Abstract
Advances in the information technologies have greatly contributed to the advent of larger datasets. These datasets often come from distributed sites, but even so, their large size usually means they cannot be handled in a centralized manner. A possible solution to this problem is to distribute the data over several processors and combine the different results. We propose a methodology to distribute feature selection processes based on selecting relevant and discarding irrelevant features. This preprocessing step is essential for current high-dimensional sets, since it allows the input dimension to be reduced. We pay particular attention to the problem of data imbalance, which occurs because the original dataset is unbalanced or because the dataset becomes unbalanced after data partitioning. Most works approach unbalanced scenarios by oversampling, while our proposal tests both over- and undersampling strategies. Experimental results demonstrate that our distributed approach to classification obtains comparable accuracy results to a centralized approach, while reducing computational time and efficiently dealing with data imbalance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Feature selection (FS) is a popular machine learning technique, whereby attributes that allow a problem to be clearly defined are selected, while irrelevant or redundant attributes are ignored [1]. Traditionally, an FS algorithm is applied in a centralized manner, i.e., a single selector is used to solve a given problem. However, in a big data scenario, data are often distributed, and a distributed learning approach allows multiple subsets of data to be processed in sequence or concurrently. While there are several ways to distribute an FS task, the two most common ways are as follows: (i) an identical FS algorithm is run on data stored together in one very large dataset and distributed into several processors and the results are combined, or (ii) an identical FS algorithm is run on data stored in different datasets in different locations and the results are combined. Several works have contributed to the development of different approaches to distributed FS. In [2], the authors proposed a method for distributing the FS process by features, i.e., using a vertical distribution. A merging procedure then updates the feature subset according to the improvements achieved in classification accuracy. In testing over several microarray datasets, a considerable reduction was obtained in the execution time, while performance was maintained or even improved with regard to the standard centralized approach. In [3], different ways of distributing the data (by features and by samples) were evaluated to determine to what extent it was possible to obtain similar results as those obtained using the whole dataset. In [4], distance correlation was used to evaluate the dependence between the classes and a given feature subset. A progressively refined feature subset model that uses a probabilistic representation was obtained through repeated extraction and evaluation steps. In [5], a different perspective for distribution was adopted that achieved distributed FS using data complexity instead of classification performance as a measure, obtaining optimal results while reducing the time required.
However, problems may appear when data are distributed into several processors, including a great imbalance between classes or even non-represented classes in some data subsets. The class imbalance problem occurs when a dataset is dominated by a class or classes with significantly more instances than the other classes. The learning algorithms would be biased toward the majority classes, since the rules to correctly predict instances are positively weighted in favor of the accuracy metric, whereas specific rules that predict examples from the minority classes are usually ignored. This means that minority class instances are more often misclassified than majority class instances [6]. To compensate for the imbalance problem, different general approaches can be followed [7]:
-
Weight adjustment This consists of assigning different weights to the instances of the dataset during the training phase, so as to prioritize less well-represented classes in the classification [8].
-
Undersampling This consists of eliminating instances of better represented classes. Available methods are used to determine the instances that contribute less as candidates for elimination, such as duplicate instances or instances close to others [9,10,11].
-
Oversampling This consists of generating more instances in the less well-represented classes, thus balancing the number of instances in all classes. This is the method we use in this research, as will be detailed below [12,13,14].
In recent years, several mixed solutions have been used that combine the basic approaches above [6]. Most of them employ the synthetic minority oversampling technique (SMOTE) to generate new samples for minority classes, followed by a cleaning step aimed at improving results. In [15] the authors propose a solution for imbalance consisting of combining the neighbor cleaning rule (NCL) and SMOTE techniques. NCL is used to remove outliers in the majority class while SMOTE is used to increase sample data in minority classes. Some authors have used combinations of over- and undersampling methods, e.g., [16], who describe the application of an editing technique based on the rough set theory (RST) as a new hybrid method with SMOTE aimed at improving results. Other combinations, such as the density-based spatial clustering of applications with noise (DBSCAN) algorithm for undersampling plus SMOTE for oversampling have performed well [17]. Other authors have combined the borderline synthetic minority oversampling technique (BSMOTE) with data cleaning techniques such as Tomek links or Wilson’s edited nearest neighbor rule (ENN) [18] to propose new support vector machine (SVM) classification algorithms for unbalanced datasets. The proposed BSMOTE-Tomek and BSMOTE-ENN hybrid preprocessing algorithms consist of first using SMOTE on minority samples in the neighborhood of the borderline and then removing redundant training samples to improve data utilization efficiency. Ensemble-based algorithms have also been popular in solving imbalanced learning [19], with some proposals adding different ways of adaptively selecting informative instances using cost-sensitive learning, among others [20].
Although a plethora of methods have separately tackled classification imbalance and FS, fewer methods have jointly considered FS and imbalance. Yang et al. [21] proposed iterative ensemble FS that combines filters and balanced sampling for multiclass classification of imbalanced microarray data using a one-versus-all (OVA) approach. Their experiments with different state-of-the-art filters—specifically, minimum redundancy maximum relevance (MRMR) and fast correlation-based filter (FCBF)—and under- and oversampling alternatives show that their proposal behaves better than the filters alone for multiclass microarrays. Another work [22] shows that in scenarios such as fraud detection, FS methods are considerably affected by unbalanced datasets, so using undersampling strategies in the preprocessing step can greatly improve the obtained results. Maldonado et al. [23] developed a family of embedded methods for backward FS using SVMs, adapted to select attributes relevant to discrimination between classes under imbalanced data conditions. The model, applicable to high-dimensional datasets and tested on several very imbalanced microarray datasets, achieved better predictions with consistently fewer relevant features.
Scalability is a challenge for both sampling-based and FS techniques and in terms of both the number of samples and the data dimensionality. One way of tackling this problem is to distribute the data into several processors or nodes. This is the approach adopted in our research as described here. Some previous works, including [4, 23] have approached FS for unbalanced high-dimensional datasets, but their approach lacked the ability to tackle datasets with large numbers of samples.
