Abstract
Urea and creatinine are commonly used as biomarkers of renal function. Abnormal concentrations of these biomarkers are indicative of pathological processes such as renal failure. This study aimed to develop a model based on Raman spectroscopy to estimate the concentration values of urea and creatinine in human serum. Blood sera from 55 clinically normal subjects and 47 patients with chronic kidney disease undergoing dialysis were collected, and concentrations of urea and creatinine were determined by spectrophotometric methods. A Raman spectrum was obtained with a high-resolution dispersive Raman spectrometer (830 nm). A spectral model was developed based on partial least squares (PLS), where the concentrations of urea and creatinine were correlated with the Raman features. Principal components analysis (PCA) was used to discriminate dialysis patients from normal subjects. The PLS model showed r = 0.97 and r = 0.93 for urea and creatinine, respectively. The root mean square errors of cross-validation (RMSECV) for the model were 17.6 and 1.94 mg/dL, respectively. PCA showed high discrimination between dialysis and normality (95 % accuracy). The Raman technique was able to determine the concentrations with low error and to discriminate dialysis from normal subjects, consistent with a rapid and low-cost test.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The kidneys are responsible for various important functions such as endocrine, metabolic, resorption, homeostasis, and blood filtration, secreting metabolic waste products and excess fluids as urine. The primary function of the kidney is to maintain homeostasis by regulating the amount of body water, controlling the blood concentration of chemical elements such as potassium, sodium, calcium, and phosphorus and eliminating drugs and toxins, in addition to the secretion of hormones [1].
The failure of renal function may affect important organs such as the heart, liver, and pancreas, making them functionally impaired [2, 3]. Physiological conditions that contribute to impaired renal function in risk groups include systemic arterial hypertension (AH), which affects more than 75 % of risk patients at any age, diabetes mellitus (DM) with a high risk of chronic kidney disease (CKD), a physiological decrease in glomerular filtration rate (GFR), a common renal alteration in the elderly, patients with cardiovascular disease, and/or parents of patients with CKD [4, 5]. Nowadays, there are few population-based studies to detect changes in renal function in the early stages and the detection of these changes is restricted, almost exclusively, to the more advanced stages, when the patients need dialysis or even kidney transplantation [3].
Urea is the major nitrogen metabolite found in the serum and is derived from the degradation of proteins. About 90 % of urea is excreted by the kidneys into urine and 10 % through the gastrointestinal tract and the skin [1]. Of the urea excreted by the kidneys approximately 75 % corresponds to nonprotein nitrogen [1, 6]. Altered values of urea, measured together with the creatinine, may indicate altered renal function [6].
Serum creatinine provides important information with which to evaluate renal function by estimating the GFR [1, 7]. Being completely excreted, it is used as a useful marker of renal function, particularly of the GFR, because it is independent of factors such as the degree of hydration and protein metabolism. In contrast, the values of creatinine are easily affected by factors such as age, sex, diet, drugs, and the biochemical method used in the assay. The creatinine concentration alone is thus used neither to assess the GFR nor to detect the presence of CKD, with clinical examination being an important component of the conclusive diagnosis [1, 7].
The quantitative assessment of urea and creatinine characterizes its clinical importance in supporting the diagnosis of renal impairment [6]. One of the major requirements for clinical analysis is a simple and accurate method for quantifying biochemical parameters such as urea and creatinine in the serum, minimally or noninvasively, directly in the fluid without the need for reagents. Physicochemical analytical methods such as infrared spectroscopy [8], nuclear magnetic resonance spectroscopy [9], and Raman spectroscopy [10–15] are being developed to measure the blood, plasma, or serum components with greater precision. The development of optical methods may bring advantages because they do not rely on chemical reactions or enzymatic analysis, allowing real-time assessment and with minimal, waste-free sample preparation [14].
Raman spectroscopy (RS) emerged as a tool to analyze the molecular components of different bio-tissues [16]. It surpasses the conventional techniques for the preservation of the sample, without the need for chemical reagents, reduced time of analysis even in low concentrations, and in the presence of interfering agent or solvents [16–18]. Due to its vibrational nature, the very narrow bands that characterize the Raman scattering correspond to the vibrations of molecular chemical bonds at different energy levels present in the sample [16, 19]. By using RS, it is possible to acquire important information about the chemical composition of the samples, a molecular “fingerprint,” allowing the identification of the biochemical components responsible for the pathological changes [16]. Modern dispersive Raman spectrometers employ near-infrared excitation (785 and 830 nm), high-resolution imaging spectrographs, and deep-cooled CCD cameras for a spectrum with lower induced fluorescence background and have been successfully applied in the characterization of biological tissues and fluids, both in vitro and in vivo in different pathologies [12, 20–22].
