Abstract
Drought is a major environmental stress factor that affects plant growth and development worldwide. Wild emmer wheat (Triticum turgidum ssp. dicoccoides), the ancestor of domesticated durum wheat (Triticum turgidum ssp. durum), has great potential for improving the understanding of the wheat drought response. MicroRNAs (miRNAs) are a recently discovered class of gene expression regulators that have also been linked to several plant stress responses; however, this relationship is just beginning to be understood. miRNA expression patterns of drought-resistant wild emmer wheat in response to drought stress were investigated using a plant miRNA microarray platform. Expression was detected to be 205 miRNAs in control and 438 miRNAs in drought-stressed leaf and root tissues. Of these miRNAs, the following 13 were differentially regulated in response to drought: miR1867, miR896, miR398, miR528, miR474, miR1450, miR396, miR1881, miR894, miR156, miR1432, miR166 and miR171. Regulation of miRNAs upon 4 and 8 h drought stress applications observed by qRT-PCR. Target transcripts of differentially regulated miRNAs were computationally predicted. In addition to miRNA microarray study, five new conserved T. turgidum miRNAs were identified through a homology-based approach, and their secondary structures and putative targets were predicted. These findings both computationally and experimentally highlight the presence of miRNAs in T. dicoccoides and further extend the role of miRNAs under shock drought stress conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
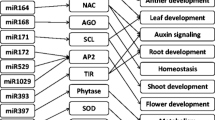
MicroRNAs (miRNAs) are a highly conserved, newly identified class of small, single stranded noncoding RNAs that range in length from 18 to 25 nucleotides. They regulate gene expression from messenger RNA (mRNA) by one of two mechanisms: post-transcriptional degradation or translational repression. In plants, most miRNAs have perfect or near perfect complementarity to their mRNA targets, and downregulate them by target cleavage (Carrington and Ambros 2003; Bartel 2004). The discovery of large numbers of miRNAs in both plants and animals has led to wide recognition of their important roles (Llave et al. 2002a; Ambros 2004). In plants, miRNAs are involved in development, signal transduction, protein degradation, response to environmental stress and pathogen invasion, and regulate their own biogenesis (Dugas and Bartel 2004; Chen 2005; Zhang et al. 2006a; Kantar et al. 2010; Unver et al. 2010). Many processes, such as leaf development, auxin signaling, phase transition, flowering, and genome maintenance, are regulated in similar ways by different miRNAs (Aukerman and Sakai 2003; Palatnik et al. 2003; Vaucheret et al. 2004; Mallory et al. 2005). Computational and experimental methods have been developed and used to identify large numbers of new miRNAs (Unver et al. 2009; Unver and Budak 2009; Kantar et al. 2010) but as yet their roles are mostly unknown.
Recently, levels of a number of miRNAs have been shown to be sensitive to abiotic or biotic stress (Sunkar and Zhu 2004; Lu et al. 2005). The first report clearly linking miRNA levels with stress tolerance concerned miR398, expression of which is transcriptionally down-regulated by oxidative stresses. In Arabidopsis thaliana, miR398 was found to target two closely related Cu/Zn superoxide dismutase coding genes, cytosolic CSD1 and chloroplastic CSD2, and a reduced level of miR398 led to improved tolerance of transgenic lines compared with the wild-type plants under oxidative stress conditions (Sunkar et al. 2006). Previously, miR395 and miR399 had been found to be induced by sulfate and inorganic phosphate starvation, respectively (Jones-Rhoades and Bartel 2004; Fujii et al. 2005). By analyzing expression patterns of their known targets, 21 miRNAs belonging to 11 miRNA families in Arabidopsis were predicted to be upregulated under UV-B stress (Zhou et al. 2007), adding to the evidence that miRNAs respond to diverse environmental stimuli.
miRNA expression profiling under drought stress conditions has now been performed in rice, Populus trichocarpa and Arabidopsis. In rice, miR169g was strongly upregulated while miR393 was transiently induced by drought (Zhao et al. 2007). In Populus, miR171l-n, miR1445, miR1446a-e and miR1447 were found to be drought responsive (Lu et al. 2008). In Arabidopsis, miR396, miR168, miR167, miR165, miR319, miR159, miR394, miR156, miR393, miR171, miR158, miR169 were shown to be drought responsive (Liu et al. 2008). Drought stress responsiveness has also been observed in a pool of Triticum aestivum small noncoding RNAs (Yao et al. 2010) and Hordeum vulgare (Kantar et al. 2010). While some of the same miRNAs have been observed to respond to drought in different species, the details of their response often differ; for example, miR169 was upregulated in rice but downregulated in Arabidopsis. Therefore, further studies are required to determine how closely miRNA expression profiles are conserved between species (Sunkar 2010).
