Abstract
Serum samples were studied using Raman spectroscopy and analyzed through the multivariate statistical methods of principal component analysis (PCA) and linear discriminant analysis (LDA). The blood samples were obtained from 11 patients who were clinically diagnosed with breast cancer and 12 healthy volunteer controls. The PCA allowed us to define the wavelength differences between the spectral bands of the control and patient groups. However, since the differences in the involved molecules were in their tertiary or quaternary structure, it was not possible to determine what molecule caused the observed differences in the spectra. The ratio of the corresponding band intensities were analyzed by calculating the p values and it was found that only seven of these band ratios were significant and corresponded to proteins, phospholipids, and polysaccharides. These specific bands might be helpful during screening for breast cancer using Raman Spectroscopy of serum samples. It is also shown that serum samples from patients with breast cancer and from the control group can be discriminated when the LDA is applied to their Raman spectra.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Clinical diagnosis of breast cancer is currently done through screening techniques such as X-rays, magnetic resonance imaging, or ultrasound [1]. However, the images obtained with these techniques offer limited information regarding the screened region and they might not offer sufficient evidence of the grade of malignancy of the tumor. To gain better knowledge of the state of malignancy, a biopsy is required, and the biopsy tissue assessment and diagnosis of breast cancer depend on the experience of the pathologist [1]. Thus, alternative techniques for clinical diagnosis of breast cancer should be implemented to reduce subjectivity to human error.
For the last three decades, several noninvasive spectroscopy techniques such as Raman, infrared, and fluorescence have been used to examine tissue. Raman spectroscopy (RS) is particularly amenable for in vivo analysis because the power and wavelength of the lasers used do not cause injury. RS can provide information about the conformation of macromolecules such as proteins, nucleic acids, and lipids [2, 3], and it has been used to assess several disease states including diabetes, atherosclerosis, and Alzheimer’s in addition to cancer [4]. Recent application of RS to breast cancer [5] indicated that normal tissue had characteristics bands at 1,079, 1,300, 1,445, and 1,651 cm−1 for benign tissue; at about 1,240, 1,445, and 1,659 cm−1 for benign breast tumors; and at about 1,445 and 1,651 cm−1 for malignant breast tumors. Furthermore, Alfano et al. found differences in the relative intensity between I 1,445 and I 1,651, where I corresponds to the intensity of the band and the subindex to the position of the band. Thus, for benign tissue they found the average characteristic value of \({I_{{1,445}} } \mathord{\left/ {\vphantom {{I_{{1,445}} } {I_{{1,651}} = 1.25 \pm 0.05}}} \right. \kern-\nulldelimiterspace} {I_{{1,651}} = 1.25 \pm 0.05}\) for benign tumors the characteristic mean value was \({I_{{1,445}} } \mathord{\left/ {\vphantom {{I_{{1,445}} } {I_{{1,651}} = 0.93 \pm 0.0}}} \right. \kern-\nulldelimiterspace} {I_{{1,651}} = 0.93 \pm 0.0}3\); and for malignant tissue the mean value was \({I_{{1,445}} } \mathord{\left/ {\vphantom {{I_{{1,445}} } {I_{{1,651}} = 0.87 \pm 0.05}}} \right. \kern-\nulldelimiterspace} {I_{{1,651}} = 0.87 \pm 0.05}\). Since this pioneering work, other groups have studied cancer of the colon, lungs, and cervix adopting Alfano’s approach [6–8].
Recently, multivariate methods (MM) have been applied to RS to classify epithelial precancers and cancers. In particular, principal component analysis (PCA) fed linear discriminate analysis (LDA) has been used to differentiate between epithelial precancers and cancers [9]. For example, Enejder et al. [10], Berger et al. [11], and Berger [12] used partial least squares regression to estimate the concentration of blood analytes from RS. Glucose, urea, cholesterol, triglycerides, total protein, albumin, and hemoglobin were measured with correlation coefficients of 0.93 except for cholesterol where the correlation coefficient was 0.66. For these reasons, Raman spectroscopy and MM seem to be very promising tools to be used in biomedical research.