The RS technique is under investigation for the quantification of analytes in serum such as glucose, total protein, urea, and creatinine [11, 13, 14]. An appropriate model based on the Raman bands can assay serum components urea, creatinine, glucose, cholesterol, triglyceride, total protein, albumin, hemoglobin, and bilirubin based on concentrations previously determined by conventional methods [23]. Studies applying RS in serum analysis have indicated the possibility of quantification of uric acid and the differentiation between HDL and LDL cholesterol [14]. The spectroscopic analysis of serum also allows the identification of pathological states, such as spectral differentiation via diagnostic models between healthy human serum and serum infected with the hepatitis C virus (HCV) [24] and the spectral discrimination of serum via the “surface-enhanced Raman spectroscopy” (SERS) technique employing hexagonal silver nanoparticles in the sera of patients with benign diseases, stomach, and colorectal cancer, as a rapid technique for monitoring cancer [25].
Due to the multivariate nature of the data obtained by RS, multivariate statistical methods such as the partial least squares technique (PLS) favor a quantitative assessment of the data, by correlating various information from the spectrum with changes in the concentration of the sample’s biochemical elements, allowing the analysis of data even in the presence of interfering agents [13, 23]. These methods consider all relevant spectral information and ignore that noncorrelated with the concentration of analytes of interest, remaining unchanged even with the inclusion of new samples [13]. The evaluation of Raman data using the PLS method has been used to measure blood analytes such as glucose, cholesterol, triglycerides, creatinine, urea, total protein, albumin, and hemoglobin [11–14, 23]. Principal components analysis (PCA), another multivariate technique, has been used in spectroscopy for classification and group discrimination [22, 24]. PCA is capable of reducing the dimensionality of the dataset by linearly transforming an m-dimensional space on a d-dimensional space (where d < m) based on the covariance of the data. The coordinates of the data in this new space are not correlated, and since they are obtained through covariance, the first variables, or principal components, preserve the greatest amount of variance within the original data [26]. PCA aims to obtain the variability of the spectral characteristic of a given material, based on repeating the experiment with different samples of the material under different conditions (in this case, data from normal and dialysis subjects). The differences between the groups (if any) appear in the first principal components. Discrimination is then achieved by correlating the principal components with known morphological or biochemical differences in the groups. PCA has been used to correlate pathological status and alterations in the principal components scores and vectors extracted from the Raman spectra of human urine [22], hemoglobin [27], skin in vitro [28] and in vivo [29, 30], breast [31], among other tissues.
This study evaluates the Raman technique as a methodology for accurately quantifying the concentrations of urea and creatinine in human serum with a unique spectral measure. To this end, we have developed a quantification model to estimate the concentrations of urea and creatinine in serum samples based on the PLS multivariate technique applied to the Raman spectra of serum. In order to develop the PLS model, the concentrations of urea and creatinine were assessed, respectively, by kinetic-spectrophotometric and enzymatic-spectrophotometric methods, commonly used in the estimation of urea and creatinine. The Raman spectra were also employed in a model to discriminate serum samples from two groups, normal healthy control volunteers, and those undergoing hemodialysis, based on PCA searching for a method of discrimination for patients with possible renal failure based on the Raman features.
Materials and methods
This study was developed with the approval of the Committee for Ethics and Research at University Camilo Castelo Branco under no. CAAE 19690113.3.0000.5494.
Serum samples
Fifty-five serum samples from clinically healthy subjects (37 women and 18 men, average age of 32.6 years—group 1), employees of the Celso Matos Laboratory, Santarém, PA, Brazil, and 47 predialysis serum samples of patients with chronic renal disease (32 women and 15 men, average age of 40.5 years—Group 2) from the Hemodialysis Center of the Municipal Hospital of Santarém, PA, Brazil, were obtained. Inclusion criteria were individuals with ages from 18 to 65 years, without a history of preexisting renal problems (group 1), and patients with chronic renal failure undergoing hemodialysis (group 2). Exclusion criteria were a history of complications or suspected renal impairment, except for those on hemodialysis, hypertension, cachexy, and diabetic patients with signs of pyelonephritis. No patient was previously diagnosed with cancer, heart, liver, degenerative, or infectious diseases. In the group 2, 27 subjects were diabetics and 32 subjects were hypertensive.