Drought stress is a major abiotic stress factor. During constant or sporadic periods of drought, crop yields are significantly reduced. Currently, the availability of water for agriculture is becoming limited, so there is growing emphasis on the need to identify novel drought-response mechanisms for the genetic improvement of crop species (Barnabás et al. 2008; Collins et al. 2008). From this point of view, naturally occuring drought-resistant relatives of domesticated crops are of great importance in understanding stress adaptation and tolerance pathways. (Tanksley and McCouch 1997). Wild emmer wheat (T. diccocoides), as an ancestor of domesticated durum and bread wheats (T. durum and T. aestivum) has considerable potential in this sense.
Recently, several studies have identified miRNAs expressed in T. aestivum by next generation sequencing of small RNA libraries (Yao et al. 2007; Wei et al. 2009a; Xin et al. 2010) while others have predicted miRNAs by computational analysis of EST databases (Jin et al. 2008; Dryanova et al. 2008). In total, members of 51 miRNA families conserved with other plant species have been found, along with 81 families that may be specific to wheat (Xin et al. 2010). However, to date there are no experimentally or computationally identified T. dicoccoides miRNAs. Analysis of T. dicoccoides miRNAs will provide insight into the conservation of miRNAs between different wheat species. In addition, identifying novel stress-regulated miRNAs and determining their expression patterns can improve our understanding of the role of miRNA regulation in drought stress adaptation. Microarrays have proved a valuable tool for genome-wide analysis of RNA expression patterns (Schena et al. 1995). Recently, microarray analysis has been applied to miRNA expression profiling (Thomson et al. 2004) and by this approach it is possible to screen all known plant miRNAs under different environmental stresses (Zhao et al. 2007). In this study, we analyzed the effects on miRNA levels of dehydration shock in leaf and root tissues, with miRNA chips representing a total of 853 miRNAs from 21 plant species. Thirteen stress-inducible miRNAs were detected, and their potential functions analyzed.
Materials and methods
Plant materials, growth conditions and dehydration stress
Wild emmer wheat [T. turgidum ssp. dicoccoides (Korn.) Thell.] drought-tolerant genotypes Tr39477 and Tr38828, originating from southeastern Turkey where the climate is characterized by long drought periods, were selected for the miRNA microarray study. The genotypes used in this study had previously been shown to have high tolerance of slow drought stress (Ergen and Budak 2009). Seeds were surface sterilized in 4% sodium hypochlorite and pre-germinated in Petri dishes for 21 days at 4°C in the dark. Seedlings of a similar developmental stage were transferred to continuously aerated Hoagland’s solution renewed every 3 days and grown under controlled conditions (16 h photoperiod, temperature 24/22°C, relative humidity 60%, and photon flux density of 600–700 μmol m−2 s−1). At the age of 3 weeks after transfer, plant seedlings had reached the four leaf stage and were dehydration shocked for 4/8 h by removing them from tanks and leaving on paper towels under the same lighting conditions, while control plants were kept in fresh hydroponic solution (Ergen et al. 2009). Root and leaf tissue samples from both stress and control plants were collected at the fourth and eighth hour of stress. Leaf and root tissues from four plants under each treatment were pooled, fast frozen in liquid nitrogen, and stored at −80°C.
RNA isolation
RNA isolation from frozen leaf tissues using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) was carried out as outlined by Ergen et al. (2007) and Budak et al. (2006). After quantification, integrity of the isolated RNA was confirmed by separating the major rRNA bands on agarose gels.
miRNA microarray chip content and hybridization of arrays
The miRNA chip included 853 miRNA probes corresponding to miRNA transcripts listed in Sanger miRBase release 12.0 (http://www.mirbase.org/) with multiple control probes. Of these miRNAs, 158 were from Arabidopsis thaliana, 24 from Brassica napus, 63 from soybean, 17 from Medicago truncatula, 7 from Mexican cotton, 114 from cottonwood, 23 from tomato, 79 from grape, 216 from rice, 39 from sorghum, 10 from sugarcane, 31 from wheat, 43 from maize, 34 from Loblolly pine, 187 from Physcomitrella patens, 60 from S. moellendorffii, and 1 each from wild cabbage, Brassica rapa, Carica papaya, Levant cotton and G. raimondii.
The miRNA microarray was synthesized in situ by LC Sciences (Houston, TX, USA; http://www.lcsciences.com/mirna.html) where chip hybridization was also performed. Leaf and root samples of 4/8 h drought shocked and control plants were collected, total RNAs of each sample were separately isolated (Cebeci and Budak 2009), and 5 μg of each was used to probe the microarray.
miRNA microarray data analysis
An Axon Gene Pix 4000B Microarray Scanner was used for data collection and ArrayProTM image analysis software (Media Cybernetics, Silver Spring, MD, USA) was used for data extraction and image processing. The signal values were derived by background subtraction and normalization performed with a LOWESS method to remove system-related variations. A miRNA signal was accepted as detectable if it met two conditions: signal intensity higher than 3× background standard deviation, and spot CV < 0.5 (CV = signal standard deviation/signal intensity). Signals from four technical replicates each of RNA derived from stressed and control plants were compared using paired, two-tailed Student’s t test; only signals with P values <0.05 and >3-fold increased or decreased differential expression were considered as significant. The clustering analysis was performed using Cluster 3.0 and heat map was visualized using Heatmap builder and TreeView (Eisen et al. 1998).
