); we detected both families. Although the fungal sequences of OTU2 and OTU9 were closely related to those of fungal sporocarps of known Endogone and Densospora species, other OTUs clustered with plant symbionts; in particular, OTU1 contained only plant symbionts distinct from all other known species. Our results imply that only a few Endogonales species are currently recognized based on the fungal sporocarps and that many symbiotic lineages remain undescribed, probably because they infrequently engage in sporocarp formation (Walker et al. 2018). The growing number of DNA sequences available from plants will reveal the phylogenetic diversity of the Mucoromycotina.
We also attempted the morphological identification of fungal hyphae in plant tissues. Mucoromycotina forms characteristic coarse intercellular hyphae in Haplomitriopsida liverworts (Duckett et al. 2006; Field et al. 2015), hornworts (Desirò et al. 2013) and the gametophytes of lycophytes (Duckett and Ligrone 1992; Schmid and Oberwinkler 1993), but such distinctive intercellular colonization was absent in fern roots (Rimington et al. 2015). FRE are distinguished from AM fungi by having much finer hyphae (0.5–4 μm diameter) in both non-vascular and vascular plants (Walker et al. 2018). Our earlier microscopic analysis of Angiopteris and Osmunda gametophytes revealed intracellular hyphal coils with arbuscules, which is typical of Glomeromycotina (Ogura-Tsujita et al. 2013), and the absence of intercellular Mucoromycotina colonization. However, our microscopic analysis found intracellular colonization by fine fungal hyphae (0.8–2.4 μm diameter) in Osmunda gametophytes (Fig. S1: Individual no. MOsmG05). This suggests that Mucoromycotina are present as FRE in fern gametophytes. Since FRE were also detected in the sporophyte roots of Equisetales (Hodson et al. 2009) and Pteridaceae and Dennstaedtiaceae of Polypodiales (Cooper 1976; Hall 1977; Turnau et al. 1999, 2005), Mucoromycotina associations may occur in a wide range of fern lineages.
In conclusion, our results demonstrate, for the first time using a molecular approach, that fern gametophytes can associate with both Glomeromycotina and Mucoromycotina. The detected Mucoromycotina fungi were closely related to those from other plant lineages. This study supports recent findings that Mucoromycotina associations are not restricted to non-vascular plants, but also occur in early diverging vascular plants (Rimington et al. 2015). Considering that FRE were detected in a wide range of vascular plant families (Orchard et al. 2017b), this association may well be widespread across the land plant phylogeny.
References
Bidartondo MI, Read DJ, Trappe JM, Merckx V, Ligrone R, Duckett JG (2011) The dawn of symbiosis between plants and fungi. Biol Lett 7:574–577
Boullard B (1957) La mycotrophie chez les pteridophytes. Sa frequence, ses caracteres, sa signification. Botaniste 41:5–185
Brundrett MC, Tedersoo L (2018) Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol 220:1108–1115
Cooper KM (1976) A field survey of mycorrhizas in New Zealand ferns. N Z J Bot 14:169–181
Desirò A, Duckett JG, Pressel S, Villarreal JC, Bidartondo MI (2013) Fungal symbioses in hornworts: a chequered history. Proc R Soc B 280:20130207
Desirò A, Rimington WR, Jacob A, Pol NV, Smith ME, Trappe JM, Bidartondo MI, Bonito G (2017) Multigene phylogeny of Endogonales, an early diverging lineage of fungi associated with plants. IMA Fungus 8:245–257