In this work we present a methodology for distributed FS which simultaneously deals with the problem of heterogeneous subsets. The method can be applied to the whole dataset, distributed in order to cope with the high dimensionality, and also when the situation is distributed in origin. In testing our proposal, we used two alternatives: (i) forcing the partitions of the dataset to maintain the balance between the classes, and (ii) applying oversampling techniques when the imbalance is inevitable.
The remainder of the paper is organized as follows: Sect. 2 provides a short description of the main characteristics of FS methods. Section 3 describes the proposed methodology to improve performance in the context of distributed FS. Section 4 provides a description of the datasets, filters and classification methods used in our study. Section 5 shows the results of applying the methodology to different datasets, distribution scenarios and resampling approach and also analyzes some research questions in detail. Finally, Sect. 6 includes some final remarks and proposals for future research.
2 Background
Feature selection (FS), which is the process of identifying and eliminating as much irrelevant and redundant information as possible [24], is typically applied as a preprocessing step before classification, although it has also proven to be useful in other learning tasks such as regression, clustering and anomaly detection.
While different procedures are used to implement FS, all of them have a subset evaluation task and a stopping criterion. Focusing on subset evaluation, there exist four main methods, as shown in Fig. 1:
-
Embedded methods With these methods feature subset evaluation is not a separate task prior to classification, but is part of the classification algorithm itself. The application can be as simple as adding or removing features in response to prediction errors regarding new instances [25, 26] or as complex as adding routines to combine features into richer descriptions, as in ID3 [27], C4.5 [28] CART [29]. The advantages of these methods are that they include interaction with the classification model, are less computationally intensive than wrappers and detect dependencies between features. As a drawback, FS depends on the classifier.
-
Wrapper methods These use a previous task to evaluate the feature subset obtained in each step before the induction phase in a classifier. Wrapper methods use a predefined algorithm to evaluate the quality of a feature subset, e.g., SVM [30], employing the estimated accuracy of the resulting classifier as a metric. Their main advantages are that they interact with the classifier and detect features dependencies. However, they are computationally expensive and incur an overfitting risk (when a model is too closely fit to a limited set of data) and the selection depends on the classifier.
-
Filter methods Like wrapper methods, these perform FS in a prior step to classification. The main difference is that filters do not use an induction algorithm to evaluate subsets of features but an independent measure such as mutual information or correlation. Since they do not use a learning algorithm, they do not inherit any bias; thus, this model is computationally more efficient than both wrapper and embedded methods. Filters usually operate in two phases: (i) they rank features based on certain criteria, and (ii) they select features with the highest rankings to induce the classification model. The main advantages are their lower cost and their capacity to handle redundant features and to generalize. A drawback is that they do not interact with the classifier.
-
Hybrid methods Hybrid models [31] combine the advantages of filters and wrappers while trying to eliminate their respective limitations as much as possible. They usually combine two or more FS algorithms of different conceptual origins in a sequential manner. A typical example consists of first applying a less computationally expensive filter to remove some features and then using a more computationally expensive wrapper for a fine-tuning.
2.1 Related work
There are several ways to distribute an FS task [32]. Here we consider data collected together in one very large dataset and then distributed randomly or homogeneously (the percentages of each class are maintained in the partitions) in different datasets. An identical FS method can be run on each partition and the partial results are combined. The authors of this work have reported previous experience with methodologies to distribute the FS process. In [33, 34] we presented a parallel framework for scaling up FS, by partitioning the data vertically and horizontally, that could be used with different filter methods. The basic idea is to split the data (either by features or by samples) and then apply a filter at each partition, performing several rounds aimed at obtaining a stable set of features. A merging procedure then combines the results into a single subset of relevant features. More recently, we have included complexity measures to assist the merging procedure [5]. As for dealing with unbalanced data, in a previous work we analyzed the usefulness of data complexity measures to evaluate the behavior of the SMOTE algorithm before and after applying FS to DNA microarray datasets [35].
The approach presented here is different than our previous works since it deals simultaneously with distributed FS and unbalanced data. As mentioned in the Introduction, there are other works in the literature that combine under and over sampling techniques to solve the imbalance problem. However, up to our knowledge, this is the first work that applies oversampling (SMOTE, in our case) also to the majority class, trying to compensate for the noise introduced by SMOTE in the minority class.
3 Proposed method
Before progressing any further, we define some key concepts related to the methodology:
-
Sample A dataset is defined as a collection of individual data, often called samples, instances or patterns.
-
Feature The information about each particular sample is given in the form of features or attributes.
-
Repetition The datasets need to be divided into training and test sets, for which we follow the standard procedure of randomly using 2/3 of samples for training and 1/3 of samples for testing. To ensure that division is not biased, this process, called repetition, is repeated a number of times.
-
Round Once the dataset is divided into training and test sets, we randomly distribute the training data to different nodes (also called packets). Again, to ensure that distribution is not biased, the process is repeated several times in what we call a round.
-
Packet Each training dataset is divided into a predefined number of nodes or packets of data to which oversampling and FS techniques are applied.
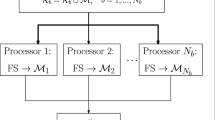
Our proposed methodology for dealing with both FS and data imbalance consists of three main steps: (i) partitioning of the data, (ii) application of an FS method to each partition, and (iii) combining the results. For each iteration (i.e., round) of our methodology, the first step is to randomly partition the training dataset into a number of disjoint packets. Repeating the process in several rounds r ensures that we have captured enough information for the combination step. We next apply an oversampling method (if the data are unbalanced) and proceed with the FS process in such a way that the features selected to be removed receive a vote. After the predefined number of rounds, features that have received votes above a certain threshold are removed, and the remaining feature subset is used in the training and test sets.
Note that, before starting, to ensure correct validation it is necessary to obtain training and test sets for each dataset to be tested. Therefore, we randomly divide each dataset into training and test sets and run several repetitions of this process (Fig. 2). After division, we start with the methodology as described below.
