Abstract
MicroRNAs (miRNAs) are endogenous small RNAs of −21 nucleotides that play an important role in diverse plant physiological processes at the post-transcriptional level by directing mRNA cleavage or translational inhibition. Previous studies have indicated that down-regulation of miR398 in response to oxidative stress allows up-regulation of the two target genes, cytosolic CSD1 and chloroplastic CSD2 (copper/zinc superoxide dismutase), resulting in protecting the plants to tolerate oxidative stress. In this study, we provide evidence that grapevine miR398 (Vv-miR398), by regulating the expression of its target genes, VvCSD1 and VvCSD2, mediates responses of grapevine to copper (Cu) stress which have been magnified due to increase in Cu-containing pesticide application. The expression of Vv-miR398 was inhibited by different concentrations of Cu stress; on the other hand, there was a steady increase in the activity of VvCSD1 and VvCSD2 genes. The function of VvCSD1 and VvCSD2 under Cu stress was thoroughly examined by overexpressing the use of the VvCSD1 and VvCSD2 in transgenic tobacco (Nicotiana tabacum). We found that both the overexpressed transgenic lines had lower Cu sensitivity and higher Cu tolerance compared with the wild type. In addition, lower levels of ROS and higher levels of SOD activities were accumulated in the transgenic lines in comparison with the wild type under the higher Cu conditions. Furthermore, these transgenic tobacco lines also recorded a higher UV and salt tolerance than the WT plants. These results suggested that overexpressing the VvCSDs will enhance the ROS-scavenging systems and protect the plant against more oxidative damage. Also, more investigations in this line are needed that would provide significant improvements in our understanding the resistance of fruit crops to environmental stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are a class of small, single-stranded, non-coding RNAs that are involved in the regulation of gene expression at the post-transcriptional level mainly by targeting mRNAs for cleavage or repressing translation (Han et al. 2014; Wang et al. 2014; Budak and Akpinar 2015; Pagliarani et al. 2017). Many miRNAs play important roles in plant growth and development, such as phase transitions, flowering, and leaf and root development (Kantar et al. 2010; Akpinar et al. 2015; Nie et al. 2015). An increasing number of evidences demonstrate that miRNAs play key regulators in plant responses to nutrient homeostasis and to biotic and abiotic stresses (Kantar et al. 2011; Khraiwesh et al. 2012; Guan et al. 2013; Budak et al. 2015a; Budak et al. 2015b; Alptekin et al. 2017). For instance, miR395 and miR399 were recently shown to be induced by sulfate and phosphate deprivation, respectively, and their induction was important for the down-regulation of certain genes under nutrient deficiency stress (Fujii et al. 2005; Chiou et al. 2006; Liang et al. 2015).
Both biotic and abiotic stresses have significant deleterious influences on plant growth and development, which causes considerable loss in crop yields worldwide. However, plants have evolved complex mechanisms to respond to environmental stresses while maintaining their growth and development (Mittler 2002; Mittler et al. 2004). A classical example is that different types of antioxidant networks including enzymatic and non-enzymatic antioxidants in higher plant cells can scavenge or detoxify stress-induced reactive oxygen species (ROS) (Birben et al. 2012). It is also reported that ROS affects many cellular functions by damaging nucleic acids, oxidizing proteins, and causing lipid peroxidation and is a major cause of loss of crop productivity (Apel and Hirt 2004; Gill and Tuteja 2010). Therefore, the effects of environmental stresses on plants depend on how a plant regulates its rate of ROS production and ROS scavenging when it is exposed to environmental stresses.
Superoxide dismutase (SOD) is one of the most important antioxidants that catalyze the conversion of superoxide radicals to H2O2 and O2, which constitutes the first line of defense against ROS (Perry et al. 2010). On the basis of the metal co-factor used, SODs are classified into three groups: iron SOD (FeSOD), manganese SOD (MnSOD), and copper-zinc SOD (Cu/Zn-SOD, also known as CSD), which are localized in different cellular compartments (Holzmeister et al. 2015). Cu/Zn-SOD is a major copper enzyme and the most important SOD in plants. The Arabidopsis genome encodes three CSD isozymes: CSD1 in the cytoplasm, CSD2 in chloroplasts, and CSD3 in peroxisomes (Guan et al. 2013). CSD1 and CSD2 are up-regulated in response to stresses that trigger ROS accumulation, including ozone, UV-B, and high light levels. It was found that over expression of the cytosolic Cu/Zn-SOD and chloroplastic Cu/Zn-SOD from pea in transgenic tobacco (Nicotiana tabacum) plants increased tolerance against ozone, high light, and low temperature (Pitcher and Zilinskas 1996; Sen Gupta et al. 1993a, b). Recently, a number of studies have revealed that miR398 in A. thaliana could target CSD1 and CSD2 genes that can detoxify ROS (Sunkar and Zhu 2004; Budak et al. 2015a; Alptekin et al. 2017). When A. thaliana plants were exposed to oxidative stress, including high light intensity and heavy metals, miR398 expression was down-regulated at transcriptional levels, and this down-regulation was important factor for post-transcriptional accumulation of CSD1 and CSD2 mRNA accumulation and oxidative tolerance (Sunkar et al. 2006).
Grapevine (Vitis) is one of the most economically important fruit crops worldwide and has nutritional and value-added properties (Han et al. 2014; Wang et al. 2014; Jia et al. 2016). The excess use of Cu-based fungicides and bactericides, waste water irrigation, and unconscionable Cu mining and Cu stress has become one of the serious environmental problems that confronts viticulture in China (Leng et al. 2015a, b; Leng et al. 2017). Recently, various researchers had demonstrated that some miRNA families directly and negatively regulated the Cu stress regulatory network (Sunkar et al. 2006; Yamasaki et al. 2007; Abdel-Ghany and Pilon 2008). Among the involved miRNAs reported, miR398 was reported to target CSD1 and CSD2 that utilize copper co-factors, and their regulation of tolerance to Cu stress in A. thaliana and rice is also well documented. However, there is hardly any literature available about the function of miR398 in grapevine. Therefore, understanding the role of Vv-miR398 and its association with the Cu stress regulatory network in grapevine is much needed. Hence, in this study, we first investigated the effects of Cu stresses on the expression of miR398 and its two putative target genes, VvCSD1 and VvCSD2. Next, we analyzed their functions in the responses of grapevine to Cu stress and other abiotic stress with transgenic tobacco plants by over-expressing VvCSD1 and VvCSD2. The results showed that the ectopic expression of grapevine VvCSD genes increased oxidative tolerance to Cu2+ toxicity. Our results suggest that Vv-miR398 and its target genes also play a key role in the responses of grapevine to abiotic and biotic stresses.