Samples corresponding to 10 mL of blood were collected from each volunteer in a dry tube without anticoagulant, stored in a plastic tube and sent to the laboratory for the biochemical assay. Sera were preprocessed by centrifugation at 5000 rpm, yielding approximately 5 mL of serum from each patient, which was used to determine the concentration of urea and creatinine using automated biochemistry equipment (model A-15, Biosystems S/A, Barcelona, Spain). After the biochemical measurement, the samples were frozen at −70 °C and stored for further Raman spectral analysis.
Serum urea was determined using the enzyme-spectrophotometric method, which has a reference (normal) range between 15 and 39 mg/dL, with detection limit of 2.5 mg/dL, linearity limit of 300 mg/dL, repeatability of 3.3 %, and reproducibility of 4.3 %, both at concentration of 42 mg/dL [32]. Serum creatinine was determined by modified Jaffe’s spectrophotometric kinetic alkaline picrate method (picric acid solution 25 mmol/L and sodium hydroxide 0.4 mol/L), based on the reaction of the picrate with the serum creatinine in alkaline medium, forming a chromogen whose speed of formation, obtained in two kinetic points, is proportional to the concentration of creatinine [33]. With the use of this modified Jaffe method, the kit’s manufacturer assures excellent correlation (r = 0.99) with the enzymatic method based on 66 standard samples. The creatinine test has reference values of 0.5–0.9 mg/dL for women and 0.7–1.2 for men, with linear measurement interval between 0.2 and 15 mg/dL, repeatability of 1.3 % and reproducibility of 3.6 %, both at lower concentrations of about 0.6 mg/dL [33].
Due to possible interferences of serum compounds in the Jaffe’s reaction, samples from subjects with hyperbilirubinemia and hyperlipidemia were excluded.
Raman spectroscopy
Spectral data was collected through a dispersive Raman spectrometer (model Dimension P-1, Lambda Solutions, Inc., MA, USA) as described elsewhere [34]. The spectrometer is composed of a laser diode (830 nm, 350 mW) for excitation, a f# 1.8 spectrometer with diffraction grating with 1200 lines/mm, providing a resolution of about 2 cm−1 in the spectral range of 400 to 1800 cm−1, and a CCD camera (back thinned, deep-depletion, 1340 × 100 pixels, Peltier-cooled down to −75 °C). The excitation of the sample and collection of the Raman signal were performed by means of a “Raman probe” fiber optic cable (model Vector Raman probe, Lambda Solutions, Inc., MA, USA). The spectral changes of biological samples could thus be accessed via optical fiber, with repeatability of the geometry of excitation and signal collection [34]. Laser power at the probe output was measured as 250 mW.
The acquisition and storage of spectra were performed via a Windows-based microcomputer using the proprietary RamanSoft software (version 1.4, Lambda Solutions, Inc., MA, USA), which controls (via USB connection) the exposure time of the detector and the number of acquisitions per sample, and storage spectra for further analysis. The total exposure time for each spectrum was 30 s (3 s, 10 accumulations). The calibration of the spectrometer was checked prior to data collection; the Raman bands used were the main bands of naphthalene, since this compound has characteristic, intense, and well-spaced bands in the spectral region of 500 to 1700 cm−1.
At the time of spectroscopic evaluation, the serum samples of normal subjects and dialysis patients were transported under dry ice, passively unfrozen, and subjected to Raman spectroscopy. Eighty microliters of each sample was pipetted into a sample holder made of aluminum with holes, and spectra were recorded by means of the Raman probe, with the excitation and collection extremity placed 10 mm apart from the sample surface. Spectra were then stored and subjected to preprocessing, consisting of the removal of background fluorescence by applying a polynomial function of seventh order using Matlab 6.0 software and the manual removal of remnant cosmic rays spikes. After that, the spectra were normalized by the intensity of the H2O band of serum proteins in 1660 cm−1.
Quantification model based on PLS
A multivariate regression model based on PLS was developed using the relevant spectral information extracted from the Raman spectra for predicting the concentrations of urea and creatinine from the concentrations obtained via the biochemical method. The PLS routine employed the leave-one-out cross-validation approach, where a sample is left out of the model and the concentration of this sample is estimated by modeling using the remnant n-1 samples with a number of latent variables [35, 36]. The process is repeated n times for predicting the concentrations of all samples. The number of latent variables to be included in the model was chosen from those with the lowest error of cross-validation for all the left-out samples. By plotting the biochemical concentration versus the predicted one, the correlation coefficient (r) and the root mean square error of cross-validation (RMSECV) of the model for the dataset are calculated, which is used to estimate the assessing prediction error in the calibration model, expressed by the equation [37]:
where conc_pred and conc_ref are, respectively, the concentration values predicted by the model and the reference values for the concentration of urea and creatinine analytes, and n is the number of samples in the regression model [37].