Quantification of micro-RNA using qRT-PCR
Stem–loop reverse transcription and RT-PCR of miRNAs were performed as outlined by Varkonyi-Gasic et al. (2007), using both the same RNA samples used for the microarray, and independent isolates. A list of the primers used is presented in Supplementary Table 1. miRNA quantification using DyNAmo SYBR Green mix (Finnzymes, Espoo, Finland) was carried out as outlined by Unver et al. (2010), along with no RNA and no RT primer controls for each sample.
Computational prediction of miRNA targets
Sequences of differentially expressed miRNA probes from the microarray study, and of newly identified T. turgidum miRNAs (below) were used to interrogate sequences for target sites on the psRNAtarget web server (http://bioinfo3.noble.org/psRNATarget/). T. dicoccoides expressed sequence tags (ESTs) were from Ergen and Budak (2009). T. durum unique transcripts were downloaded from PlantGDB (http://www.plantgdb.org/).
miRNA identification by homology in T. turgidum
Homology-based computational identification of T. turgidum miRNAs was previously performed (Dryanova et al. 2008) using 769 known plant miRNA sequences deposited in miRBase version 8.0, February 2006 (Griffiths-Jones et al. 2006, 2008). We performed a similar analysis using miRBase version 14.0, September 2009, containing an additional 457 entries. T. turgidum ESTs were compared with previously known plant mature miRNA sequences using the strategy developed by Zhang et al. (2008) focusing on (1) sequence homology/conservation of ESTs of T. turgidum with known miRNAs, (2) features of secondary structure of pre-miRNAs. Mature miRNA sequences were used as queries for BLASTn algorithm v2.2.21 (Zhang et al. 2000) with an E-value cutoff of 1,000, window size of 7 and other parameters automatically adjusted for short input sequences, to search ESTs of T. turgidum recorded at the NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). EST sequences showing only 0–3 nucleotide mismatches compared with the miRNA sequence queries were chosen manually. For second filtering, BLASTn results were subjected to the Zuker folding algorithm using MFOLD 3.2 software (Mathews et al. 1999; Zuker 2003) to predict secondary structures of T. turgidum pre-miRNAs. EST sequences classified as mature and pre-miRNAs met all the following criteria: (1) mature miRNA length from 19 to 24 nucleotides, (2) not more than 3 base mismatches between candidate mature miRNA and query miRNA, (3) minimum folding free energy ındex (MFEI) >0.67 as determined by Zhang et al. (2006b), (4) no loops or large breaks in the mature miRNA sequence, (5) mature miRNA sequence located in the arms of stem–loop structure, (6) not more than 6 nucleotide mismatches between the miRNA and its opposite sequence in the RNA structure (miRNA*). To validate secondary structures and other criteria of predicted T. turgidum pre-miRNAs including free energy, size and symmetry of internal loops within arms, and miRNA-like helicity, we used RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
Results
miRNAs identified in root and leaf tissues by miRNA microarray
A plant miRNA chip microarray was used to detect differentially regulated miRNAs at 4 and 8 h drought shock conditions in leaf and root tissues of T. diccocoides. Dehydration shock was selected as the stress condition because, as reported by Ergen et al. (2009), for profiling the majority of drought-responsive genes. The microarray was probed with pooled RNA isolated from 4 T. diccocoides seedlings under each condition. In addition to 56 hybridization controls, for each miRNA probe, 8 technical replicas were tested for the control condition and 4 replicas for each stress condition.
A total of 205 miRNAs out of 853 were detected in unstressed tissues. Of these, 46 were found to be leaf specific and 21 were found to be root specific, the remaining 138 miRNAs being detected in both leaf and root tissues (Table 1).
In drought-stressed tissues, a total of 438 miRNAs out of 853 were detected. Of these, 78 were specific to 4-h stressed leaves; 43 were specific to 8-h stressed leaves; and 20 were leaf specific and detected after both stress periods. Among root-specific miRNAs, 48 were specific to 4-h stressed roots, 7 were detected only after 8-h stress, and 15 were found to be present at both time points. Expression of 19 miRNAs was detected in both leaf and root tissues only after 4 h, and 8 in both tissues only after 8 h of drought stress. Finally, 143 miRNAs were expressed in both tissues under all drought treatments, the majority (135) of which had also been detected in unstressed tissues (Table 2).