Results
Experimental verification of miR398-guided cleavage of target mRNAs in grapevine
It was well documented that miR398 identified in A. thaliana and rice play key roles in response both to abiotic and biotic stresses by mediating cleavage of the CSD1 and CSD2 transcripts, the two target genes encoding two Cu/Zn-SODs (Sunkar et al. 2006; Jagadeeswaran et al. 2009; Lu et al. 2011). Our results showed that Vv-miR398 family was highly conserved and were encoded by three loci: miR398a (located on the No. 1 chromosomes), miR398b, and mIR398c (located on the No. 6 chromosomes). As is the case in A. thaliana and rice, bioinformatics analysis result showed that the target genes of VvmiR398 were also two CSD genes, VvCSD1 and VvCSD2. Further, RT-PCR and RACE were employed to amplify the cDNA sequences of VvCSD1 and VvCSD2, which was deposited in gene Bank as JQ692111 and JQ692112. The cDNA sequences of VvCSD1 and VvCSD2 genes were 749 and 884 bp in length with open reading frames of 459 bp and 666 bp, respectively. They were highly complementary with Vv-miR398 (Fig. 1a, b), which suggested that these two CSD genes might be the targets of Vv-miR398.
Mapping of the mRNA cleavage site by 5′-RACE. The accession numbers for VvCSD1 (JQ692111) and VvCSD2 (JQ692112) are found in NCBI database, and the nucleotide sequence alignment of two Vv-miR398 sequences is from miRBase database. Each bottom strand (black) depicts a miRNA complementary site, and each top strand depicts the miRNA (red). Watson-Crick pairing (vertical dashes) and G/U wobble pairing (circles) are indicated. The arrows indicate the 5′ termini of mRNA fragments isolated from grape, as identified by cloned 5′-RACE products, with the frequency of clones shown. Only the cloned sequences that matched the correct gene and had 5′ ends within a 100 nt window centered on the miRNA validation are included. The partial mRNA sequences from the target genes were aligned with the miRNAs. The numbers indicate the fraction of cloned PCR products terminating at different positions
To verify whether the two cloned VvCSD genes can be cleaved by Vv-miR398 in grapevine and to identify their cleavage sites on VvCSDs, RLM-RACE (RNA ligase-mediated 5′ rapid amplification of cDNA ends) was employed. Usually, when the fragments with predicted size are detected, it could be inferred that the miRNA regulates its target through the degradation mode. And the further sequencing of the cleavage products can tell us the cleaving site. All the anticipated RLM-RACE products could be visualized on agarose gels, isolated, and sequenced. The RLM-RACE products revealed that the two VvCSD mRNAs were indeed the targets of VvmiR398 (Fig. 1a, b), and their specific cleavage sites could be clearly found. Among them, two cleavage sites were mapped at the nucleotide complementary to the 10th nucleotide of the 5′-end of corresponding miRNAs (Fig. 1). Like most plant miRNA targets, the Vv-miR398 complementarity site in the VvCSD2 mRNA is within the coding sequence. In contrast, the complementary site with Vv-miR398 in VvCSD1 transcript was located in the 5′-UTR.
The spatiotemporal expression patterns of miR398 and its target genes in grapevine
The spatiotemporal expression profiles of miRNAs and their corresponding target genes are important for understanding their physiological function, which contribute to the elucidation of mechanism responsive for miRNAs’ interaction with their targets. The differential expressions of miRNAs are expected to have opposite effects on its target gene(s) expression. Negative correlations have been observed between grapevine miRNAs and their target mRNAs (Wang et al. 2013), as shown for miR164 and target mRNA named NAC1 and NAC2 in grapevine (Sun et al. 2012). In our study, both two VvCSDs transcripts and Vv-miR398 were detected by qRT-PCR analysis. A high expression of Vv-miR398 was found in young leaves and young berries, whereas low expression level of Vv-miR398 was detected in inflorescences, flowers, and mature leaves (Fig. 2a). On the contrary, the VvCSD1 and VvCSD2 transcripts showed a clear opposite expression pattern compared with Vv-miR398 (Fig. 2b, c). Further, tissues with high levels of Vv-miR398 (young leaves and young berries) resulted in low levels of VvCSD1 and VvCSD2 mRNAs, whereas tissues with low expression of Vv-miR398 (inflorescences, flowers, and mature leaves) recorded high levels of VvCSD1 and VvCSD2. Furthermore, the expression levels of Vv-miR398 exhibited a typical negative correlation (the correlation coefficients: −0.8825 and −0.8472) with their target genes (VvCSD1 and VvCSD2) in corresponding organs or growth stages.
The expression patterns and correlation of Vv-miR398 and corresponding target genes in different tissues of grapevine. R, YL, ML, IF, F, YB, and MB are samples of root, young leaves (2 weeks old), mature leaves (8 weeks old), inflorescences (2 weeks old), flowers (fully open), young berries (15 days after flowering), mature berries (60 days after flowering), respectively. “r” denoted the correlation coefficient. Each reaction was repeated three times and the standard error plotted
Effects of Cu stresses on expression of Vv-miR398 and its two CSD genes
Both abiotic and biotic stresses like heavy metals usually exert their deleterious influences on plants through inducing ROS accumulation, which is harmful to plant cells (Mittler et al. 2004). The problem due to Cu stress in viticulture has become more serious due to the intensive use of copper-based bactericides and fungicides, such as Bordeaux mixture. Plants have evolved several antioxidant mechanisms to cope with the possible imbalance of ROS within the cells, among which the SOD-catalyzed scavenging of ROS is considered as the most important of antioxidant enzyme (Mittler 2002). However, the detailed molecular mechanisms of this system are largely unknown in response to Cu stress in grapevine.