Outliers can distort the results and the accuracy of a regression curve. A criterion that is commonly used to detect outliers in a regression is based on the Cook’s distance (CD) [38]. The CD evaluates the influence that the exclusion of a certain outlier i can have on regression parameters such as the root mean square error (RMSE) of the regression and r. Given the regression of an estimated y based on the estimator x using the dataset [(x1 y1), (x2 y2), …, (xn yn)], the Cook’s distance of the observation i is calculated by the equation [38]:
where ŷ is the average value of the predicted values obtained by regression, ŷ(i) is the average value of the predicted values after removing the ith observation (the outlier), and p is the number of coefficients of the regression model. Intuitively, CD is the normalized measure of the influence of a certain data point i in the predicted average values. The criterion for a value to be considered an outlier is DC ≥ 4/n [38]. The Cook’s distance was applied to the data to determine possible outliers in the regressions obtained by the PLS models of urea and creatinine. The quantitative model was developed using the software Matlab 6.0, PLS toolbox.
Discrimination based on PCA
PCA was applied to the spectra in order to determine spectral variables that could be used to discriminate normal and dialysis groups, and the principal components that provided the greatest differences between the two groups, tested by t test (5 % significance level), were used in a discrimination model to separate the data set into two classes (normal and dialysis) based on the Euclidean distance [39], calculated using the software Matlab 6.0, Statistics toolbox.
Results
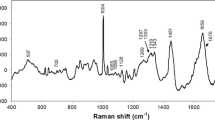
The mean Raman spectra of the serum of normal and dialysis patients are presented in Fig. 1. The serum spectrum is dominated by peaks related to serum proteins (mainly albumin), as per the major peaks assigned in recent literature [40–42]. The compounds of interest, urea and creatinine, present peaks at 1004 cm−1 (urea) [42, 43] and 680 and 846 cm−1 (creatinine) [42, 44]. These peaks, highlighted in Fig. 1, are overlapped by the peaks of other serum components, and thus, it is not possible to clearly distinguish them.
Mean Raman spectra from serum of normal and dialysis volunteers. The Raman peak positions of urea at 1004 cm−1 and creatinine at 680 and 846 cm−1 (evidenced with dashed lines) are overlapped with peaks of other compounds, especially proteins. Laser parameters 830 nm, 250 mW at probe tip, resolution 2 cm−1, exposure time 30 s
PLS has been used to develop quantification models for urea and creatinine, using the leave-one-out cross-validation approach. These multivariate regression models aimed to find a correlation between the concentrations of these serum compounds, obtained by biochemical methods, with the Raman spectral features, modeled by PLS, giving the predicted concentrations as outputs. The number of latent variables for each model was determined by the lowest RMSECV for the left-out samples [35, 36]. It was found that four latent variables for urea and five for creatinine provided the lowest RMSECV. These PLS quantitative models are presented in Fig. 2. Correlation coefficients were r = 0.97 for urea and r = 0.93 for creatinine. The RMSECV values were 17.6 mg/dL for urea and 1.94 mg/dL for creatinine. The data presented in Fig. 2 excluded the outliers determined by CD.
Plot of the concentration of urea and creatinine obtained by the biochemical method versus the concentrations predicted by the PLS models using four latent variables for urea and five for creatinine. Gray shaded intervals represent the range of the reference values according to the biochemical assay kit manufacturers [37, 38]. Values of r = 0.97 and 0.93 and RMSECV = 17.6 and 1.94 mg/dL for urea and creatinine, respectively
Despite the small spectral difference between normal and dialysis groups, PCA has been used as a possibility for discriminating the Raman spectra into normal and dialysis groups. The PCA variables, called principal components 2 and 3 (PC2 and PC3), showed statistically significant differences between normal and dialysis groups (t test, p < 0.05), indicating that these variables (PC2 and PC3) could be effective in discriminating normal and dialysis groups. Figure 3 shows the binary plot of PC2 versus PC3 for normal and dialysis groups and the discrimination line between the normal and dialysis groups based on Euclidean distance. This approach could discriminate dialysis subjects from normal ones with a sensitivity of 91 % (43 out of 47 dialysis subjects), specificity of 98 % (55 out of 56 normal subjects), and overall accuracy of 95 %. Figure 4 shows the PC2 versus the biochemical concentrations of urea (Fig. 4a) and creatinine (Fig. 4b) and the discrimination line between the groups based on Euclidean distance. The differentiation of both samples was evident either using both PC2 and PC3 or using PC2, separating the spectra between normal and dialysis.