Stress responsive miRNAs identified by miRNA microarray analysis
The microarray results showed that the expression of 5 of the 205 miRNAs expressed in control tissues was not detected during drought stress, whereas 236 out of 438 miRNAs expressed in stressed tissues were not detected in control tissues. This indicates that drought stress causes an overall increase in the number of miRNAs expressed.
Differential expression due to drought stress was analyzed for those 141 miRNAs that were detected both in controls and in all drought stress conditions, and showed >3-fold increased or decreased expression at a P value <0.05; results are summarized in Table 3. In leaf tissue, miR1867 and miR896 were found to be differentially regulated under both drought conditions; miR1867 was upregulated at both timepoints while miR896 expression was downregulated after 4 h but upregulated after 8-h stress. Also after 8-h stress, probes for 2 different miR398 family members were upregulated while miR528 was downregulated.
In root tissue, miR474 was strongly induced under both drought treatments, with miR398 also upregulated. After 4 h of drought, levels of miR396 transiently decreased while miR1450 increased, but both had returned to control levels by 8-h post-stress induction. Roots that had been drought-stressed for 8 h showed the most changes in miRNA levels: in addition to miR474 and miR398, miR156, miR894, miR1432 and miR1881 were induced, while miRNA166 and miRNA171 were downregulated. The distribution of differentially expressed miRNAs by tissue and drought condition is displayed by Venn diagram (Fig. 1). In several cases multiple probes from the same family were detected as differentially expressed (miR398, miR474, miR156 and miR166). As miRNA family members usually differ from each other by only 2 or 3 bases, it is difficult to tell whether these multiple signals come from a single T. dicoccoides miRNA or several closely related ones. However, some indication is given by comparing their expression levels and cluster analysis (Fig. 2). For example, all three miR474 probes are upregulated to a similar degree, suggesting that they are detecting the same T. dicoccoides miR474, or several miR474s that are regulated similarly. For miRNA156 and 166 family members, although mean expression levels vary, they group together in the cluster analysis again suggesting a single corresponding T. dicoccoides miRNA or miRNA family. However, the different miR398 probes do not consistently cluster together, and show different levels of induction from each other (or no induction) depending on the drought condition. Therefore, it is probable that T. dicoccoides contains 2 or more miR398 family members that are induced under drought conditions but to different amounts depending on the tissue and drought conditions.
Heat map of microarray data showing differentially expressed miRNAs by tissue and drought treatment, clustered according to expression pattern. a Clustering performed by the SOM (self-organizing map) method using Euclidean distance. Green indicates low signal intensity and red high signal intensity. b Complete hierarchical clustering carried out using Euclidean distance, color coding according to the scale given
miRNA quantification
In order to confirm the differential expression of these miRNAs under drought stress (Table 3), we carried out qRT-PCR for one member of each differentially regulated miRNA family as described by Varkonyi-Gasic et al. (2007). However, we were unable to consistently amplify specific products from some miRNAs. As this PCR strategy relies on precise base-pairing between the RT primer and the 6 bases at the 3′ end of the target miRNA sequence, it is likely that the T. dicoccoides miRNAs detected by hybridization to the microarray have some differences in the last 6 bases from the probes to which they bound. These findings were also observed by Unver et al. (2010). However, the qRT-PCR data for miRNAs 474, 156 and 398 did show reproducible correlation with the microarray data (Fig. 4). In particular, miR156 was three- to fourfold upregulated in 8-h stressed roots compared to controls, while miR398 was fourfold upregulated in 8-h stressed leaves, and fivefold upregulated in 4-h stressed roots, closely correlating with the microarray data for ath-miR398b. However, we were not able to detect the expected upregulation in 8-h stressed roots. Similarly, miR474 was shown to be upregulated after 8-h but not 4-h stress in roots, and not by as large a factor as expected. We believe this is due to the tendency of stem-loop RT primers to produce nonspecific products at high cycle numbers as reported by Varkonyi-Gasic et al. (2007), meaning that the very low expression of some miRNAs in control plants may be artificially inflated in this assay.
Prediction of targets of differentially expressed miRNAs
Several of the differentially expressed miRNAs have experimentally confirmed targets in model plant species (Table 4). However, as the majority have not, we computationally predicted targets for all the differentially expressed miRNAs as outlined by Unver et al. (2009). EST resources for T. dicoccoides are currently limited; we previously sequenced 13,000 ESTs corresponding to 2,376 unique sequences from drought-stressed T. dicoccoides and T. durum. In this dataset, only one predicted target was found for a differentially expressed miRNA; a putative heat-shock protein containing thioredoxin and DnaJ domains, which is a predicted target of miR396. Therefore, the analysis was expanded to the set of 8,460 PlantGDB-assembled unique transcripts from T. durum, and the T. aestivum gene index (release 9.0) from bread wheat (Lee et al. 2005); results are listed in Table 5. For miR156, miR166, miR396 and miR398, targets were predicted in Triticeae that matched those experimentally verified in model species (Table 4). For all the differentially expressed miRNAs except for miR896, there was either a experimentally verified target in model plant species, and/or a predicted target from Triticum ESTs or gene sequences.