To understand the roles of Vv-miR398 in the expression of the two VvCSD genes, we first compared the expression patterns of Vv-miR398 and its two target genes in response to various Cu stresses with different concentrations of Cu2+. The semi-quantitative RT-PCR (RT-PCR) and qRT-PCR (Fig. 3) revealed that when the grapevine leaves were treated with either 100 μM (Fig. 3a, d) or 200 μM (Fig. 3b, e) CuSO4, the expression of Vv-miR398 was steadily down-regulated over 24 h. Consistently, we observed an enhancement in the mRNA expression of VvCSD1 and VvCSD2. These opposite expression profiles confirmed the reverse correlation between Vv-miR398 and VvCSD1 and VvCSD2 (Fig. 3a, b, d, e). Nevertheless, the data also showed that Vv-miR398 and the two VvCSD genes were subjected to a deviate stable regulation under 400 μM CuSO4. The expression levels of two VvCSD genes were found to be the highest at 6 h, and there after it gradually decreased over the treatment time (Fig. 3c, f). This could be due to the fact that balance between the formation and removal of ROS was destroyed, and the defense system could be overwhelmed when presented with increased ROS formation under 400 μM Cu stress conditions. These results demonstrated that the expression patterns of Vv-miR398 and the target gene were similarly regulated by Cu stress as the same to Arabidopsis.
Effects of Cu stress treatments on expression of Vv-miR398, VvCSD1, and VvCSD2 genes in grapevine. Three-year-old grapevine were treated with different concentrations of CuSO4 and total RNA was extracted from stressed leaves at different time points (0, 6, 12,and 24 h) (a–c). Effect of 100, 200, and 400 μM CuSO4 treatment on the expression of miR398 and two target genes analyzed by RT-PCR with 30 cycles, respectively (d–f). Effect of 100, 200, and 400 μM CuSO4 treatment on the expression of miR398 and two target genes analyzed by qRT-PCR, respectively
Cu concentration, MDA content, H2O2 level, and SOD activity in grapevine under copper stress
As shown above, the expression of both Vv-miR398 and the VvCSD genes was significantly affected by Cu stress. Therefore, it was reasonable to assume that grape leaves inevitably absorb exogenous Cu ions in a short period and lead to an increased accumulation of ROS when copper solutions were sprayed later. As shown in Fig. 4a, there was a remarkable increase in Cu concentration in grapevine leaves with the spray concentration and treatment time increased compared with the control treatment (Fig. 4a). After treatments with 400 μM CuSO4, the Cu concentration was significantly increased to 30 μg/g DW. This concentration was a critical toxicity level of Cu in the leaves (Leng et al. 2015a). MDA, a product of lipid peroxidation, has often been considered an indicator of oxidative stress (Shalata et al. 2001). All the three concentrations of Cu treatments during the experimental period led to a stable and significant increase in the levels of MDA contents in leaves (Fig. 4b), which indicated a high degree of lipid peroxidation under Cu stress. Furthermore, all Cu stress treatments significantly enhanced H2O2 accumulation, and the effect of the 400 μM CuSO4 treatments was the highest in this study (Fig. 4c). The increased MDA and H2O2 level indicated that Cu caused severe oxidative stress by stimulating ROS generation. Finally, exposure to 100 and 200 μM Cu could cause a relatively stable increase of SOD activity in grapevine leaves. However, a dynamic regulation of SOD activity was also detected under 400 μM Cu treatment (Fig. 4d), and also recorded a two-fold increase in the SOD activity at 6 h, and this activity was reduced drastically after 6 h (Fig. 3d). Our results are in agreement with previous deduction that 400 μM Cu stress treatment destroyed the balance between the formation and removal of ROS.
Effects of three concentrations of Cu treatments (100, 200, and 400 μM CuSO4) on four physiological parameters in grapevine. a Changes of Cu concentration. b Changes of MDA content. c Changes of cellular hydrogen peroxide levels. d Changes of SOD activity. Each data represents the average of two experiments with three replicates (n = 6). Vertical bars indicate measurements ± SD
Generation and characterization of transgenic tobacco overexpressing two VvCSD genes
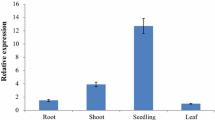
Usually, plant miRNAs function through suppressing the expression of their targeted genes. Therefore, in order to reveal the role of Vv-miR398, the functions of the two VvCSD genes were first illustrated. Based on the reports about the miR398 in other plants, the roles of two VvCSD genes in responses to oxidative stresses were extensively studied using transgenic tobacco plants overexpressing with two VvCSD genes. The two VvCSD ORFs without the stop codon were linked to the upstream of the GFP gene in the vector of pCAMBIA1302. Thereafter, the constructed vectors were used to generate the transformed tobacco plants via Agrobacterium-mediated leaf disk transformation. After several cycles of kanamycin-resistance selection, transgenic tobacco plants expressing VvCSDs were produced. More than 14 independent transgenic lines were confirmed by PCR analysis of genomic DNA using the primers specific to VvCSDs and GFP (Fig. 5a). Further, the RT-PCR data recorded that VvCSD1 and VvCSD2 mRNA could be detected in the transgenic plants, but not in the WT (Fig. 5b).
Generation and characterization of VvCSD1 and VvCSD2 gene in transgenic tobacco. a PCR analysis of the kanamycin-resistant tobacco plants using GFP-specific primers and two VvCSD gene primers. WT wild type, lane P positive control, and the numbers show the regenerated lines. b Transgene expression analysis in the transgenic tobacco plants and WT based on semi-quantitative RT-PCR assay. c Fusion protein expression analysis in the transgenic tobacco plants and WT based on Western blotting assay. Lane 6 highly expressed transgenic lines in VvCSD1; lane 4 highly expressed transgenic lines in VvCSD2. Two GFP tag fusion protein in lanes 6 and 4 encoded for a 43 and 50 kD protein, respectively
The expression levels of VvCSD1 and VvCSD2 in putative lines were significant differences. Two highly expressed transgenic lines (OE-6 and OE-4) with the transferred VvCSD1 and VvCSD2, respectively, were used for further verification about whether the transgenic plants could express the foreign protein, for which Western blotting was performed. According to our finding that VvCSD1 and VvCSD2 encoded 15 and 22 kD products, respectively (Leng et al. 2015a), and green fluorescent protein gene (GFP) from pCAMBIA1302 vector encoded 28kD product. Our data showed that two protein products of 43 and 50 kD were detected in the two transformed tobacco plants, suggesting the existence of the anticipated GFP tag fusion proteins (GFP-VvCSD1 and GFP-VvCSD2) in the transgenic plants (Fig. 5c). However, the wild plants did not produce any protein signal.