Discussion
The use of RS as an analytical tool for quantifying urea and creatinine in human serum has advantages in reduced run time, being reagent-free and an acceptable accuracy for the monitoring and management of patients with risk of renal failure, with high reliability. A diagnostic method based on RS may contribute to reducing the cost to governments of screening large populations, favoring the detection and monitoring of patients with risk of renal failure.
One of the markers of renal damage is the increased creatinine in serum and urine [1–3, 6, 7]. The diagnostic methods routinely used for the biochemical evaluation of urea and creatinine are based on kinetic-spectrophotometric and enzyme-spectrophotometric measurements [32, 33]. These photometric methods, particularly the kinetic, have limitations mainly due to limits in the linearity and interferences related to pathologic hyperpigmentation, sample turbidity, and the use of drugs that interfere with the metabolism of the measured substances. The RS has the potential to quantify blood analytes in physiological concentrations for medical applications using multivariate technique PLS [13]. Interesting is the emerging application of RS for cancer diagnosis based on spectral analysis of serum through multivariate analysis PCA [45].
The spectra of normal subjects and those undergoing dialysis exhibit very similar peak intensities and positions. The difference in the concentrations of substances due to the nonsufficient filtration process in the kidney, particularly urea and creatinine, may be detected by RS and multivariate methods.
Cook’s distance revealed five outliers present in urea, all in dialysis patients. It is interesting to observe that three samples were outliers for both urea and creatinine. The occurrence of these outliers may be due to a high noise level in the Raman spectrum, which hinders the determination of spectral features using the PLS model, as well as the presence of other serum compounds that may interfere in both photometric and PLS biochemical assays.
The intense peaks at 1006 cm−1 for urea and 680 and 846 cm−1 for creatinine are superimposed by peaks from other components of serum, mainly proteins. Multivariate methods such as PLS and PCA are capable of revealing spectral variations that are correlated with the biochemical concentrations of urea and creatinine. The reliability of the PLS model depends on the number of samples used in the model, accuracy of the calibration procedure, spectrometer stability, and band overlap in a multicomponent sample. Changes in variables such as the laser power, sample position, and spectrograph calibration may lead to spectral changes that can be improperly modeled by the PLS. On the other hand, spectral changes that are uncorrelated with the concentrations of the compounds of interest, such as other intervening agents, are not modeled [26]. In this study, the PLS model was developed using 114 samples, which increases the robustness of the model, as it is able to determine the spectral variables most correlated with changes in concentration of the analytes of interest. Four latent variables for urea and five for creatinine have also been employed in order to find the lowest cross-validation error.
The employed colorimetric method has a reproducibility of 4.3 % at a concentration of 42 mg/dL for urea and reproducibility of <1.0 % at a concentration of 6.0 mg/dL for creatinine [32, 33]. The PLS model using the Raman spectra of serum presented a RMSECV of 17.6 mg/dL for urea and of 1.94 mg/dL for creatinine. These values suggest that RS may be developed as a tool for the population screening of renal failure, with an advantage of the fact that the quantitative analysis of both variables can be obtained in a single measurement, reducing the time necessary for examination. For serum samples with both urea and creatinine in the normal range, these error values are of concern, since the lower concentrations of these compounds result in low Raman scattering and such spectral features cannot be easily modeled by PLS.
Results obtained by Stosch et al. [46], where SERS employing colloidal silver was used along with the multivariate analysis, showed a standard deviation of less than 2 % for the quantitative determination of creatinine in human serum in preestablished dilutions. This reinforces the results presented in this study, suggesting the potential of Raman as a test that determines the concentrations of urea and creatinine in serum, rapidly, with acceptable error, and without the need for reagents, thus reducing costs.
Discrimination of the normal and dialysis groups through PC2 and PC3 scores demonstrated a high rate (95 % overall accuracy). It revealed the presence of a normal subject in the dialysis group, which was a case of altered serum assay without clinical symptoms. This subject was found to be in the stage preceding renal failure, where the clinical symptoms are not present, and only laboratory analysis can detect the risk of developing a renal lesion. This is an important finding in the early detection of renal changes, because, even with absent clinical symptoms, spectral discrimination is possible, leading to earlier diagnosis, before irreversible damage occurs and the patient requires dialysis.