Duckett JG, Ligrone R (1992) A light and electron microscope study of the fungal endophytes in the sporophyte and gametophyte of Lycopodium cernuum with observations on the gametophyte–sporophyte junction. Can J Bot 70:58–72
Duckett JG, Carafa A, Ligrone R (2006) A highly differentiated glomeromycotean association with the mucilage-secreting, primitive antipodean liverwort Treubia (Treubiaceae): clues to the origins of mycorrhizas. Am J Bot 93:797–813
Ebihara A, Iwatsuki K, Kurita S, Ito M (2002) Systematic position of Hymenophyllum rolandi-principis Rosenst. or a monotypic genus Rosenstockia Copel. (Hymenophyllaceae) endemic to New Caledonia. Acta Phytotax Geobot 53:35–49
Ebihara A, Farrar DR, Ito M (2008) The sporophyte-less filmy fern of eastern North America Trichomanes intricatum (Hymenophyllaceae) has the chloroplast genome of an Asian species. Am J Bot 95:1645–1651
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Field KJ, Pressel S (2018) Unity in diversity: structural and functional insights into the ancient partnerships between plants and fungi. New Phytol 220:996–1011
Field KJ, Rimington WR, Bidartondo MI, Allinson KE, Beerling DJ, Cameron DD, Duckett JG, Leake JR, Pressel S (2015) First evidence of mutualism between ancient plant lineages (Haplomitriopsida liverworts) and Mucoromycotina fungi and its response to simulated Palaeozoic changes in atmospheric CO2. New Phytol 205:743–756
Field KJ, Rimington WR, Bidartondo MI, Allinson KE, Beerling DJ, Cameron DD, Duckett JG, Leake JR, Pressel S (2016) Functional analysis of liverworts in dual symbiosis with Glomeromycota and Mucoromycotina fungi under a simulated Palaeozoic CO2 decline. ISME J 10:1514–1526
Gemma JN, Koske RE, Flynn T (1992) Mycorrhizae in Hawaiian pteridophytes: occurrence and evolutionary significance. Am J Bot 79:843–852
Hall IR (1977) Species and mycorrhizal infections of New Zealand Endogonaceae. Trans Br Mycol Soc 68:341–356
Hasebe M, Wolf PG, Pryer KM, Ueda K, Ito M, Sano R, Gastony GJ, Yokoyama J, Manhart JR, Murakami N, Crane EH, Haufler CH, Hauk WD (1995) Fern phylogeny based on rbcL nucleotide sequences. Am Fern J 85:134–181
Hodson E, Shahid F, Basinger J, Kaminskyj S (2009) Fungal endorhizal associates of Equisetum species from Western and Arctic Canada. Mycol Progr 8:19–27
Hoysted GA, Kowal J, Jacob A, Rimington WR, Duckett JG, Pressel S, Orchard S, Ryan MH, Field KJ, Bidartondo MI (2018) A mycorrhizal revolution. Curr Opin Plant Biol 44:1–6
Kessler M, Jonas R, Cicuzza D, Kluge J, Piątek K, Naks P, Lehnert M (2010) A survey of the mycorrhization of Southeast Asian ferns and lycophytes. Plant Biol 12:788–793
Lee J, Lee S, Young JPW (2008) Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol Ecol 65:339–349
Ogura-Tsujita Y, Sakoda A, Ebihara A, Yukawa T, Imaichi R (2013) Arbuscular mycorrhiza formation in cordate gametophytes of two ferns, Angiopteris lygodiifolia and Osmunda japonica. J Plant Res 126:41–50
Ogura-Tsujita Y, Hirayama Y, Sakoda A, Suzuki A, Ebihara A, Morita N, Imaichi R (2016) Arbuscular mycorrhizal colonization in field-collected terrestrial cordate gametophytes of pre-polypod leptosporangiate ferns (Osmundaceae, Gleicheniaceae, Plagiogyriaceae, Cyatheaceae). Mycorrhiza 26:87–97
Öpik M, Vanatoa A, Vanatoa E, Moora M, Davison J, Kalwij JM, Reier U, Zobel M (2010) The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol 188:223–241
Orchard S, Hilton S, Bending GD, Dickie IA, Standish RJ, Gleeson DB, Jeffery RP, Powell JR, Walker C, Bass D, Monk J, Simonin A, Ryan MH (2017a) Fine endophytes (Glomus tenue) are related to Mucoromycotina, not Glomeromycota. New Phytol 213:481–486