In this study, Raman spectroscopy and MM (PC-DA) were used to highlight differences in the chemical composition of serum samples from patients with a clinical diagnosis of breast cancer vs healthy control subjects. Furthermore, Alfano’s concept of investigating the band ratio values was applied, and we show that the serum samples from both groups can be discriminated when LDA is applied to their RS.
Statistical methods
Principal component analysis
PCA is a multivariate technique acting in an unsupervised manner and is used to analyze the inherent structure of the data. PCA reduces the dimensionality of the data set by finding an alternative set of coordinates: principal components (PCs). PCs are linear combinations of the original variables, which are orthogonal to each other and designed in such a way that each one successively accounts for the maximum variability of the data set. When the principal component scores are plotted, they reveal relations existing between the samples, such as natural data clustering or outliers. Also, when the principal component loadings are plotted as a function of different variables, they reveal which variable accounts for the greatest difference.
Linear discriminate analysis
LDA is a multivariate technique acting in a supervised manner, meaning that we know apriori how many groups there are and which samples correspond to each group. Sometimes, though not as a general rule, the PC scores are analyzed with LDA. Then, LDA reduces the dimensionality of the data set by finding an alternative set of coordinates named canonical components, or DAs. The DAs are linear combinations of the original variables (PC scores). The alternative set is obtained by maximizing the variability between the samples of different groups and minimizing the variability between samples of the same group. When the canonical component scores are plotted, they reveal relationships existing between the samples such as natural clustering of the data. This technique provides insight into how effective a pattern recognition algorithm is in classifying the data. Both the PCA and LDA methods are reviewed in [13] and [14].
Experimental
Subjects and protocol
Our study contained two groups: 11 patients with a confirmed clinical and histopathological diagnosis of breast cancer (including four patients with metastases) and 12 healthy control subjects. All patients were from the central region of Mexico and had similar ethnic and socioeconomic backgrounds. The mean age for the cancer group was 48.0 ± 10.0 years and for the control group was 34.0 ± 9.0 years. Subjects were recruited through the Human Ethical Committee of the Rodolfo Padilla-Padilla Foundation (León, Guanajuato, Mexico). Written consent was obtained from the subjects and the study was conducted according to the Declaration of Helsinki. Table 1 presents the most relevant clinical information for each patient.
Serum preparation and Raman spectroscopy analysis
After 10–12 h of overnight fasting, a single 10-ml non-heparinized peripheral blood sample was obtained between 8:00 and 9:00 a.m. and was centrifuged to get the serum specimens. Aliquots of the samples were frozen at −70°C before Raman spectroscopy analysis was performed. The RS of the solid residues from serum samples were measured by placing a drop of serum from the aliquots onto an aluminum substrate, which was subsequently examined using a Leica microscope (DMLM) integrated to the Raman system (Reni-Shaw 1000B). Multiple scans were conducted on the solid residues by moving the substrate on an X–Y stage. The Raman system was calibrated with a silicon semiconductor using the Raman peak at 520 cm−1. The wavelength of excitation was 830 nm and the laser beam was focused on the surface of the sample with a 50× objective. The radius of the beam was 2.0 μm and the laser power irradiation over the samples was 65 mW. Each spectrum was taken with an exposure of 10 s and collected in the region from 450 to 1,780 cm−1, with a resolution of 2 cm−1, and all spectra were obtained on the same day.
Data preprocessing
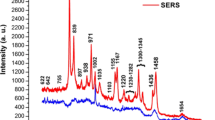
A total of 254 RS were measured from the serum blood samples of breast cancer patients and control group, with an average of 5.0 and 17.7 RS per patient in each group, respectively. The samples were preprocessed through a filter based on the Savitzky–Golay algorithm [15] using a third degree polynomial function with a window with 11 points. The fluorescence contribution was removed using a cubic spline interpolation method with eight points. Figure 1a shows an example of the recorded RS of serum blood of the control group, Fig. 1b shows the same data with smoothing, and Fig. 1c shows the same data with both smoothing and baseline correction. All the algorithms for data analysis were implemented in MatLab commercial software.