The PLS quantitative method has a disadvantage in that the strength and reliability of the model depend on a large number of experimental measurements [26], which in some cases may become difficult to obtain in clinical practice, where the identification of subjects with altered serology is more important than the exact laboratory value. In these situations, discriminatory assessment via PCA may be a very promising alternative through which to discriminate subjects with altered serology. Used as screening method, Raman-based serological assay or discrimination would rapidly direct patients with abnormal results for more specific exams with smaller errors, without additional costs for nonaltered patients. For population screening, the results of discrimination by PCA are encouraging.
The estimated cost for a single colorimetric assay in Brazil is around US$0.50 for the consumables used for routine testing. Considering the high frequency in which these tests are performed, the overall cost is high for many countries such as in Brazil and the time and additional infrastructure needed may not be disregarded. On the other hand, Raman-based analysis would be an effective alternative to reduce the dependence on these inputs primarily for remote and low resources regions, as the modern Raman equipments are portable and allows real-time analysis and consequently reducing the time for the results.
Despite the method based on Jaffe’s reaction being the routine for serum creatinine assay, it presents limitation due to the reaction of the picrate with some serum compounds, notably glucose, bilirubin, ascorbic acid, proteins including hemoglobin, and kenotic bodies [47]. Most of the commercial kits employ modifications in the Jaffe’s reaction to minimize these interferences, mainly the bilirubin and proteins [47]. The colorimetric kit used in this study presents no interference from common serum compounds in the reference range (ascorbic acid below 30 mg/dL, bilirubin below 4 mg/dL, hemoglobin below 500 mg/dL, and triglycerides below 2000 mg/dL). Therefore, the possible interferences in the Jaffe method caused by these compounds were minimized in the present study. For the subjects in the group 2 (dialysis), the presence of glucosuria in diabetic patients may lead to an overestimation of the concentration of creatinine when glucose is present, as demonstrated by Weykamp et al., in experiments were glucose were added to serum samples [48]. Therefore, the correlation between the Raman spectra and the biochemical method might be biased. It is important to report the efforts to standardize the creatinine measurements aiming improvement in the diagnosis of CKD, with appropriate choice of assay methods that are creatinine-specific such as the enzymatic methods [48, 49]. This might improve the results in a Raman-based system for creatinine measurements, raising the correlation and reducing the prediction error.
Conclusion
The quantitative analysis of human serum via the PLS multivariate model, using the Raman spectral variables and concentrations of urea and creatinine obtained by standard biochemical methods, showed that the concentrations of these compounds can be obtained using a single spectral measurement, with an error of 17.6 mg/dL for urea and of 1.94 mg/dL for creatinine. Discrimination based on PCA was also possible, with the principal components PC2 and PC3 effective in discriminating between normal and dialysis groups with a sensitivity of 91 % and specificity of 98 %.
References
Alpern RJ, Caplan MJ, Moe OW (2013) Seldin and Giebisch’s the kidney—physiology and pathophysiology, vol 1. Academic Press, Waltham
About chronic kidney disease: a guide for patients and their families (2010) National Kidney Foundation. http://www.kidney.org/atoz/atozcopy.cfm?pdflink=11-50-0160_jai_patbro_aboutckdv2lr.pdf. Accessed 10 Mar 2014.
Bastos MG, Bregman R, Kirsztajn GM (2010) Chronic kidney diseases: common and harmful, but also preventable and treatable. Rev Assoc Med Bras 56:248–53
Von Schacky C (2009) Cardiovascular disease prevention and treatment. Prostaglandins Leukot Essent Fatty Acids 81(2-3):193–8
Centers for Disease Control (CDC) (1992) Incidence of treatment for end-stage renal disease attributed to diabetes mellitus, by race/ethnicity-Colorado, 1982-1989. Morb Mortal Wkly Rep 41(44):845–8
Haase-Fielitz A, Bellomo R, Devarajan P et al (2009) Novel and conventional serum biomarkers predicting acute kidney injury in adult cardiac surgery—a prospective cohort study. Crit Care Med 37(2):553–60
Gabriel IC, Nishidal SK, Kirsztajn GM (2011) Serum cystatin C: a practical alternative for renal function evaluation? J Bras Nefrol 33:261–7
Perez-Guaita D, Ventura-Gayete P-RC, Sancho-Andreu M, Garrigues S, de la Guardia M (2013) Evaluation of infrared spectroscopy as a screening tool for serum analysis: impact of the nature of samples included in the calibration set. Microchem J 106:202–11
Prat R, Galeano I, Lucas A et al (2010) Relative value of magnetic resonance spectroscopy, magnetic resonance perfusion, and 2-(18F) fluoro-2-deoxy-D-glucose positron emission tomography for detection of recurrence or grade increase in gliomas. J Clin Neurosci 17(1):50–3
Pilotto S, Pacheco MTT, Silveira L, Villaverde AB, Zângaro RA (2001) Analysis of near-infrared Raman spectroscopy as a new technique for a transcutaneous non-invasive diagnosis of blood components. Lasers Med Sci 16(1):2–9
Rohleder D, Kocherscheidt G, Gerber K et al (2005) Comparison of mid-infrared and Raman spectroscopy in the quantitative analysis of serum. J Biomed Opt 10(3):031108
Chaiken J, Finney WF, Knudson PE et al (2001) Noninvasive in-vivo tissue-modulated Raman spectroscopy of human blood: microcirculation and viscosity effects. Proc. SPIE 4254, Biomedical Diagnostic, Guidance, and Surgical-Assist Systems III, 106. doi: 10.1117/12.427952.