Computational identification of T. turgidum miRNAs
Dryanova et al. (2008) previously searched for conserved miRNAs and their targets by sequence homology with Triticeae ESTs deposited in Genbank, including those from T. turgidum ssp. durum and dicoccoides, predicting 11 members of 7 conserved miRNA families in these species. We extended the analysis by including 457 new plant miRNA sequences that have been reported since the original study was carried out. A total of five new putative conserved miRNAs have been identified from T. turgidum ESTs on the basis of sequence similarity to known miRNAs and RNA folding characteristics. Stem–loop pre-miRNA structures for the putative miRNAs are shown in Fig. 3; see Supplementary data for the predicted pri-miRNA structures. These include miR397, miR1127, miR818a, miR2275-5 and miR-2275-3, the latter two deriving from different arms of the same pri-miRNA hairpin with slightly different base-pairing. Experimental confirmation is required to determine which of these is processed to mature miRNA in vivo, or whether they both are. While the sequences used to predict these T. turgidum miRNAs were present on our microarray, several other miR397 family members were present and gave positive signals, providing indirect confirmation that this miRNA is expressed in T. dicoccoides. Similarly, all seven of the putative T. turgidum miRNA families previously predicted by Dryanova et al. (2008) were detected by our microarray. Potential targets for the new miRNAs within Triticeae sequences were also predicted by the same procedure described above (Table 6), although these also await experimental confirmation.
Discussion
The current literature suggests that plant genes involved in responses to stresses such as drought may be regulated at the post-transcriptional level, and particularly in plants, miRNAs have a major role in post-transcriptional regulation. miRNAs are involved in the regulation of numerous cellular events (reviewed by Jones-Rhoades et al. 2006). More particularly, several elements related to miRNA metabolism have been shown to be involved in abscisic acid (ABA) signaling (Xiong et al. 2001; Nishimura et al. 2005) which is an important component of the plant response to abiotic stress. Following these findings, microarray-based methods have been used to identify systematically drought-responsive miRNAs in Arabidopsis thaliana (Liu et al. 2008), Oryza sativa (Zhao et al. 2007), and Hordeum vulgare (Kantar et al. 2010).
In this study, we carried out the first systematic identification of miRNAs in T. dicoccoides, the wild progenitor of durum and bread wheats, under normal growth and dehydration shock conditions. Based on previous studies in wild emmer wheat, dehydration shock produces detectable changes in the expression profile of a higher proportion of drought-responsive genes than less acute drought treatments (Ergen and Budak 2009; Ergen et al. 2009). However, there are likely to be other genes involved in long-term drought acclimation that would be detected using a soil-drying type experiment, which would be a valuable complement to this study.
Over half of the probes (443 out of 853) on the microarray hybridized to miRNAs present in T. dicoccoides, as expected from the high conservation of miRNAs between plant species. Furthermore, more than twice as many miRNAs were detected in plants undergoing drought stress than in controls (438 compared to 205). This suggests that miRNAs are generally upregulated and have an important role in abiotic stress responses. By acting at the post-transcriptional level, they enable the plant rapidly to shut down nonessential gene expression without waiting for new proteins to be produced.
Using a cross-species miRNA microarray enabled us to identify miRNAs expressed in a species for which there is very little genome or EST sequence available. The disadvantage of this approach is that the small RNA binding to each probe may differ by one or more nucleotides from the miRNA it was designed against; so only the family, not the exact sequence of the putative miRNA can be deduced. Although as Thomson et al. (2004) noted, miRNAs that hybridize to more than one probe may also well hybridize to multiple target sequences in vivo. Additionally, there are likely to be some false positives; for example, probes for miR158 and miR163, which are believed to be specific to A. thaliana and its close relatives, gave a positive signal, although only under one of the stress conditions and not in control plants (Table 2). However, we verified the data for several miRNAs using qRT-PCR analysis (Fig. 4); moreover, a high proportion of the miRNA probes that gave positive signals here (90.2% of those in control tissues, and 62.6% of those in stressed plants) belong to families that are expressed in T. aestivum (Yao et al. 2007; Wei et al. 2009a; Xin et al. 2010) and therefore are likely to be present in T. dicoccoides as well. The plants used in these studies had not been subjected to water stress, which may explain why a higher proportion of the miRNA probes that hybridized RNA from drought-stressed tissues have not previously been identified in wheat. These newly detected drought-specific miRNAs are potentially particularly interesting, but require experimental verification by an independent technique. To reduce the risk of selecting false positives, we only examined drought-sensitive miRNAs that were detected in both stressed and control plants, but showed differential expression.