3.1 Step 1: data partitioning
The first step (the core of this work) consists of splitting the data without replacement, assigning groups of n samples to each subset of data. This division can be done in one of two ways, horizontally or vertically [2, 5]. Horizontal partitioning divides the dataset by instances while keeping all the features, whereas vertical partitioning divides the dataset by features while keeping all the instances. Horizontal partitioning is useful when the number of instances is much larger than the number of features, while vertical partitioning is preferred when the number of features is larger than the number of samples. The number p of packets (or subsets of data) depends on the features/instances ratio (ensuring a minimum of three packets, for our experiments, as we will see in subsequent sections). As mentioned above, this first step is repeated over several rounds (r, in this work set to five). Figure 3 illustrates the procedure of different rounds in which several packets are formed from the training set.
One of two main approaches may be followed when partitioning the data: random partitioning, in data are distributed randomly in the different nodes, or homogeneous partitioning, in which the proportions of each class in the original dataset are maintained in each of the newly generated subsets (see an example in Fig. 4, including the centralized scenario, in which all the data are together).
After partitioning, there are two situations in which new subsets of data may be unbalanced. The first one is when, for a balanced dataset, partitioning is random, leaving the majority of the samples belonging to the same class in some of the nodes by chance and thus producing data imbalance. The second situation is when the dataset is unbalanced before partitioning and the new subsets of data in each node continue to be unbalanced. In these two cases, applying the SMOTE oversampling method [12] adds synthetic minority class examples to the original dataset until the class distribution becomes balanced. The SMOTE algorithm generates the synthetic minority class examples using original minority class examples in the following way: it searches for the k nearest neighbors of the minority class sample to be used as the basis for the new synthetic sample. Next, in the segment that unites the minority class sample with one or all of its neighbors, a synthetic sample is randomly taken and is added to the new oversampled dataset. Figure 5 shows an example of SMOTE synthetic sample generation in the minority class.
Note that SMOTE is typically used to generate synthetic samples in minority classes only, whereas we propose to also generate synthetic samples in the majority class, as a means to improve the method and its results.
3.2 Step 2: FS application
An FS method is applied to each packet p to reduce the dimensionality of the problem. The features selected to be removed receive votes, a new round is performed leading to a new partitioning of the dataset and other iterations of voting are completed, until the predefined number of rounds is reached (set to five in this work). The set of all rounds can be seen as an ensemble, and so a combination of the partial results is needed, as detailed in the next step.
3.3 Step 3: results combination
The features that receive votes above a predefined threshold have to be removed. Thus, a unique set of features is obtained to train a classifier C and to test its performance on a new set of samples (test dataset). As described in [33, 34], choosing the threshold of votes is not an easy matter, since it depends on each dataset. Following the recommendations in [36], selecting the number of votes must take into account two criteria: the training classification error and the percentage of features retained. Both values should be minimized to the extent possible, in accordance with minimization of the following criterion:
where a term \(\gamma \), introduced to measure the relative importance of both values, is set to \(\gamma = 0.75\) as suggested in [36], giving more importance to the classification error. Note that the maximum number of votes is the number of rounds r when dividing by features and \(r*p\) when dividing by samples, so all the threshold values from 1 to r or 1 to \(r*p\) are evaluated. The features with votes above the obtained threshold are removed from the final subset of features. See an example in Fig. 6.
This is an example in which we have our data divided by samples (in three packets), so we have the same set of features in each packet (all the original features, in this case five features) and feature 1 was marked as not relevant for packets 1, 2 and 3; feature 2 was marked as not relevant for packet 1, feature 3 was marked as not relevant for packet 2, and features 4 and 5 are not marked as irrelevant for any packet. Our intuition says that, in this case, features 4 and 5 should be relevant and feature 1 should be not relevant, and we will have doubts about features 2 and 3 (this is why threshold of votes is needed)
After obtaining the final subset of features to be selected from the whole training set, SMOTE is applied again if the dataset is still unbalanced and a classifier is then applied.
To sum up, a pseudo-code for the proposed methodology is given in Algorithm 1.

4 Experimental setup
The goal of the experimental study is twofold. First, we aim to check if our distributed FS methodology obtains comparable results to those achieved by a centralized methodology. Second, we want to test if oversampling methods can improve performance results when FS is applied in a distributed environment and to check the effect of creating some synthetic samples from the majority class.
Described below are the datasets chosen for testing the distributed approaches, as well as the filters to perform the FS process. To test the adequacy of our distributed approach, we use four well-known supervised classifiers, of different conceptual origins. All the classifiers and filters are executed using the Weka tool,Footnote 1 using the default values for their parameters. Experiments are performed on an Intel®Core™i5-3470 CPU @ 3.20 GHz with RAM 4 GB. The code and the results of the experiments can be found in GitHub.Footnote 2
4.1 Datasets
An important issue to correctly validate the proposed methodology is to choose the appropriate benchmark of datasets. Because we present a distributed methodology which can partition the data both by filters and by samples, we consider it necessary to use datasets with high numbers of samples (standard datasets) and others with high numbers of features (microarray datasets). The standard datasets are seven datasets as described in Table 1, available for download from the UCI Machine Learning Repository.Footnote 3 The microarray datasets, from cancer classification research [2], are five datasets as described in Table 2. Those particular datasets were chosen because they represent different sizes, different numbers and different distribution of classes, and are also widely used in other research, including in our own previous work [2, 5].
In the standard datasets, partitioning in packets is done horizontally by samples, while in microarray datasets, partitioning is done vertically by features. In the latter case, homogeneous partitioning is not applied because all the packets have the same class distribution (they have all the samples). In both cases, splitting is made without repetition, so each element is in only one packet.
To determine the number of packets in each dataset (we set three as the minimum because we consider that fewer than three packets/nodes is not worthwhile for a distributed methodology), we calculate proportions, computed as the number of samples divided by the number of features for the horizontal split, and the number of features divided by the number of samples for the vertical split. If the proportion is greater than or equal to three packets, the process stops; otherwise, the denominator (number of elements in each packet) is increased by multiplying it successively by a factor from a predetermined array [100, 50, 20, 10, 5, 2, 1, 0.5, 0.25] until the condition of three or more packets is met. The pseudo-code of this process is given in Algorithm 2, and an example is shown in Fig. 7.