Overexpression of two VvCSD genes conferred enhanced tolerance to cu stress
To know whether the two VvCSD genes play a critical role in Cu stress tolerance, the transgenic tobacco lines overexpressing two VvCSDs along with the untransformed wild type (WT) were analyzed. There was no apparent difference in plant visible growth and morphology between the WT and the transgenic plants under natural (non-stressed) conditions. When tobacco seedlings were exposed to 200 μM CuSO4 for 12 h, the leaves of WT plants began to wilt, whereas no obviously adverse phenotype was observed in the transgenic tobacco seedlings. After 48 h of treatment, more serious leaf wilting was visualized in the WT, whereas the transgenic plants were less affected (Fig. 6a). More analysis indicated that the expression level of VvCSD1 and VvCSD2 in the transgenic lines gradually increased during the experimental period, whereas this did not happen in WT plants (Fig. 6b), displaying that the transcriptional levels of VvCSD1 and VvCSD2 genes were induced by Cu stress. In addition, Cu stress also led to excessive production of MDA in the tobacco leaves. As shown in Fig. 6c, OE-VvCSD1 (from 6.98 to 11.54 μmol/g) and OE-VvCSD2 (from 6.34 to 10.28 μmol/g) had lower MDA content and when compared with the WT (from 7.76 to 15.36 μmol/g) which recorded an increase in the MDA content during the treatment. H2O2 has been implicated as a key factor mediating programmed cell death. The increasing trend of H2O2 content in two transgenic lines were also obviously lower than that in WT plants (Fig. 6d). Chlorophyll levels were used as an indicator of chloroplast damage, and the chlorophyll content decreased under oxidative stress (Khayatnezhad et al. 2011). The higher chlorophyll contents in OE-VvCSD1 and OE-VvCSD2 indicated that chlorophyll was less damaged than the WT did after 48 h Cu treatment (Fig. 6e).
Overexpression of VvCSD1 and VvCSD2 enhances Cu tolerance in transgenic tobacco. a Phenotypes of 40-day-old seedlings of WT and transgenic plants (OE-VvCSD1 and OE-VvCSD2) subjected to 200 μM CuSO4 treatment. b Analysis of expression levels of VvCSD1 and VvCSD2 by semi-quantitative RT-PCR in the WT and transgenic lines (OE-VvCSD1 and OE-VvCSD2) after Cu treatment. c–f The MDA content, H2O2 content, chlorophyll content, and SOD activity during the experimental period, respectively. FW fresh weight. Data represent the means ± SE of at least three replicates
Furthermore, Cu/Zn-SODs, encoded by CSD1 and CSD2, are the most important antioxidant enzymes. Thus, SOD activity in the tested plants was measured and compared. Under normal conditions, SOD activities in OE-VvCSD1 and OE-VvCSD2 were 80.76 and 83.48 U/g, being slightly higher than that in WT (71.93 U/g) (Fig. 5e). During 24-h exposure to Cu stress, SOD activities were notably enhanced both in the transgenic lines and WT, and the activity in the transgenic lines was much higher than that in the WT plant. After 48-h treatment, the SOD activity in WT plants (67.36 U/g) decreased, whereas two transgenic lines still maintained much higher SOD activity (110.14 and 123.43 U/g, respectively) (Fig. 6f). All the findings above suggested that overexpression of VvCSD1 and VvCSD2 led to dramatic improvement of Cu tolerance in the transgenic tobacco plants.
Transgenic tobacco lines over-expressing Vv-CSD genes showed more tolerance to other abiotic stresses
In order to further explore the function of VvCSD1 and VvCSD2, we also analyzed the phenotypes and expression level of two CSDs gene in responses to UV and salt stress between transgenic tobacco lines and WT plants. As shown in Figs. 7 and 8a, a remarkable improvement of UV and salt tolerance were observed in both the transgenic tobacco lines compared with those of WT plants. The growth of the WT plants was significantly inhibited and their leaves were severely crimped and etiolated after UV treatment (Fig. 7a). Also, the leaves of WT plants began to dehydration and wilt during the phase of salt stress (Fig. 8a), whereas a low symptom was observed in both the two transgenic lines. The expression level of VvCSD1 and VvCSD2 in two transgenic lines also gradually increased under UV and salt treatment (Figs. 7b and 8b), displaying that the VvCSD1 and VvCSD2 played an important role in various stress responses. Furthermore, both under UV and salt treatments, a lower MDA and H2O2 contents was recorded in two transgenic lines than WT plants at all time intervals (Fig. 7c, d and Fig. 8c, d). In the contrary, the WT plants recorded a slight increase in the SOD activity at 12 h, and then decreased rapidly, whereas the SOD activity in two transgenic lines increased markedly and remained at high levels under UV and salt treatment (Figs. 7f and 8f). Moreover, the chlorophyll contents of the two transgenic lines were also higher than those of the WT in responses to UV and salt treatment (Figs. 7e and 8e).
Overexpression of VvCSD1 and VvCSD2 enhance UV tolerance in transgenic tobacco. a Phenotypes of 40-day-old seedlings of WT and transgenic plants (OE-VvCSD1 and OE-VvCSD2) treated with 1 h UV (100 J/m2). b Analysis of expression levels of VvCSD1 and VvCSD2 by semi-quantitative RT-PCR in the WT and transgenic lines (OE-VvCSD1 and OE-VvCSD2) after UV treatment. c–f The MDA content, H2O2 content, chlorophyll content, and SOD activity during the experimental period, respectively. FW fresh weight. Data represent the means ± SE of at least three replicates
Overexpression of VvCSD1 and VvCSD2 enhance salt tolerance in transgenic tobacco. a Phenotypes of 40-day-old seedlings of WT and transgenic plants (OE-VvCSD1 and OE-VvCSD2) treated with 400 mM NaCl. b Analysis of expression levels of VvCSD1 and VvCSD2 by semi-quantitative RT-PCR in the WT and transgenic lines (OE-VvCSD1 and OE-VvCSD2) after NaCl treatment. c–f The MDA content, H2O2 content, chlorophyll content, and SOD activity during the experimental period, respectively. FW fresh weight. Data represent the means ± SE of at least three replicates
Discussion
Plants are extremely sensitive and sophisticated in responses to various environmental stresses, but are under stringent regulation. Various environmental stresses, such as heavy metals, salinity, water-logging, drought, and cold, can lead a rapid and excessive accumulation of ROS causing progressive oxidative damage and ultimately cell death. It is well known fact that plants have evolved complicated mechanisms to regulate the delicate balance between ROS production and scavenging through a sophisticated ROS detoxification system (enzymatic and non-enzymatic) in response to various oxidative stresses (Foyer and Noctor 2003; Suzuki and Mittler 2006). Previous studies have shown that miRNAs play an important roles in biotic and abiotic stress responses, including drought, salinity, and heavy metal stress (Mendoza-Soto et al. 2012; Budak et al. 2015b; Alptekin et al. 2017).