Amplification curves and bar graph of miRNAs in control and stress samples. LC leaf control, LS leaf stress, RC root control, RS root stress. a miRNA474 expression upregulated in 8 h stress treatments relative to 4 h stress and controls. b miRNA156 expression upregulated in 8 h stress treatment in root. c miRNA398 expression upregulated in 8 h stressed leaf and 4 h stressed root relative to corresponding control tissues. d Bar graph showing relative expression levels of the three miRNAs as a fold increase above arbitrary baseline. Error bars are the standard deviation of two separate PCRs each performed in triplicate
The majority (8 out of 13) of these drought-responsive miRNAs were upregulated (Fig. 2). These included miR1867, miR474, miR398, miR1450, miR1881, miR894, miR156, and miR1432. As miRNAs are negative regulators of their target mRNAs, we would expect miRNAs that shut down processes involved in normal metabolism and growth to be upregulated during drought stress, in order to conserve water and protect the cell. One example would be miR156, which downregulates transcription factors involved in development and flowering. Xin et al. (2010) found similarly that T.aestivum miR156 is upregulated in response to heat stress. In contrast, we can predict that miRNAs that are downregulated during drought stress have targets that have positive roles in stress responses. From this perspective it is interesting to note that miR396, which has been shown to target growth factor-like (GRL) transcription factors, was downregulated in drought-stressed roots, suggesting that its target would be upregulated. Although GRLs do not have a known function in roots or in stress responses, we identified as a putative alternative target a T. dicoccoides EST that encodes a heat-shock protein predicted to help protect other proteins from degradation during stress conditions. This EST has been shown to be upregulated during drought stress (Ergen and Budak 2009), correlating with the downregulation of miR396, although as different drought stresses were applied in the two studies further experiments are required to verify this reciprocal relationship. In any case, this finding highlights the possibility that the same miRNA may be re-used to downregulate different targets in different tissues and environmental conditions. We would therefore predict that miR166 and miR171, which like miR396 target transcription factor families involved in development but were downregulated upon drought stress, may have additional targets that are yet to be identified.
Similarly, the confirmed targets of miR474, which was the most strongly upregulated miRNA, do not have an obvious role in abiotic stress. However, Wei et al. (2009b) recently reported that this miRNA is also upregulated in drought-stressed maize seedlings, and proposed that it could target proline dehydrogenase (PDH), an essential enzyme in proline catabolism. As proline is a key osmoprotectant, shutting down PDH expression to allow proline accumulation would be highly favorable in drought conditions. In the same study, miR528 was found to be downregulated, as was also demonstrated here.
Known targets of miR398, which was found to be upregulated in both tissues, are involved in respiration and oxidative stress. Under drought conditions it is plausible to expect reduction in respiration, but as oxidative stress is one component of drought stress, this result is surprising. Similarly, miR1450 was predicted to target Mn superoxide dismutases, but was upregulated on drought stress. Previously, miR398 has not been shown to be drought-responsive, but was downregulated in response to ABA, oxidative stress and salt stress (Sunkar et al. 2006; Liu et al. 2008; Jia et al. 2009). The opposite result shown here may be due to the precise nature of the stress treatment, and suggests that oxidative stress is not the determining factor in miR398 expression under these conditions.
From the microarray data, it was also notable that while both control and stressed leaves contained a larger variety of miRNAs than control roots (184 leaf:159 root in controls; 311 leaf:240 root in stressed tissue), the majority of the drought stress-regulated miRNAs were specific to root tissue. This is understandable as the root tissue would be the first to be affected by drought conditions. It would be interesting in further studies to find out whether a larger number of miRNAs are up- or downregulated in leaves after prolonged drought.
We have shown that miR1867, miR896, and miR528 are responsive to drought specifically in T. dicoccoides leaves while miR156, miR166, miR171, miR396, miR474, miR894, miR1432, miR1450, and miR1881 are drought responsive in root tissue. Yao et al. (2007) previously found miR156 and miR171 to be more highly expressed in wheat roots than leaves, highlighting the fact that even miRNAs that are ubiquitously expressed may be differentially regulated in different tissues.
Of these only miR156, miR171 and miR396 have previously been reported to be drought induced. These three were all found to be upregulated on mannitol treatment of Arabidopsis thaliana plants along with miR167 and miR168 (Liu et al. 2008) which were not induced in our study. Furthermore, while miR156 was upregulated in both cases, miR171 and miR396 were upregulated in A. thaliana but downregulated in our study. Xin et al. (2010) recently reported that miR166 is upregulated on heat stress in T. aestivum Chinese spring, but not in a heat resistant genotype. Our data suggest that it can be downregulated during water stress in drought-tolerant T. dicoccoides; while the two stresses are not the same, both results imply that limiting the expression of miR166 may be a characteristic of wheat lines that tolerate abiotic stress. In the same study miR156 was also upregulated under stress, but the other seven heat-responsive miRNAs do not overlap with the dehydration-responsive miRNAs described here. These discrepancies could be explained by the different type of stress applied (osmotic pressure or heat vs. dehydration), or to differences in miRNA regulation between species.