Tables 1 and 2 show, for each dataset, the number of samples, number of features, number of different classes, percentage for the top (one) majority class, percentage for the bottom (one) minority class, imbalance ratio (IR) as the majority class percentage divided by the minority class percentage, and number of packets in the distributed split. We consider that a dataset is unbalanced when IR > 1.
4.2 Feature selection filters
From the different categories of FS methods reviewed in Sect. 2, for this work we use filters because they are less computationally expensive and are independent of the induction algorithms. We used several popular filter methods: two ranker algorithms (InfoGain and ReliefF), a subset method based on correlation between features (CFS) and, finally, a subset method based on consistency (Consistency). We thus have representative filters based on mutual information, distance, correlation and consistency. All these filters are available in the popular Weka tool and are widely used by FS researchers.
-
InfoGain: Information Gain [37] is one of the best known FS methods. It is a univariate ranker, i.e., it evaluates only one feature at a time. It is based on the information gain equation by Quinlan [28]:
$$\begin{aligned} \textit{IG}(A|B)=H(A)-H(A|B) \end{aligned}$$(1)where H is the entropy, an uncertainty measure of a random feature associated with information theory [38]. InfoGain computes the information gain (IG) of each feature with respect to the class. This method obtains an ordered classification of all features and selects the first \(\delta \) features, where \(\delta \) is a defined threshold, set in this work, to the number of features selected by CFS.
-
ReliefF: ReliefF [39] is another very commonly used ranker algorithm in FS. ReliefF derives from Relief [40], which evaluates features by randomly selecting one instance, searching for the nearest neighbor for the same class and the opposite class (Relief is valid only in binary classification) and evaluating the feature values with respect to another closer neighbor’s feature values. The idea is that relevant features have similar values when they belong to the same class and different values when they belong to different classes. ReliefF improves Relief by handling multiclass and noisy data. Again we used as a threshold the number of features selected by CFS.
-
CFS: Correlation-based feature selection [24, 41] is a FS method that selects the best feature subset instead of returning a ranking of all the features. The goal is to obtain the feature subset that is best correlated with the class containing features that are not correlated among themselves. Irrelevant features are ignored because they have little correlation with the class. Redundant features are also ignored because they have high correlation with one or more features in the candidate subset of features. Each feature is evaluated individually and so CFS cannot identify strong relationships, like those reflected, for example, in parity problems.
-
Consistency: Like CFS, this FS method [42] selects an optimal feature subset, using consistency interpreted as zero inconsistency. The inconsistency rate compares feature subsets according to the level of consistency with the class. The consistency measure is monotonic, is always equal or greater for each iteration, is fast, is capable of removing redundant and/or irrelevant features and is capable of handling some noise. Different feature search strategies can be followed (exhaustive, complete, heuristic, probabilistic or hybrid), depending on the number of relevant/irrelevant/redundant/total features.
4.3 Classifiers
Classification is an important machine learning task. Essentially it involves dividing up elements so that each one is assigned to one of a number of mutually exhaustive and exclusive categories known as classes [43]. Of the several classification algorithms in the literature, for this study we use four classifiers from different families: two linear (Naïve Bayes and SVM using a linear kernel) and two nonlinear (C4.5 and IB1).
-
Naïve Bayes uses the Bayes rule defined by Reverend Thomas Bayes (1702–1761) [43]. Bayesian learning can be seen as the process of finding the most probable hypothesis given a set of training examples and a priori knowledge about the probability of each hypothesis. The application of Bayes theorem to classification consists of calculating the hypothesis with a higher a posteriori probability. It is said to be naïve because it assumes that the attributes are conditionally independent on each other given the class. Advantages are its simplicity, efficiency and robustness to noise and irrelevant features. Besides, it only needs a few instances to estimate the classification parameters. As disadvantages, this method requires a priori knowledge and is computationally expensive.
-
SVM [44] is a type of classifier based on statistical learning techniques. The original idea of the algorithm is to transform a set of input features X into a set of Y vectors of a higher dimension (even of infinite dimension) in which the problem can be solved linearly, i.e., linear models are used by SVM to implement nonlinear class boundaries. The SVM algorithm performs very well, partly because it allows the construction of flexible decision boundaries and also due to its good generalization capacity. A weak point is its high computational cost, due to the fact that it needs to handle a large number of coefficients which greatly increase when the input set is large.
-
C4.5 was developed by Quinlan [28] as an extension of Iterative Dichotomiser 3 (ID3), based on decision trees. To construct the best tree, C4.5 iterates using the feature with the best gain rate, i.e., the relation between the information gain of a feature and class and the entropy of the feature. Each feature value makes a new branch where instances are distributed until all the instances have the same class (leaf). To overcome a possible problem of overfitting with this strategy, C4.5 uses post pruning after constructing the tree, whereby a confidence factor deletes branches with little accuracy gain. In this study the factor is set to 0.5. Afterward, if so desired, the tree can be converted to easily understandable rules.
-
IB1 is an algorithm part of the instance-based learning (IBL) family. In IBL the training examples are stored verbatim while a distance function is used to determine which member of the training set is closest to an unknown test instance. Once the nearest training instance has been located, its class is predicted for the test instance. The only remaining problem is defining the distance function, which is quite easy, particularly if the attributes are numeric [45]. IB1 is limited to looking for the case stored closest to the example to be classified, generally using an Euclidean distance metric. Like Naïve Bayes, it is simple and works very well, but may be slow because of the need to store all training instances.
4.4 Evaluation metrics
The metrics used to evaluate the performance of the different models are classification accuracy, kappa statistic, packet filter time and classification time.