Among environmental stress, Cu stress has become one of the serious environmental crises with high risk in viticulture production due to the intensive use of copper-based bactericides and fungicides (Leng et al. 2017). However, much attention has been taken, and so far, detailed study is not done. With more studies on miRNAs in plants, the miR398 family was shown to determine the expression pattern of CSD1 and CSD2 in response to various environmental stress, especially in Cu stress (Sunkar et al. 2006; Yamasaki et al. 2007; Abdel-Ghany and Pilon 2008; Jagadeeswaran et al. 2009). The studies on the crucial role of miR398 and its targets have mainly been focused on the model plant, A. thaliana. Till now, not much research on functions of grapevine miR398 has been carried out. In our study, RLM-RACE assays showed that the two VvCSD genes had specific cleavage sites corresponding to the Vv-miR398 complementary sequences. The expression profiles of Vv-miR398 and its two targeted genes, VvCSD1 and VvCSD2, displayed a clear negative correlation under natural conditions, implying that Vv-miR398 plays a key role in controlling the accumulation of VvCSD1 and VvCSD2 transcripts in grapevine organs by cleavage of the two VvCSDs targets mRNA. In addition, high expression levels of Vv-miR398 were detected in young organs, such as young leaves and young berries, indicating that Vv-miR398 also plays a role in plant growth and development. Furthermore, the negative correlation between the expression patterns of Vv-miR398 and two VvCSD genes was further verified by comparing the expression profiles in response to Cu stress. Vv-miR398 transcript accumulation was significantly inhibited by Cu stress. On the contrary, VvCSD1 and VvCSD2 transcript accumulation under the same stress conditions showed reverse patterns (Fig. 3). The expression profile of Vv-miR398, VvCSD1, and VvCSD2 in the same RNA samples indicated a clear negative correlation under different concentrations of Cu stress and suggested a critical role for miR398 in controlling the CSD1 and CSD2 mRNA levels in response to Cu stress in grapevine.
In order to validate the roles of Vv-miR398 and two VvCSDs in responses to environmental stress, we successfully constructed the transgenic tobacco plants which over-expressed VvCSD1 and VvCSD2 and analyzed the expression profiles of both the two VvCSDs and response to environmental stress. The two VvCSDs-overexpressing transgenic tobacco lines displayed increased resistance to the Cu, salt, and UV tolerance, compared with wild type. This result is consistent with previous studies that modification of SOD expression in transgenic tobacco plants can improve plant stress tolerance (Sen Gupta et al. 1993a, b). Exposed to abiotic stresses, the oxidative burst, a transient increase in ROS, predominantly superoxide (O2 −), and hydrogen peroxide (H2O2), is among the first biochemical responses of plants to abiotic stress (Joo et al. 2005). In our study, the two transgenic lines overexpressing the VvCSD genes accumulated remarkably less MDA and H2O2 and more chlorophyll than WT under abiotic stressful conditions. The decreasing of chlorophyll content under oxidative stress may be the result of chlorophyll degradation or be due to chlorophyll synthesis deficiency or changes of thylakoid membrane structure. The results suggested lower levels of ROS and reduced oxidative damage in the transgenic plants. As ROS level during stresses greatly relies on the homeostasis between generation and removal (Miller et al. 2010), accumulation of lower ROS in the transgenic lines seems to indicate that scavenging systems in these plants might work more effectively compared with WT, and several ROS-scavenging enzymes play a positive role in scavenging ROS and protecting the cells against oxidative stress. Among the enzymes, SODs are the most important, because these are the first enzymes to act on highly toxic superoxide radicals (O2 −) and convert them into less toxic hydrogen peroxide (Lu et al. 2011). In our study, the activity of SOD was slightly higher in the transgenic lines than in the WT plants under normal conditions, which sounds reasonable because under normal conditions, ROS production remained at low levels and oxidative stress was not serious (Huang et al. 2010). However, under Cu, salt, and UV stress, activity of SOD was significantly higher in the two transgenic plants than WT, demonstrating that the two transgenic plants had better ROS-scavenging capability during stress and keep the ROS accumulation balance. Furthermore, the increased SOD activity was consistent with the accumulation of two VvCSD gene transcripts in two transgenic lines, suggesting that the enzyme may be regulated at transcriptional levels. These works suggested that a better ROS-scavenging system could protect plants against more oxidative damage, and this point might be an integral part of abiotic stress tolerance in the transgenic plants overexpressing VvCSDs. Because we have shown that two VvCSDs mRNA were cleaved by Vv-miR398 in our previous studies, our results also shed light on the possibility of manipulating miRNA or small RNA to improve the tolerance of grapevine to environmental stresses. We remain hopeful that investigations of this type will eventually provide significant improvement resistance to environmental stress in fruit crops.
Conclusion
Our results shed light on the possibility of Vv-miR398 to improve the tolerance of grapevine plants to Cu stress. Several attempts have been made to improve plant stress tolerance by overproduction of Cu/Zn-SODs in transgenic plants. Physiological and molecular analysis showed that Vv-miR398 and its target genes can take part in the grapevine’s response to Cu stress. Overexpressing VvCSD1 and VvCSD2 conferred enhanced tolerance to Cu, UV, and salt stress. These results suggest that miR398-regulation triggers a regulatory loop that is critical to manipulate stress tolerance in grapevine, and our findings offer an improved strategy to fruit crop plants with enhanced stress tolerance.
Materials and methods
Plant materials and stress treatments
Plant materials were sampled at the different developmental stages with respect to their growth period. Root, young leaves (2 weeks old), mature leaves (8 weeks old), inflorescences (2 weeks old), flowers (fully open), young berries (15 days after flowering), and mature berries (60 days after flowering) were collected during the summer of 2015 from 3-year-old table grapevine ‘Fujiminori’ grown under standard grapevine cultivation conditions at the fruit experimental farm, Nanjing Agricultural University, Nanjing, China. For Cu treatment, uniform and healthy 3-year-old ‘Fujiminori’ grapevine was sprayed with different concentrations such as 100, 200, and 400 μM CuSO4; the treated leaves were then sampled at 0, 6, 12, and 24 h. After collection, all the samples were immediately frozen in liquid nitrogen and stored at −80 °C for further experiments.