Results
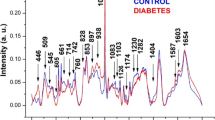
Figure 2 shows the mean spectra of the control and breast cancer groups, demonstrating that the mean spectra look similar to one another, but some peaks show differences in intensity. For example, looking at 1,003 cm−1 for phenylalanine, the peak intensity of the control group is higher than the peak intensity of the breast cancer group. Figure 2 also shows that the number of peaks in each RS is the same for both groups. Some authors have reported that some peaks in the RS of tissue from malignant tumors are shifted when compared with the spectra of tissue from benign tumors [5, 6, 18]. In this study, a Gaussian decomposition was used to localize the position of the bands in the RS using the commercial software Microcal Origin. For each group, the average position, the standard deviation, and the statistical p value were calculated using the Student t test for each band. The p values show that the average positions of ten bands from breast cancer patients are shifted when compared with those of the control group with p < 0.05 (see Table 2 for more details).
Two of the shifted bands correspond to 1,660 ± 1.0 cm−1 (proteins and phospholipids) and 1,445 ± 1.0 cm−1 (phospholipid, C–H scissor in CH2). The latter bands are close to the characteristic bands reported by Alfano et al. for the case of benign tumors. Applying Alfano’s idea to these specific bands, the mean value of the band ratio for the control group was \({\left( {{I_{{1,446}} } \mathord{\left/ {\vphantom {{I_{{1,446}} } {I_{{1,658}} }}} \right. \kern-\nulldelimiterspace} {I_{{1,658}} }} \right)} = 0.91 \pm 0.04\), and it was \({\left( {{I_{{1,446}} } \mathord{\left/ {\vphantom {{I_{{1,446}} } {I_{{1,658}} }}} \right. \kern-\nulldelimiterspace} {I_{{1,658}} }} \right)} = 0.83 \pm 0.10\) for the breast cancer patients. The statistical error and the p value (p > 0.05) do not allow this information to be used to discriminate between control and breast cancer patient groups. Therefore, the analysis suggested by Alfano et al. does not show clear distinctive differences between these groups. Thus, two multivariate statistical methods were performed and applied to our data: PCA and LDA.
PCA results
The RS were analyzed in the region from 450 to 1,780 cm−1 with a resolution of 2 cm−1.
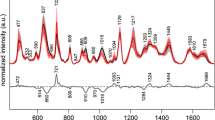
The PCA was carried out after smoothing the RS for the A and B cases with and without baseline correction, respectively. The main information obtained from the PCA is described by the first ten principal components for case A (98.2% of variance) and by the first seven principal components for case B (99.5 % of variance). By plotting the loading vectors as a function of the wave number, the position of relevant differences [17] between the control and breast cancer groups could be determined.