From the confusion matrix shown in Table 3, classification accuracy is defined as the percentage of instances that are classified correctly:
Kappa statistic [46] is an index that compares the agreement between several experts, considering agreement obtained by chance: values close to 1 indicate total agreement (the ideal value) and values close to 0 indicate agreement similar to that obtained by chance:
where
and
Classification accuracy and the kappa statistic value are obtained from the results of the classification process (line 13 of Algorithm 1), the filtering time by packet, measured in seconds, is obtained from the sum of the oversampling time for FS plus the FS method application time for each partition (lines 6 to 11 of Algorithm 1) and, finally, classification time, measured in seconds, is the summed time to remove features with votes above the threshold, applying SMOTE if data are unbalanced (lines 12 and 13 of Algorithm 1). The reason to include the kappa value is because it assesses the quality of the learning, taking into account situations in which the dataset is unbalanced and the classifier correctly learns majority class instances but systematically misclassifies minority class instances. In the case of accuracy and kappa, the higher the value, the better the results, whereas for filter time by packet and classification time, low values are preferred.
5 Experimental results and discussion
To evaluate our methodology we compare the results of a centralized scenario with the distributed (random and homogeneous, see Sect. 3.1) approaches. The results for each scenario are also compared for use of SMOTE in the different approaches. For the standard datasets, the percentage of SMOTE applied in the minority class is [0, 100, 300, 600 and Auto (i.e., the option to balance minority and majority classes)] and in the majority class is [0, 20, 40, 100]. For the microarray datasets with small samples, the percentage in both classes is [0, 20, 40, 100] plus the Auto option for the minority class. Random undersampling (RUS) was tested applying SMOTE on the minority class for percentages of [0, 100, 300, 600] for standard datasets and [0, 20, 40, 100] for microarray datasets with RUS in the majority class to balance both classes. Because of the high number of combinations being tested and the different metrics employed, the results of the experiments are difficult to analyze exhaustively; however, below we analyze results with a view to answer certain research questions.
5.1 Exploring the accuracy of the different approaches
A summary of accuracy results is summarized in Table 4, which shows the number of best cases in each distribution and the SMOTE approach. Overall, we can see that the centralized distribution functions better for the microarrays, while the distributed approach is a good option for standard datasets and especially the homogeneous distribution.
Tables 6, 7 and 8 in Appendix show more specific results in the form of detailed accuracy results for data distributions in a very unbalanced standard dataset (Musk2—Random), a balanced standard dataset (Isolet—Homogeneous) and a microarray dataset (Brain—Random), respectively. These tables show the prediction accuracy for the four classifiers and four filters, comparing different levels of oversampling (both in minority and majority classes) with the option to apply oversampling in the minority class and random undersampling in the majority class. In the case of Musk2 (Table 6), there are some cases for which the highest accuracy is obtained for the combination of filter, classifier and SMOTE percentage when no oversampling is applied; however, the highest accuracy for all combinations (97.36% with random distribution, Consistency filter and C4.5 classifier) is obtained when SMOTE is applied in both minority and majority classes. As for Isolet (Table 7), there is no combination of filter and classifier for which not applying SMOTE performed better than applying SMOTE; in particular, the highest accuracy for all combinations (96.01%) is obtained for CFS and SVM, again applying SMOTE in both minority and majority classes. Finally, for the Brain dataset (Table 8), the highest accuracy (91.43%, with Consistency filter and C4.5 classifier) is also achieved after applying SMOTE in both minority and majority classes. Moreover, there is no case in which not using SMOTE was better than using SMOTE, for all combinations of FS method and classifier.
In the interest of brevity, the rest of the results for datasets are summarized in Tables 9, 10, 11, 12, 13 and 14 in Appendix (for complete results see please GitHub repositoryFootnote 4 classification accuracy (Tables 9 and 10), kappa (Tables 11 and 12) and filter time by packet in seconds (Tables 13 and 14).
Recall that for all measures except filter time by packet higher values are better. For each dataset and evaluation metric, the best value for each scenario is indicated in bold font and the best value of all scenarios is indicated by underlining. Remember that, for the microarray datasets, homogeneous partitioning is not applied because the split in data is by features.
These results are analyzed in more detail below, but in general it can be observed as follows: (i) for accuracy, the distributed scenarios obtain comparable results to the centralized scenario (in some cases even improving on them, probably because of the application of a divide-and-conquer strategy, since, in some cases, the result obtained by the learner is more accurate if focused on a local region of the data), (ii) for kappa, SMOTE is useful in improving performance for unbalanced datasets, and (iii) for packet time, the time used was significantly reduced for the distributed scenarios.
To get more insights on the results, the statistical Friedman test [47] is used to check for significant differences between distributions and between SMOTE scenarios. Used for the Friedman test are mean results for all combinations of dataset, filter, classifier, distribution approach and SMOTE percentages. Executed for the level of significance \(\alpha =0.05\), if \(p<0.05\), the null hypothesis is rejected and at least one of the variables (distribution or scenario) is statistically significant, whereas if \(p>0.05\), the variables are not significantly different. To graphically summarize these statistical accuracy and kappa comparisons for distributions and scenarios in standard and microarray datasets, we use a critical difference (CD) diagram [48]. The axis of a CD diagram represents the average rank of best values of variables in each combination, with the lowest ranks (1 is the best rank) to the right side of the figure. Groups of variables that are not significantly different are connected by a bold line, while the CD value is shown above the graph.
Figures 8, 9, 10, 11, 1213 and 14 show statistical comparisons of accuracy and kappa for different distributions and SMOTE applications, while Fig. 11 shows a comparison of different SMOTE scenarios. In addition, Figs. 12 and 13 show detailed CD diagrams with different combinations of SMOTE percentages in minority and majority classes. Below we explain the unexpected result obtained by considering only the average value for the accuracy ranking.