Isolation of full-length VvCSD1 and VvCSD2 cDNA
Based on the available grapevine genome database, there were two sequences with a complete opening reading frame, a high degree of similarity with AtCSD1 (AT1G08830.1) and AtCSD2 (AT2G28190.1). The primers GSP1 and GSP8 were designed for amplifying the open reading frame (ORF) in ‘Fujiminori’ (all of the primers used in this study are listed in Table 1). The PCR mixture (50 μL) contained 300 ng cDNA, 1 × TransStart FastPfu buffer, 0.25 mM deoxyribonucleotide (dNTP), 0.4 μM of each primer, and 2.5 units of TransStart FastPfu DNA polymerase. PCR program was performed as follows: initial denaturation at 95 °C for 2 min, 40 cycles of 95 °C for 20 s, 55 °C for 20 s, 72 °C for 60 s, and 72 °C for 5 min. To obtain the full length cDNA of VvCSD1 and VvCSD2, 5′ and 3′ rapid amplification of cDNA ends (RACE) was carried out. For the 3′ RACE, the Full RACE Core Set Ver.2.0 Kit was used, while SMARTerTM RACEcDNA Amplification Kit (Clontech, CA) was used for 5′ RACE, both of which were done following the manufacturer’s instructions. GSP2 and GSP9 were designed for 3′ RACE, and GSP3 was designed for 5′ RACE of VvCSD1. The PCR products were cloned into pMD19-Tvector (Takara, Japan) and were sequenced.
High, low molecular weight RNA extraction and construction of complementary DNA libraries
Total RNA was isolated from 200 mg of the grapevine tissues mentioned above using the modified CTAB method (Wang et al. 2011) and treated with RNase-free DNase I (Thermo) to remove DNA contamination. Low molecular weight RNA (LMW RNA) and high molecular weight RNA (HMW RNA) were separated with 4 M LiCl (Adai et al. 2005; Wang et al. 2011) and the small RNA fraction was then dissolved in 30 μL of RNase free water. The concentration of RNA was measured by a UV-1800 spectrophotometer (Shimadzu, Japan) and visually ascertained in a 2.5% agarose gel.
The larger molecular weight RNA samples were used to study the expression patterns of the target genes of Vv-miRNAs. First-strand cDNA was synthesized from total RNA with ReverTra Ace qPCR RT Kit (Toyobo, Shanghai, China) according to the manufacturer’s instructions. The low molecular weight RNA samples were used to study the expression patterns of Vv-miRNAs. Small RNAs were polyadenylated at 37 °C for 60 min in a 50-μL reaction mixture with 1.5 μg of total RNA, 1 mM ATP, 2.5 mM MgCl2, and 4 U poly(A) polymerase (Ambion, Austin, TX). Poly(A)-tailed small RNA was recovered by phenol/chloroform extraction and ethanol precipitation. 5′ adapter (5′-CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA-3′) was ligated to the poly(A)-tailed RNA using T4 RNA ligase (Invitrogen, Carlsbad, CA), and the ligation products recovered by phenol/chloroform extraction followed by ethanol precipitation. Reverse transcription was performed using 1.5 μg of small RNA and 1 μg of (dT)30 RT primer (ATTCTAGAGGCCGAGGCGGCCGACATG-d(T)30 (A, G, or C) (A, G, C, or T)) with 200 U of SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). Poly(A)-tailed small RNA (10 μL total volume) was incubated with 1 μL of (dT)30 RT primer and 1 μL dNTP mix (10 mM each) at 65 °C for 5 min to remove any RNA secondary structures. The reactions were chilled on ice for at least 2 min; then, the remaining reagents [5× buffers, dithiothreitol (DTT), RNaseout, SuperScript III] were added as specified in the SuperScript III manual, and the reaction left to proceed for 60 min at 50 °C. Finally, the reverse transcriptase was inactivated by incubation for 15 min at 70 °C.
Mapping of mRNA cleavage sites using RLM-RACE
RLM-RACE was used to map miRNA-mediated cleavage products. After construction of libraries of poly(A)-tailed HMW RNA and adapter-ligated HMW RNA, the productions of reverse transcription of poly(A)-tailed HMW RNA and adapter-ligated HMW RNA were performed with RLM-RACE using corresponding common primer and specific primers, respectively. GSP4 and GSP10 were designed for RLM-RACE of two target genes, respectively (Table 1). The amplification products were gel purified, cloned, and sequenced, and at least eight independent clones were sequenced.
miRNA and its target gene expression analysis by RT-PCR and qRT-PCR
The qRT-PCR was employed for measuring the transcript levels of Vv-miR398 and two target genes under different tissues. The RT-PCR and qRT-PCR were employed for measuring transcript levels of Vv-miR398 and two target genes under different concentration of treatments. The template used for qPCR of Vv-miR398 expression was the miRNA-enriched library mentioned above. To amplify the Vv-miRNAs from the reverse transcribed cDNAs, the qPCRs were performed using the Vv-miR398 precise sequences (TGTGTTCTCAGGTCACCCCTT) as the forward primer and the R16328 (ATTCTAGAGGCCGAGGCGGCCGACATG) as the reverse primer (Wang et al. 2013). The 20-μL qPCR reactions contained 10 μL of SYBR-Green PCR Master Mix (SYBR Premix EX Taq TM, TaKaRa), 5 pmol each primer, 100 pg of total RNA, and nuclease-free water. The conditions for the qPCR amplification were as follows: polymerase activation at 95 °C for 1 min, then 95 °C for 1 min, followed by 50 cycles of 95 °C for 15 s, 95 °C for 15 s, 60 °C for 20 s, and 72 °C for 20 s. The relative quantification procedure was used to determine relative expression levels. The 5.8S rRNA was used as a reference gene in the qPCR detection of miRNAs as done previously in Arabidopsis (Shi and Chiang 2005). The expression of experimentally verified target genes using qPCR followed the report of Wilson et al. (2005). The reverse transcription product was amplified by gene-specific primers in Table 1 (GSP5 and GSP11). Data were normalized to UBI which was the homologous gene of AT5G08290 (Sun et al. 2012) and analyzed using a comparative quantification procedure. UBI was used as a reference gene in the qPCR detection of mRNAs in grapevine. qRT-PCR was conducted with the Rotor-Gene 3000 (Corbett Robotics, Australia) and the Rotor-Gene software version 6.1, and the relative gene expression data were calculated by the 2−ΔΔCT method (Livak and Schmittgen 2001).