A further eight miRNA families are reported to respond to drought in various plant species (reviewed by Covarrubias and Reyes 2010). The miR169 family also shows different responses to drought depending on the species: in Oryza sativa miR169g was shown to be induced in whole plants, shoots and roots in response to PEG (Zhao et al. 2007), while miRNA169a/c were found to be drought downregulated in Arabidopsis thaliana (Li et al. 2008).
In a previous report, seven miRNAs, miR156, miR159, miR160, miR164, miR169, miR172, miR395 and miR444 were computationally identified in T. turgidum ESTs (Dryanova et al. 2008). We added five more computationally identified miRNAs to this pool using the updated miRBase, namely miR397, miR1127, miR818, miR2275-5 and miR2275-3. In the microarray study, probes based on five different miR397 family members gave positive signals under one or more of the conditions, confirming the expression of this family in T. dicoccoides. We were also able to predict putative targets among Triticum genes for all of these miRNAs except miR2275-5, with functions such as metabolism and disease resistance. Of these miR397 has been shown to downregulate copper-containing plantocyanin and laccase proteins in Arabidopsis (Abdel-Ghany and Pilon 2008); our prediction of its action on a protein related to ribonucleoprotein 1 suggests, subject to experimental confirmation, a possible second role for this miRNA family in nucleic acid regulation.
In conclusion, T. dicoccoides miRNAs responsive to initial and ongoing drought stress were identified and quantified with the miRNA microarray. Furthermore, additional T. turgidum miRNAs were predicted computationally. A large number of miRNAs were induced or upregulated on drought treatment, showing that miRNAs have an important role in abiotic stress responses. To our knowledge this is the first systematic identification of drought-responsive miRNAs in a wheat species. As the T. dicoccoides lines used here are known to be drought resistant, this study will be a valuable comparison for future studies on domesticated wheats, to identify differences in miRNA expression that correlate with drought tolerance. Also, those miRNAs that were downregulated indicate targets that are likely to have positive effects on the drought response.
Abbreviations
- miRNA:
-
MicroRNA
- mRNA:
-
Messenger RNA
- MFEI:
-
Minimum folding free energy index
- EST:
-
Expressed seqeunce tag
- ABA:
-
Abscisic acid
- GRL:
-
Growth factor-like transcription factor
- PDH:
-
Proline dehydrogenase
References
Abdel-Ghany SE, Pilon M (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem 283:15932–15945
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741
Barnabás B, Jäger K, Fehér A (2008) The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ 31:11–38
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Budak H, Kasap Z, Shearman RC, Dweikat I, Sezerman U, Mahmood A (2006) Molecular characterization of cDNA encoding resistance gene-like sequences in Buchloe dactyloides. Mol Biotech 34:293–301
Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301:336–338
Cebeci O, Budak H (2009) Global expression patterns of three Festuca species exposed to different doses of glyphosate using the affymetrix genechip wheat genome array. Comp Funct Genomics 2009:505701
Chen X (2005) MicroRNA biogenesis and function in plants. FEBS Lett 579:5923–5931
Collins NC, Tardieu F, Tuberosa R (2008) Quantitative trait loci and crop performance under abiotic stress: where do we stand? Plant Physiol 147:469–486
Covarrubias AA, Reyes JL (2010) Post-transcriptional gene regulation of salinity and drought responses by plant microRNAs. Plant Cell Environ 33:481–489
Dryanova A, Zekharov A, Gulick PJ (2008) Data mining for miRNAs and their targets in the Triticeae. Genome 51:433–443
Dugas DV, Bartel B (2004) MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol 7:512–520
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95:14863–14868
Ergen NZ, Budak H (2009) Sequencing over 13,000 expressed sequence tags from six subtractive cDNA libraries of wild and modern wheats following slow drought stress. Plant Cell Environ 32:220–236
Ergen NZ, Dinler G, Shearman RC, Budak H (2007) Identifiying, cloning and structural analysis of differentially expressed genes upon Puccinia infection of Festuca rubra var rubra. Gene 393:145–152
Ergen NZ, Thimmapuram J, Bohnert HJ, Budak H (2009) Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct Integr Genomics 9:377–396
Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15:2038–2043
Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D140–D144
Jia X, Wang WX, Ren L, Chen QJ, Mendu V, Willcut B, Dinkins R, Tang X, Tang G (2009) Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant Mol Biol 71:51–59
Jin W, Li N, Zhang B, Wu F, Li W, Guo A, Deng Z (2008) Identification and verification of microRNA in wheat (Triticum aestivum). J Plant Res 121:351–355
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MCP (2004) Micro-RNA mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428:84–88