5.2 Exploring the distributed partitioning consequences for classification
Comparing the two (random and homogeneous) distributed approaches with the centralized scenario, for the standard datasets, in general, the distributed approaches have better accuracy averages, as can be seen in Tables 4 and 9 and Figs. 8a, 9a and 14a . The homogeneous approach seems to achieve higher accuracy and more stable results (better kappa values), as illustrated in Tables 4 and 11 and Figs. 9b and 14b. As shown in Table 13, the distributed approaches considerably reduce the time required to filter the best features; however, there is an additional cost in calculating a votes threshold to remove irrelevant features and this increases the total classification time comparatively to the centralized approach (Table 5). The random and homogeneous partitioning approaches are a good solution to reducing computation time (especially if executed concurrently) without degrading classification performance. For the microarray datasets in some cases, classification accuracy slightly worsens with respect to centralized distribution after applying random distribution (Tables 4 and 10 and Figs. 8c, 9c and 14c), e.g., for Ovarian and Gli 85. This results from the fact that a high accuracy value is obtained in the centralized distribution, probably due to overfitting in datasets with few instances and many features. On the other hand, on applying a vertical split in random distribution, it is possible that average accuracy is reduced when features are marked as relevant/irrelevant in one but not in another packet (because of feature interactions). On the positive side, in random distribution, computational time is considerably reduced with respect to the centralized approach, since for this distribution, the reduction in filtering time (Table 13) is much greater than the increase in classification time (Table 5), while the kappa value is maintained or even increased (Tables 4 and 12 and Figs. 8d, 9d and 14d), thus making the solution more stable.
5.3 SMOTE use: minority class only versus both minority and majority classes
To analyze the advantages of using versus not using SMOTE, we compare the accuracy results in Tables 9 and 10. In standard datasets, except for Spambase and Weight, the best average accuracy is obtained without applying SMOTE. Figure 11 shows the ranking of best-case SMOTE scenarios for the different combinations, while Fig. 11a shows the ranking for accuracy in standard datasets. Not using SMOTE obtains the best results, although those results are not significantly different from those for SMOTE applied in both minority and majority classes. Decomposing the CD diagram with the different combinations of SMOTE in minority class and majority class, as shown in Fig. 12, gives more insights about these results; it is easily seen that distributions using SMOTE in both classes obtain a better ranking than not using SMOTE in any class. Note that combinations with high percentages of SMOTE (e.g., 600) are usually in the last positions of the ranking (left in the figure).
This difference in the ranking when SMOTE is applied in different percentages explains why the average results of applying SMOTE are poorer than those without SMOTE.
For example, focusing on the results in Table 8, the average value obtained without SMOTE is 78.57, compared to an average value of 77.35 for the rest of combinations of filter–classifier–SMOTE minority class–SMOTE majority class for the Brain dataset and random distribution, which is apparently a poorer result. However, if we consider the maximum value for each option, using SMOTE usually obtains better results than not using SMOTE (even 100 in some cases). For the microarray datasets, the behavior is similar, as can be seen in Fig. 13.
Comparing the accuracy percentages for SMOTE used in the minority class and SMOTE used in both the minority and majority classes (Tables 9 and 10 and Figs. 12 and 13), SMOTE for both classes usually obtains good results for both types of datasets. For standard datasets, a percentage of 100 in the minority class generally offers better results, so it is not necessary to increase this percentage to improve the method, as this may even deteriorate performance. This percentage in the minority class combined with different percentages of SMOTE in the majority class yields a good ranking position for all distributions, while a high proportion of SMOTE in the minority class or the balanced Auto option yield poorer rankings. In contrasts, for microarray datasets, the Auto option yields a better ranking that is the same as the higher values for SMOTE, even though the trend is not as clear as for the standard datasets.
The kappa value (Tables 11 and 12 and Fig. 11b, d) is also higher when SMOTE is applied in both classes, and sometimes even in the minority class only, but is never higher when SMOTE is not used; this would confirm that the use of this oversampling method produces more reliable results. This is a significant issue for large unbalanced datasets like Connect-4 and Ozone, where the use of SMOTE versus non-use of SMOTE practically doubles the kappa value. In contrast, for balanced datasets like Isolet, kappa values and accuracy averages hardly vary, as expected. The use of SMOTE implies that more cases have better kappa values (Fig. 11b, d); both distributed approaches are statistically similar, with very good p statistic values.
The filtering time (Tables 13 and 14) is always shorter without using SMOTE, as expected, due to the time consumed by the extra layer of SMOTE processing and especially in large datasets with many features or samples. As expected, and for the same reason, SMOTE applied to both classes results in a higher filtering time than SMOTE applied only in the minority class; furthermore, depending on the dataset and combination, it can be very computationally expensive, e.g., for Connect4 and Gli85. Classification time on average (Table 5) shows similar behavior as filtering time, with time increasing when distribution is applied, due to the search for a threshold, and when SMOTE is applied, due to the additional time required to apply SMOTE.
Finally, the best type of distribution for each case does not change whether or not SMOTE is applied.
5.4 Split: horizontal versus vertical
As can be seen in Table 4, with respect to the centralized approach, the distributed approaches with horizontal split (standard datasets) obtain a higher number of better combinations than the random distribution approaches with vertical split (microarray datasets). The reason is that, with a vertical split, redundant features can go in different packages (both would be selected); also it may happen that two features that are relevant together but not separately (if they fall into different packages) will not be selected (such as in the classic XOR problem). One solution is to group the features considering their relevance, ranking the original features before generating the packets and grouping features sequentially over the ranking, in such a way that features with a similar ranking will be in the same packet (as tested by us in a previous work [2]).
Of the two horizontal split approaches, homogeneous partitioning yielded better results than random partitioning, with more combinations of FS methods obtaining better accuracy results and with less deviation.
5.5 Exploring how filtering and classification methods behave in different scenarios
Regarding the different FS methods tested, for filtering methods, consistency and CFS generally obtain the best results for the different distributions and SMOTE approaches, while only InfoGain shows similar behavior when SMOTE is applied in both classes. For classification methods, C4.5 achieves the best average accuracy results, while IB1 is the most stable (with the best kappa value) for almost all distributions and SMOTE approaches. Finally, for the packet filter time, for horizontal division (standard datasets) and for vertical division (microarray datasets), the most computationally expensive methods are ReliefF and CFS, with both filtering methods having quadratic complexity.