Orchard S, Standish RJ, Dickie IA, Renton M, Walker C, Moot D, Ryan MH (2017b) Fine root endophytes under scrutiny: a review of the literature on arbuscule-producing fungi recently suggested to belong to the Mucoromycotina. Mycorrhiza 27:619–638
Pirozynski KA, Malloch DW (1975) The origin of land plants: a matter of mycotrophism. Biosystems 6:153–164
Pressel S, Bidartondo MI, Field KJ, Rimington WR, Duckett JG (2016) Pteridophyte fungal associations: current knowledge and future perspectives. J Syst Evol 54:666–678
Remy W, Taylor TN, Hass H, Kerp H (1994) Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA 91:11841–11843
Rimington WR, Pressel S, Duckett JG, Bidartondo MI (2015) Fungal associations of basal vascular plants: reopening a closed book? New Phytol 205:1394–1398
Schmid E, Oberwinkler F (1993) Mycorrhiza-like interaction between the achlorophyllous gametophyte of Lycopodium clavatum L. and its fungal endophyte studied by light and electron microscopy. New Phytol 124:69–81
Silvestro D, Michalak I (2012) raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 12:335–337
Simon L, Lalonde M, Bruns TD (1992) Specific amplification of 18S fungal ribosomal genes from vesicular–arbuscular endomycorrhizal fungi colonizing roots. Appl Environ Microbiol 58:291–295
Spatafora JW, Chang Y, Benny GL et al (2016) A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108:1028–1046
Strullu-Derrien C, Kenrick P, Pressel S, Duckett JG, Rioult JP, Strullu DG (2014) Fungal associations in Horneophyton ligneri from the Rhynie Chert (c. 407 million year old) closely resemble those in extant lower land plants: novel insights into ancestral plant–fungus symbioses. New Phytol 203:964–979
Strullu-Derrien C, Selosse MA, Kenrick P, Martin FM (2018) The origin and evolution of mycorrhizal symbioses: from palaeomycology to phylogenomics. New Phytol 220:1012–1030
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Taylor TN, Remy W, Hass H, Kerp H (1995) Fossil arbuscular mycorrhizae from the Early Devonian. Mycologia 87:560–573
Turnau K, Ronikier M, Unrug J (1999) Role of mycorrhizal links between plants in establishment of liverworts thalli in natural habitats. Acta Soc Bot Pol 68:63–68
Turnau K, Anielska T, Jurkiewicz A (2005) Mycothallic/mycorrhizal symbiosis of chlorophyllous gametophytes and sporophytes of a fern, Pellaea viridis (Forsk.) Prantl (Pellaeaceae, Pteridales). Mycorrhiza 15:121–128
van der Heijden MGA, Martin FM, Selosse MA, Sanders IR (2015) Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol 205:1406–1423
Walker C, Gollotte A, Redecker D (2018) A new genus, Planticonsortium (Mucoromycotina), and new combination (P. tenue), for the fine root endophyte, Glomus tenue (basionym Rhizophagus tenuis). Mycorrhiza 28:213–219
Wang B, Qiu YL (2006) Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16:299–363
Acknowledgements
The authors thank K. Hashimoto, A. Sakoda, and A. Suzuki for collecting samples and for their technical assistance. We are grateful to two anonymous reviewers for their comments on the manuscript. This study was supported by JSPS KAKENHI Grants 17K07536 and 18K06391.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ogura-Tsujita, Y., Yamamoto, K., Hirayama, Y. et al. Fern gametophytes of Angiopteris lygodiifolia and Osmunda japonica harbor diverse Mucoromycotina fungi. J Plant Res 132, 581–588 (2019). https://doi.org/10.1007/s10265-019-01121-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-019-01121-x