Berger AJ, Koo TW, Itzkan I, Horowitz G, Feld MS (1999) Multicomponent blood analysis by near-infrared Raman spectroscopy. Appl Opt 38(13):2916–26
Rohleder D, Kiefer W, Petrich W (2004) Quantitative analysis of serum and serum ultrafiltrate by means of Raman spectroscopy. Analyst 129(10):906–11
Qi D, Berger AJ (2007) Chemical concentration measurement in blood serum and urine samples using liquid-core optical fiber Raman spectroscopy. Appl Opt 46(10):1726–34
Hanlon EB, Manoharan R, Koo TW et al (2000) Prospects for in vivo Raman spectroscopy. Phys Med Biol 45(2):R1–59
González-Solís JL, Martínez-Espinosa JC, Torres-González LA, Aguilar-Lemarroy A, Jave-Suárez LF, Palomares-Anda P (2014) Cervical cancer detection based on serum sample Raman spectroscopy. Lasers Med Sci 29(3):979–85
Premasiri WR, Clarke RH, Womble ME (2001) Urine analysis by laser Raman spectroscopy. Laser Surg Med 28:330–4
Smith E, Dent G (2005) Modern Raman spectroscopy: a practical approach. John Wiley & Sons, Chiclester
Guimarães AE, Pacheco MTT, Silveira L et al (2006) Near infrared Raman spectroscopy (NIRS): a technique for doping control. Spectrosc Int J 20:185–94
Silveira L, Sathaiah S, Zângaro RA, Pacheco MT, Chavantes MC, Pasqualucci CA (2002) Correlation between near-infrared Raman spectroscopy and the histopathological analysis of atherosclerosis in human coronary arteries. Laser Surg Med 30:290–7
Bispo JAM, de Sousa Vieira EE, Silveira L, Fernandes AB (2013) Correlating the amount of urea, creatinine, and glucose in urine from patients with diabetes mellitus and hypertension with the risk of developing renal lesions by means of Raman spectroscopy and principal component analysis. J Biomed Opt 18(8):87004. doi:10.1117/1.JBO.18.8.087004
Enejder AM, Koo TW, Oh J et al (2002) Blood analysis by Raman spectroscopy. Opt Lett 27:2004–6
Saade J, Pacheco MTT, Rodrigues MR, Silveira L (2008) Identification of hepatitis C in human blood serum by near-infrared Raman spectroscopy. Spectroscopy Int J 22:387–95. doi:10.3233/SPE-2008-0344
Ito H, Inoue H, Hasegawa K et al (2014) Use of surface-enhanced Raman scattering for detection of cancer-related serum-constituents in gastrointestinal cancer patients. Nanomedicine 10(3):599–608
Chatfield C, Collins AJ (1980) Introduction to multivariate statistics. Chapman & Hall, London
Bueno Filho AC, Silveira L, Yanai ALS, Fernandes AB (2015) Raman spectroscopy for a rapid diagnosis of sickle cell disease in human blood samples: a preliminary study. Lasers Med Sci 30(1):247–53
Bodanese B, Silveira FL, Zângaro RA, Pacheco MTT, Pasqualucci CA, Silveira L (2012) Discrimination of basal cell carcinoma and melanoma from normal skin biopsies in vitro through Raman spectroscopy and principal component analysis. Photomed Laser Surg 30(7):381–7
Lui H, Zhao J, McLean D, Zeng H (2012) Real-time Raman spectroscopy for in vivo skin cancer diagnosis. Cancer Res 72(10):2491–500
Silveira FL, Pacheco MTT, Bodanese B, Pasqualucci CA, Zângaro RA, Silveira L (2015) Discrimination of non-melanoma skin lesions from non-tumor human skin tissues in vivo using Raman spectroscopy and multivariate statistics. Lasers Surg Med 47(1):6–16
Chowdary MV, Kumar KK, Kurien J, Mathew S, Krishna CM (2006) Discrimination of normal, benign, and malignant breast tissues by Raman spectroscopy. Biopolymers 83(5):556–69
Biosystems S/A (2011) UREA/BUN - UV (Urease/Glutamato Deshidrogenase). http://www.imunotech.com.br/bulas/ureia.pdf. Accessed 12 Mar 2014.