5.6 Exploring the benefits to combining undersampling and oversampling approaches
Finally, we compare the application of SMOTE in both the majority and minority classes with the application of SMOTE in the minority class and RUS in the majority class. In this experiment, the samples in the minority class are first increased by applying SMOTE and then RUS is applied to the samples of the majority class to balance both classes. Analyzing Fig. 11 and Tables 9, 10, 11 and 12, the application of oversampling combined with undersampling yields poorer accuracy results than the other scenarios. This is due to the fact that undersampling removes a large number of samples in order to balance the classes and this causes relevant information to be lost in obtaining the best features, subsequently affecting the classification. The kappa value behaves differently for standard and microarray datasets. For standard datasets, it yields the worst result for all scenarios (the same as accuracy), whereas the opposite happens for the microarray datasets, because the number of samples to be removed is small and the classes are completely balanced, obtaining more robust results. This issue requires further study and undoubtedly constitutes an interesting line of future research.
6 Conclusions and future work
Our methodology for distributed FS tries to solve the problem of the heterogeneity of data in different partitions. We force partitions to maintain the same class distribution as the original dataset and apply the SMOTE oversampling technique to both minority and majority classes. From experimental results for seven standard datasets with a horizontal split and five microarray datasets with a vertical split, we draw the following conclusions:
-
The distributed approach with either random or homogeneous partitioning is competitive compared with the standard centralized approach and even improves the classification performance in some cases.
-
The packet filter time obtains the lowest value for random distributions running concurrently, thereby considerably reducing processing time, especially for large datasets with many samples or features.
-
Although the classification time is greater for the distributed approach due to the search for a threshold to eliminate irrelevant features, this increase is compensated for by the reduction in the filtering time, with the outcome that the total time decreases for the distributed approaches compared to the centralized approach.
-
Homogeneous partitioning usually obtains better kappa values (more stable results) than random partitioning and than the centralized distribution.
-
Applying SMOTE in the minority class (i.e., the way SMOTE is normally used) can improve the quality of learning in unbalanced datasets, but this depends on the percentage of synthetic samples generated, since in some cases the contrary effect of a slight decrease in general accuracy is obtained.
-
Applying SMOTE in both minority and majority classes (i.e., an innovative use of SMOTE) can improve on the standard approach, but, as before, this depends on the percentage applied. Using SMOTE for the majority class as well as the minority class results in better classification accuracy values irrespective of dataset type and partitioning approach. This new approach obtains higher kappa values, which indicates that the method has greater robustness.
-
The application of SMOTE in the minority class or SMOTE in both classes obtains better results than the combination of SMOTE in the minority class and RUS in the majority class in terms of balancing classes, especially for unbalanced standard datasets, where the elimination of a large amount of relevant samples prevents best features from being obtained, with the consequent reduction in accuracy.
As future work, we plan to test other methods to deal with heterogeneity, such as further undersampling techniques and weighting. We also plan to explore how to improve vertical divisions and the kind of distribution that simultaneously makes subsets of samples and features, representing a more challenging scenario.
Notes
https://github.com/jlmorillo/Heterogeneity_distributed_features. In the tables for standard and microarray datasets in Appendix, the following information is provided: mean and standard deviation (top row), maximum value (second row) and (in the lowest row) the combination that obtained the maximum value for:
References
Guyon I (2006) Feature extraction: foundations and applications, vol 207. Springer, Berlin
Bolón-Canedo V, Sánchez-Maroño N, Alonso-Betanzos A, Brown G (2015) Distributed feature selection: an application to microarray data classification. Appl Soft Comput 30:136–150
Bolón-Canedo V, Sechidis K, Sánchez-Maroño N, Alonso-Betanzos A, Brown G (2019) Insights into distributed feature ranking. Inf Sci 496:378–398
Brankovic A, Hosseini M, Piroddi L (2019) A distributed feature selection algorithm based on distance correlation with an application to microarrays. IEEE/ACM Trans Comput Biol Bioinf 16(6):1802–1815
Morán-Fernández L, Bolón-Canedo V, Alonso-Betanzos A (2017) Centralized vs. distributed feature selection methods based on data complexity measures. Knowl Based Syst 117:27–45
He H, Garcia EA (2009) Learning from imbalanced data. IEEE Trans Knowl Data Eng 21(9):1263–1284
Haixiang G, Yijing L, Shang J, Mingyun G, Yuanyue H, Bing G (2017) Learning from class-imbalanced data: review of methods and applications. Expert Syst Appl 73:220–239
Murphy P, Pazzani M, Merz C, Brunk C (1994) Reducing misclassification costs. In: International conference of machine learning. Morgan Kauffman, New Brunswick, pp 217–225
Tahir MA, Kittler J, Mikolajczyk K, Yan F (2009) A multiple expert approach to the class imbalance problem using inverse random under sampling. In: Multiple Classifier Systems, pp 82–91
Solberg AH, Solberg R (1996) A large-scale evaluation of features for automatic detection of oil spills in ERS SAR images. In: International geoscience and remote sensing symposium. Lincoln, NE, pp 1484–1486
Chawla NV, Herrera F, Garcia S, Fernandez A (2018) Smote for learning from imbalanced data: progress and challenges, marking the 15-year anniversary. J Artif Intell Res 61:863–905
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2011) Smote: synthetic minority over-sampling technique. arXiv:1106.1813
He H, Bai Y, Garcia EA, Li S (2008) Adasyn: adaptive synthetic sampling approach for imbalanced learning. In: IEEE joint conference in neural networks, IJCNN 2008