Semi-quantitative reverse transcription PCR (RT-PCR) of two targets was carried out as described (Yu et al. 2009) and RT-PCR program was 94 °C for 5 min, followed by 30 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 62.0 °C, and extension for 60 s at 72 °C and a final extension for 10 min at 72 °C. Each reaction was repeated three times. A control PCR with primers that amplify a 180-bp fragment of UBI was performed in parallel to verify similar amounts of cDNA in each sample. The PCR products were analyzed by electrophoresis on a 2.0% agarose gel containing ethidium bromide and photographed under UV light. RT-PCR was also employed, as described above, to detect transcript levels for miR398. The 5.8S rRNA was used as a reference gene.
Vector construction and generation of transgenic tobacco by agrobacterium-mediated transformation
The specific primers (GSP1) containing the restriction sites of BglII and SpeI were used to amplify VvCSD1 and VvCSD2 cDNA. The confirmed plasmid was double-digested with BglII and SpeI and then ligated into the pCAMBIA1302 vector driven by the CaMV 35S promoter. Subsequently, the two recombinant plasmids, pCAMBIA1302-VvCSDs, were transferred into an Agrobacterium tumefaciens strain GV3101 and used for tobacco (Nicotiana tabacum) transformation based on using leaf disks. The methods of co-culture and selection of explants by kanamycin-resistance were described by Huang et al. (2010).
PCR and Western blot verification of the transgenic plants
The genomic DNA and total RNA were extracted from the young leaves of Kanamycin-resistant T0 transgenic plants and wild type using the CTAB method. PCR amplifies for detection of the two VvCSDs gene from the genomic DNA were performed using the GSP7 and GSP13 (VvCSDs and GFP) primers. RT-PCR amplifies for the detection of two VvCSDs genes from RNA level with specific primers of GSP6 and GSP12. Only adopting the PCR target fragments by the two pairs of primers were considered as putative transgenic plants. For Western blotting, soluble proteins from tobacco were extracted by grounding using liquid nitrogen and resuspended in 500 μl of extraction buffer (200 mM Tris-HCl (pH 8.0), 100 mM NaCl, 10 mM EDTA, 2 mM PMSF). Twenty micrograms of soluble protein was resolved on 1D SDS/PAGE and transferred onto nitrocellulose membrane. The specific position of the antigen-antibody complex on the membrane was visualized by using alkaline phosphatase linked to secondary antibodies (Singla-Pareek et al. 2003).
Analysis of Cu, MDA, H2O2, chlorophyll contents, and SOD activity of the two transgenic plants under oxidative stress
Putative two transgenic plants and WT were planted in a greenhouse. To test Cu, UV, and salt tolerance, 40-day old tobacco plants were directly exposed to 200 μM CuSO4, 1 h UV (100 J/m2), and 400 mM NaCl treatment, respectively. The concentrations of copper in digested samples were determined by inductively coupled plasma-optical emission spectrometry (ICP-OES) with Perkin-Elmer Optima 2100 DV ICP-OES instrument at 324.75 nm wavelength. The instrument was controlled by the ICP WinLab32 Perkin Elmer software. The concentration of each element in the sample was determined in triplicate. MDA content was measured as previously described by thiobarbituric acid method (Schmedes and Holmer 1989) and expressed in μmol per gram freshweight (μmol/g FW). The content of H2O2 was measured according to the modified method of Patterson (1984). The chlorophyll content was measured by using acetone method as previously described (Huang et al. 2010). The absorbance of the supernatant was measured at 663, 645, and 480 nm in a spectrophotometer. SOD activities were determined by monitoring its ability to inhibit photochemical reduction of nitroblue tetrazolium (NBT) at 560 nm (Chen and Pan 1996). One SOD unit was taken to be the amount of enzyme causing 50% inhibition of NBT reduction and expressed in units per gram fresh weight (U/g FW).
Statistical analysis
Three stress treatments of the wild-type and transgenic lines were repeated at least triplicate, all experimental results are at least three independent replicates, shown as mean ± SE. The data were analyzed using SPSS software.
References
Abdel-Ghany SE, Pilon M (2008) MicroRNA-mediated systemic downregulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem 283:15932–15945
Adai A, Johnson C, Mlotshwa S, Archer-Evans S, Manocha V, Vance V et al (2005) Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res 15:78–91
Akpinar BA, Kantar M, Budak H (2015) Root precursors of microRNAs in wild emmer and modern wheats show major differences in response to drought stress. Funct Integr Genomics 15(5):587–598
Alptekin B, Langridge P, Budak H (2017) Abiotic stress miRNomes in the Triticeae. Funct Integr Genomics 17(2–3):145–170
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu rev Plant Biol 55:373–399
Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O (2012) Oxidative stress and antioxidant defense. World Allergy Organ J 5(1):9–19
Budak H, Akpinar BA (2015) Plant miRNAs: biogenesis, organization and origins. Funct Integr Genomics 15(5):523–531
Budak H, Hussain B, Khan Z, Ozturk NZ, Ullah N (2015a) From genetics to functional genomics: improvement in drought signaling and tolerance in wheat. Front Plant Sci 6:1012
Budak H, Kantar M, Bulut R, Akpinar BA (2015b) Stress responsive miRNAs and isomiRs in cereals. Plant Sci 235:1–13
Chen CN, Pan SM (1996) Assay of superoxide dismutase activity by combining electrophoresis and densitometry. Bot Bull Acad Sin 37:107–111
Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18:412–421
Foyer CH, Noctor G (2003) Redox sensing and signaling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Plant 119:355–364
Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15:2038–2043
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48(12):909–930
Guan QM, Lu XY, Zeng HT, Zhang YY, Zhu JH (2013) Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J 74:840–851
Han J, Fang JG, Wang C, Yin YL, Sun X, Leng XP et al (2014) Grapevine microRNAs responsive to exogenous gibberellin. BMC Genomics 15:111
Holzmeister C, Gaupels F, Geerlof A, Sarioglu H, Sattler M, Durner J, Lindermayr C (2015) Differential inhibition of Arabidopsis superoxide dismutases by peroxynitrite-mediated tyrosine nitration. J Exp bot 66(3):989–999
Huang XS, Liu JH, Chen XJ (2010) Overexpression of PtrABF gene, a bZIP transcription factor isolated from Poncirus trifoliata, enhances dehydration and drought tolerance in tobacco via scavenging ROS and modulating expression of stress-responsive genes. BMC Plant Biol 10:230