Eurotech Prod. Laboratoriais e Serviços (2012) Creatinina “Mod. Jaffe”. Reagente diagnóstico para determinação quantitativa in vitro da creatinina em soro humano, plasma ou urina em sistemas fotométricos. http://www.imunotech.com.br/bulas/rea_5073240e63694.pdf. Accessed 12 Mar 2014.
Silveira L, Silveira FL, Bodanese B, Zângaro RA, Pacheco MT (2012) Discriminating model for diagnosis of basal cell carcinoma and melanoma in vitro based on the Raman spectra of selected biochemicals. J Biomed Opt 17:077003
Martins JPA, Ferreira M (2013) QSAR modeling: a new open source computational package to generate and validate QSAR model. Quim Nov. 36(4):554–60. doi:10.1590/S0100-40422013000400013
Geladi P, Kowalski BR (1986) Partial least-squares regression: a tutorial. Anal Chim Acta 185:1–17
Brereton RG (2000) Introduction to multivariate calibration in analytical chemistry. Analyst 125(11):2125–54
Nurunnabi AAM, Hadi AS, Imon AHMR (2014) Procedures for the identification of multiple influential observations in linear regression. J Appl Stat 41(6):1315–31
Black PE (2004) Euclidean Distance. U.S. National Institute of Standards and Technology. http://www.nist.gov/dads/HTML/euclidndstnc.html. Accessed 07 May 2014.
Ivanov AI, Zhbankov RG, Korolenko EA et al (1994) Infrared and Raman spectroscopic studies of the structure of human serum albumin under various ligand loads. J Appl Spectrosc 60(5-6):305–9
Saha A, Yakovlev VV (2010) Structural changes of human serum albumin in response to a low concentration of heavy ions. J Biophotonics 3(10-11):670–7
Movasaghi Z, Rehman S, Rehman IU (2007) Raman spectroscopy of biological tissues. Appl Spectrosc Rev 42(5):493–541
Bayrak C, Bayari SH (2010) Vibrational and DFT studies of creatinine and its metal complexes. Hacett J Biol Chem 38(2):107–18
Keuleers R, Desseyn HO, Rousseau B, Van Alsenoy C (1999) Vibrational analysis of urea. J Phys Chem 103(24):4621–30
Pichardo-Molina JL, Frausto-Reyes C, Barbosa-García O et al (2007) Raman spectroscopy and multivariate analysis of serum samples from breast cancer patients. Lasers Med Sci 22(4):229–36
Stosch R, Henrion A, Schiel D, Güttler B (2005) Surface-enhanced Raman scattering based approach for quantitative determination of creatinine in human serum. Anal Chem 77:7386–92
Peake M, Whiting M (2006) Measurement of serum creatinine—current status and future goals. Clin Biochem Rev 27(4):173–84
Weykamp C, Kuypers A, Bakkeren D, Franck P, van Loon D, Klein Gunnewiek J, de Jonge R, Steigstra H, Cobbaert C (2015) Creatinine, Jaffe, and glucose: another inconvenient truth. Clin Chem Lab Med 53(12):e347–9
Panteghini M (2008) Enzymatic assays for creatinine: time for action. Clin Chem Lab Med 46(4):567–72
Acknowledgments
L. Silveira Jr. thanks São Paulo Research Foundation (FAPESP) for supporting the acquisition of the Raman spectrometer (Process no. 2009/01788-5). C. J. Saatkamp and M. L. Almeida thank Fundação Esperança, sponsor of the Instituto Esperança de Educação Superior (IESPES) for partly funding this research (Process no. 05/2012). The authors acknowledge the support of the Laboratório Celso Matos in this survey.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de Almeida, M.L., Saatkamp, C.J., Fernandes, A.B. et al. Estimating the concentration of urea and creatinine in the human serum of normal and dialysis patients through Raman spectroscopy. Lasers Med Sci 31, 1415–1423 (2016). https://doi.org/10.1007/s10103-016-2003-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10103-016-2003-y