Kantar M, Unver T, Budak H (2010) Regulation of barley miRNA upon dehydration stress correlated with target gene expression. Funct Integr Genomics. doi:10.1007/s10142-010-0181-4
Lee Y, Tsai J, Sunkara S, Karamycheva S, Pertea G, Sultana R, Antonescu V, Chan A, Cheung F, Quackenbush J (2005) The TIGR gene ındices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res 33:D71–D74
Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20:2238–2251
Liu D, Yu D (2009) MicroRNA (miR396) negatively regulates the expression of ceramidase genes in Arabidopsis. Prog Nat Sci 19:781–785
Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14:836–843
Llave C, Kasschau KD, Rector MA, Carrington JC (2002a) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14:1605–1619
Llave C, Xie Z, Kasschau KD, Carrington JC (2002b) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056
Lu S, SunYH Shi R, Clark C, Li L, Chiang VL (2005) Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17:2186–2203
Lu S, Sun YH, Chiang VL (2008) Stress-responsive microRNAs in Populus. Plant J 55:131–151
Mallory AC, Bartel DP, Bartel B (2005) MicroRNA-directed regulation of Arabidopsis auxin response factor 17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17:1360–1375
Mathews DH, Sabina J, Zuker M, Turner DH (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288:911–940
Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, Asami T, Shinozaki K, Hirayama T (2005) Analysis of ABA Hypersensitive Germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J 44:972–984
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263
Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467–470
Sunkar R (2010) MicroRNAs with macro-effects on plant stress responses. Semin Cell Dev Biol. doi:10.1016/j.semcdb.2010.04.001
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miRNA398 and important for oxidative stress tolerance. Plant Cell 18:2051–2065
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Thomson JM, Parker J, Perou CM, Hammond SM (2004) A custom microarray platform for analysis of microRNA gene expression. Nat Methods 1:47–53
Unver T, Budak H (2009) Conserved microRNAs and their targets in model grass species Bracyhpodium distachyon. Planta 230:659–669
Unver T, Namuth-Covert DM, Budak H (2009) Review of current methodological approaches for characterizing microRNAs in plants. Int J Plant Genomics. doi:10.1155/2009/262463
Unver T, Bakar M, Shearman RC, Budak H (2010) Genome-wide profiling and analysis of Festuca arundinacea miRNAs and transcriptomes in response to foliar glyphosate application. Mol Genet Genomics 283:397–413
Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12. doi:10.1186/1746-4811-3-12
Vaucheret H, Vazquez F, Crété P, Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18:1187–1197
Wei B, Cai T, Zhang R, Li A, Huo N, Li S, Gu YQ, Vogel J, Jia J, Qi Y, Mao L (2009a) Novel microRNAs uncovered by deep sequencing of small RNA transcriptomes in bread wheat (Triticum aestivum L.) and Brachypodium distachyon (L.) Beauv. Funct Integr Genomics 9:499–511
Wei L, Zhang D, Xiang F, Zhang Z (2009b) Differentially expressed miRNAs potentially involved in the regulation of defense mechanism to drought stress in maize seedlings. Int J Plant Sci 170:979–989
Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133:3539–3547
Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z, Sun Q (2010) Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol 10:123
Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK (2001) Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 1:771–781
Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q (2007) Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol 8:R96
Yao Y, Ni Z, Peng H, Sun F, Xin M, Sunkar R, Zhu JK, Sun Q (2010) Non-coding small RNAs responsive to abiotic stress in wheat (Triticum aestivum L.). Funct Integr Genomics 10:187–190
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Zhang BH, Pan XP, Cobb GP, Anderson TA (2006a) Plant microRNA: a small regulatory molecule with a big impact. Dev Biol 289:3–16
Zhang BH, Pan XP, Cobb GP, Anderson TA (2006b) Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci 63:246–254
Zhang BH, Pan XP, Stellwag EJ (2008) Identification of soybean microRNAs and their targets. Planta 229:161–182
Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, Ruan K, Jin Y (2007) Identification of drought-induced microRNAs in rice. Biochem Bioph Res Commun 354:585–590
Zhou X, Wang G, Zhang W (2007) UV-B responsive microRNA genes in Arabidopsis thaliana. Mol Syst Biol 3:103. doi:10.1038/msb4100143
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Author information
Authors and Affiliations
Corresponding author
Additional information
M. Kantar and S. J. Lucas regarded as joint first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kantar, M., Lucas, S.J. & Budak, H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 233, 471–484 (2011). https://doi.org/10.1007/s00425-010-1309-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1309-4