Jagadeeswaran G, Saini A, Sunkar R (2009) Biotic and abiotic stress downregulate miR398 expression in Arabidopsis. Planta 229(4):1009–1014
Jia HF, Zhang C, Pervaiz T, Zhao PC, Liu ZJ, Wang BJ et al (2016) Jasmonic acid involves in grape fruit ripening and resistant against Botrytis Cinerea. Funct Integr Genomics 16(1):79–94
Joo JH, Wang S, Chen JG, Jones AM, Fedoroff NV (2005) Different signaling and cell death roles of heterotrimeric G protein alpha and beta subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17(3):957–970
Kantar M, Unver T, Budak H (2010) Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct Integr Genomics 10(4):493–507
Kantar M, Lucas SJ, Budak H (2011) miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 233(3):471–484
Khayatnezhad M, Gholamin R, Somarin SJ, Mahmoodabad RZ (2011) The leaf chlorophyll content and stress resistance relationship considering in corn cultivars (Zea mays). Adv Environ Bio 5:118–122
Khraiwesh B, Zhu JK, Zhu J (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819:137–148
Leng XP, Mu Q, Wang XM, Li XP, Zhu XD, Shangguan LF et al (2015a) Transporters, chaperones, and P-type ATPases controlling grapevine copper homeostasis. Funct Integr Genomics 15:673–684
Leng XP, Jia HF, Sun X, Shangguan LF, Mu Q, Wang BJ et al (2015b) Comparative transcriptome analysis of grapevine in response to copper stress. Sci Rep 5:17749
Leng XP, Wang PP, Zhao PC, Wang MQ, Cui LW, Shangguan LF et al (2017) Conservation of microRNA-mediated regulatory networks in response to copper stress in grapevine. Plant Growth Regul 82:293–304
Liang G, Ai Q, Yu D (2015) Uncovering miRNAs involved in crosstalk between nutrient deficiencies in Arabidopsis. Sci Rep 5:11813
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4):402–408
Lu YZ, Feng Z, Bian LY, Xie H, Liang JS (2011) miR398 regulation in rice of the responses to abiotic and biotic stresses depends on CSD1 and CSD2 expression. Funct Plant Biol 38:44–53
Mendoza-Soto AB, Sánchez F, Hernández G (2012) MicroRNAs as regulators in plant metal toxicity response. Front Plant Sci 3:105
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33(4):453–467
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9(10):490–498
Nie SS, Xu L, Wang Y, Huang DQ, Muleke EM, Sun XC et al (2015) Identification of bolting-related microRNAs and their targets reveals complex miRNA-mediated flowering-time regulatory networks in radish (Raphanus sativus L.) Sci Rep 5:14034
Pagliarani C, Vitali M, Ferrero M, Vitulo N, Incarbone M, Lovisolo C et al (2017) The accumulation of miRNAs differentially modulated by drought stress is affected by grafting in grapevine. Plant Physiol 173(4):2180–2195
Patterson BD (1984) Estimation of hydrogen peroxide in plant extracts using titanium (IV). Anal Biochem 139:487–492
Perry JJ, Shin DS, Getzoff ED, Tainer JA (2010) The structural biochemistry of the superoxide dismutases. Biochim Biophys Acta 1804(2):245–262
Pitcher LH, Zilinskas BA (1996) Overexpression of copper/zinc superoxide dismutase in the cytosol of transgenic tobacco confers partial resistance to ozone-induced foliar necrosis. Plant Physiol 110:583–588
Schmedes A, Holmer G (1989) A new thiobarbituric acid (TBA) method for determining free malondialdehyde (MDA) and hydroperoxides selectively as a measure of lipid peroxidation. J am oil Chem Soc 66:813–817
Sen Gupta A, Heinen JL, Holaday AS, Burke JJ, Allen RD (1993a) Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic cu/Zn superoxide dismutase. PNAS 90:1629–1633
Sen Gupta A, Webb RP, Holaday AS, Allen RD (1993b) Over-expression of superoxide dismutase protects plants from oxidative stress: induction of ascorbate peroxidase in superoxide dismutase-overexpressing plants. Plant Physiol 103:1067–1073
Shalata A, Mittova V, Volokita M, Guy M, Tal M (2001) Response of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii to salt-dependent oxidative stress: the root antioxidative system. Physiol Plant 112:487–494
Shi R, Chiang VL (2005) Facile means for quantifying microRNA expression by real-time PCR. BioTechniques 39:519–525
Singla-Pareek SL, Reddy MK, Sopory SK (2003) Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. PNAS 100(25):14672–14677
Sun X, Kibet NK, Han J, Shangguan LF, Emrul K, Leng XP et al (2012) Characterization of grapevine microR164 and its target genes. Mol Biol Rep 39:9463–9472
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18:2051–2065
Suzuki N, Mittler R (2006) Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol Plant 126:45–51
Wang C, Wang X, Nicholas KK, Song C, Zhang C, Li X et al (2011) Deep sequencing of grapevine flower and berry short RNA library for discovery of new microRNAs and validation of precise sequences of grapevine microRNAs deposited in miRBase. Physiol Plant 143:64–81
Wang C, Han J, Kibet NK, Wang XC, Liu H, Li XY et al (2013) Characterization of target mRNAs for grapevine microRNAs with an integrated strategy of modified RLM-RACE, newly developed PPM-RACE and qPCRs. J Plant Physiol 170:943–957
Wang C, Leng XP, Zhang YY, Kayesh E, Zhang YP, Sun X et al (2014) Transcriptome-wide analysis of dynamic variations in regulation modes of grapevine microRNAs on their target genes during grapevine development. Plant Mol Biol 84:269–285
Wilson DN, Chung H, Elliott RC, Bremer E, George D, Koh S (2005) Microarray analysis of postictal transcriptional regulation of neuropeptides. J Mol Neurosci 25:285–298
Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M (2007) Regulation of copper homeostasis by microRNA in Arabidopsis. J Biol Chem 282:16369–16378
Yu Y, Li Y, Li L, Lin J, Zheng C, Zhang L (2009) Overexpression of PwTUA1, a pollen-specific tubulin gene, increases pollen tube elongation by altering the distribution of alpha-tubulin and promoting vesicle transport. J Exp Bot 60:2737–2749
Acknowledgments
This work was supported by grants from China Postdoctoral Science Foundation (2015M581811), the Natural Science Foundation of China (NSFC) (No. 31401846), the Fundamental Research Funds for the Central Universities (KJQN201540), the Postdoc Foundation of China (2014M561664), the Postdoc Foundation of Jiangsu Province (1401051B).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Leng, X., Wang, P., Zhu, X. et al. Ectopic expression of CSD1 and CSD2 targeting genes of miR398 in grapevine is associated with oxidative stress tolerance. Funct Integr Genomics 17, 697–710 (2017). https://doi.org/10.1007/s10142-017-0565-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-017-0565-9