Ling C, Li CX (1998) Data mining for direct marketing: problems and solutions. In: Proceedings of the fourth international conference on knowledge discovery and data mining, KDD’98, vol 98, pp 73–79
Junsomboon N, Phienthrakul T (2017) Combining over-sampling and under-sampling techniques for imbalance dataset. In: ICMLC 2017: proceedings of the 9th international conference on machine learning and computing (ICMLC), pp 243–247
Ramentol E, Caballero Y, Bello R, Herrera F (2012) SMOTE-RSB*: a hybrid preprocessing approach based on oversampling and undersampling for high imbalanced data-sets using SMOTE and rough sets theory Knowledge Inform Syst 33(2): 245–265
Sanguanmak Y, Hanskunatai A (2016) DBSM: the combination of DBSCAN and SMOTE for imbalanced data classification. In: 13th international joint conference on computer science and software engineering (JCSSE), pp 1–5
Wang Q, Xin J, Wu J, Zheng N (2017) SVM classification of microaneurysms with imbalanced dataset based on borderline-SMOTE and data cleaning techniques. In: Verikas A, Radeva P, Nikolaev DP, Zhang W, Zhou J (eds) Ninth international conference on machine vision (ICMV 2016), vol 10341. International Society for Optics and Photonics, SPIE, pp 355–361
Zhang C, Gao W, Song J, Jiang J (2016) An imbalanced data classification algorithm of improved autoencoder neural network. In: 2016 Eighth international conference on advanced computational intelligence (ICACI). IEEE, pp 95–99
Krawczyk B, Woźniak M, Schaefer G (2014) Cost-sensitive decision tree ensembles for effective imbalanced classification. Appl Soft Comput 14:554–562
Yang J, Zhou J, Zhu Z, Ma X, Ji Z (2016) Iterative ensemble feature selection for multiclass classification of imbalanced microarray data. J Biol Res (Thessalon) 23(Suppl 1):13
A fraud detection model based on feature selection and undersampling applied to web payment systems. In: IEEE/WIC/ACM International conference on web intelligence and intelligent agent technology (WI-IAT)
Maldonado S, Weber R, Famili F (2014) Feature selection for high-dimensional class-imbalanced datasets using support vector machines. Inf Sci 286:228–246
Hall M (1999) Correlation-based feature selection for machine learning. PhD thesis, The University of Waikato
Mitchell TM (1982) Generalization as search. Artif Intell 18:203–226. Reprinted in Shavlik JW, Dietterich TG (eds) (1990) Readings in machine learning. Morgan Kaufmann, San Francisco
Winston PH (1975) Learning structural description from examples. In: Winston PH (ed) The psychology of computer vision. McGraw-Hill, New York
Quinlan JR (1983) Learning efficient classification procedures and their application to chess end games. In: Michalski RS, Carbonell JG, Mitchell TM (eds) Machine learning: an artificial intelligence approach. Morgan Kaufmann, San Francisco
Quinlan JR (1993) C4.5: programs for machine learning. Morgan Kaufmann, San Francisco
Breiman L, Friedman JH, Olshen RA, Stone CJ (1984) Classification and regression trees. Wadsworth, Belmont
Cortes C, Vapnik VN (1995) Support-vector networks. Mach Learn 20(3):273–297
Guyon I, Gunn S, Nikravesh M, Zadeh L (2006) Feature extraction. Foundations and applications. Springer, Berlin
Hand DJ, Mannila H, Smyth P (2001) Principles of data mining. MIT press, Cambridge
Bolón-Canedo V, Sánchez-Maroño N, Cerviño-Rabuñal J (2013) Scaling up feature selection: a distributed filter approach. In: Conference of the Spanish Association for artificial intelligence. Springer, Berlin, pp 121–130
Bolón-Canedo V, Sánchez-Marono N, Cervino-Rabunal J (2014) Toward parallel feature selection from vertically partitioned data. In: Proceedings of ESANN 2014, pp 395–400
Morán-Fernández L, Bolón-Canedo V, Alonso-Betanzos A (2016) Data complexity measures for analyzing the effect of smote over microarrays. In: ESANN
de Haro Garcia A (2011) Scaling data mining algorithms. Application to instance and feature selection. PhD thesis, Universidad de Granada
Hall MA, Smith LA (1998) Practical feature subset selection for machine learning. In: Proceedings of the 21st Australasian computer science conference ACSC 98. Springer, Berlin, pp 181–191
Shannon CE (1948) Mathematical theory of communication. Bell Syst Tech J 27:379–423
Kononenko I (1994) Estimating attributes: analysis and extensions of relief. In: Machine learning: ECML-94, pp 171–182
Kira K, Rendell LA (1992) A practical approach to feature selection. In: Proceedings of the ninth international workshop on Machine learning. Morgan Kaufmann Publishers Inc., Los Altos, pp 249–256
Hall MA (2000) Correlation-based feature selection for discrete and numeric class machine learning. In: Proceedings of the 17th international conference on machine learning, pp 856–863
Dash M, Liu H, Moto H (2003) Consistency-based search in feature selection. Artif Intell 151(1–2):155–176
Bramer M (2007) Principles of data mining. Springer, Berlin
Vapnik V (1999) The nature of statistical learning theory. Springer, Berlin
Witten IH, Frank E (2005) Data mining practical machine learning tools and techniques. Morgan Kaufmann Publishers Inc., Los Altos
Altman DG (1991) Practical statistics for medical research. Chapman & Hall, London
Hollander M, Wolfe DA (1973) Nonparametric statistical methods. John Wiley, New York
Demšar J (2006) Statistical comparisons of classifiers over multiple datasets. J Mach Learn Res 7:1–30
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This research has been financially supported in part by the Spanish Ministerio de Economía y Competitividad (research projects TIN2015-65069-C2-1-R and PID2019-109238GB-C22), by European Union FEDER funds and by the Consellería de Industria of the Xunta de Galicia (research project ED431C 2018/34). Financial support from the Xunta de Galicia (Centro singular de investigación de Galicia accreditation 2016–2019) and the European Union (European Regional Development Fund—ERDF), is gratefully acknowledged (research project ED431G 2019/01).
Appendix
Appendix
See Appendix Tables 6, 7, 8, 9, 10, 11, 12, 13 and 14.
Rights and permissions
About this article
Cite this article
Morillo-Salas, J.L., Bolón-Canedo, V. & Alonso-Betanzos, A. Dealing with heterogeneity in the context of distributed feature selection for classification. Knowl Inf Syst 63, 233–276 (2021). https://doi.org/10.1007/s10115-020-01526-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10115-020-01526-4