Figures 3 and 4 correspond to second loading (PC2) for case A and to third loading (PC3) for case B. These figures show the position of some of the major differences between both groups, which are seen for the more intense peaks. For case A, these differences appear at 731, 851 (protein, Tyr), 1,002 (Phe), 1,157 (beta carotene, C–C skeletal stretch), 1,318 (adenine), 1,338(Trp, adenine, α helix, and phospholipids), 1,450 (β sheet and phospholipids), 1,523 (beta carotene), and 1,656 cm−1 (C=O stretch, Trp, adenine, phospholipids), which come from the loading vectors of PC2, PC3, and PC4. For case B, differences are observed at 939, 1002 (Phen), 1,315 (adenine), 1,448 (β sheet and phospholipids), and 1,658 cm−1 (C=O stretch, α helix, phospholipids) which come from the loading vectors of PC2 and PC3. As can be seen, Fig. 2 shows more clearly the differences in intensity observed by the loading vectors of PC2 for case A. For example, the band at 1,002 cm−1 is one of the bands that show significant differences between the control group and breast cancer patients. The rest of the bands including 731, 1,318, or 939 cm−1 show small differences in shape and position, and the nature of these differences probably comes from the intensity variation, shifting of the bands, or a mixture of both features. For further analysis of the peaks in case A, the RS (original data) were normalized using the band positions 1,656, 1,523, 1,450, 1,338, 1,318, 1,157, 1,002, 851, and 731 cm−1 (positions found by loadings). The result of normalization of the spectra using band ratios shows statistical differences between the mean values of control and breast cancer groups for seven bands. Table 3 shows the mean values, the standard deviation of each band ratio, and the p values. The p values show that all of these band ratios are statistically significant, so they can be useful to discriminate between controls and breast cancer patients. The band ratios labeled with letters a and b for the breast cancer group in Table 3 have larger statistical error when compared with the same band ratios of the control group. This is because the serum samples came from patients at different stages of disease, including four metastatic patients. Furthermore, the bands at 1,340, 714, and 1,523 cm−1 correspond to protein, phospholipids, polysaccharides, and beta carotene. Beta carotene has been reported as a possible marker of breast cancer, and polysaccharides and genistein have been used for cancer inhibition. Nevertheless, significant variations in the intensity of seven band ratios were observed. However, it is difficult to conclude from these results whether the observed variations in intensity come only from changes in the concentrations of some of the above-mentioned molecules. Scatter plots of the score vectors were observed carefully for cases A and B. However, the scatter plots of PC1 vs PC2 and PC1 vs PC3 for both cases do not show clear discrimination between patients and control, except that points related to metastatic patients are separate from those of controls. To obtain a real discrimination between the groups, LDA was used.
So far, we used PCA to reduce the number of variables for LDA, and we also used the preliminary results obtained from loading vectors to find seven band ratios to discriminate between the spectra of the control and breast cancer groups. These bands correspond to proteins, phospholipids, and polysaccharides. In the next section, we will input the new reduced variable data into LDA to discriminate between the RS of the control group and breast cancer patients.
PC-DA
LDA was applied to discriminate between the RS of serum from control and breast cancer patients using cross-validation. In cross-validation, a portion of the data is set aside as training data, leaving the remainder as testing data. In this approach, one sample (testing data) at a time was left out. LDA was conducted after data reduction through PCA (PC-DA). Seven components for smoothing without baseline correction spectra and ten components for smoothing with baseline correction spectra were considered for this analysis. Figures 5 and 6 show the results of these calculations, and in both we were able to discriminate between the spectra of controls and breast cancer patients. The sensitivity of PC-DA is 92.2 % of the total spectra of breast cancer for data with smoothing and baseline correction, and 86.0% of the total spectra of breast cancer for smoothed data without baseline correction. The calculated linear border decision is represented by the continuous line that separates both groups in each figure. The LDA study is mainly based on the differences due to changes in the intensity of the Raman peaks, principally those bands observed in the plots of the PCA loading vectors.
Thus, PC-DA was able to discriminate very well the spectra of control groups from those of breast cancer patients. The sensitivity, specificity, positive predicted power (PP+), and negative predictive power (PP−) are summarized in Table 4. These results show that case A has the better results for all the parameters mentioned above. PC-DA provides efficient discrimination when the baseline correction is made on the RS data.
PC-DA was also applied to discriminate between patients with and without metastases; however, the PC-DA scatter plots did not show a clear discrimination, which probably reflects the small sample size.
Discussion
Alfano et al. reported that the average value of the intensity ratio of two characteristic bands (I 445 and I 1,651) could be used to identify benign and malignant tissue in breast biopsies [5, 7, 16]. These authors reported average values of \({\left( {{I_{{445}} } \mathord{\left/ {\vphantom {{I_{{445}} } {I_{{1,651}} }}} \right. \kern-\nulldelimiterspace} {I_{{1,651}} }} \right)} = 1.25 \pm 0.05\) for healthy tissue, 0.93 ± 0.03 for benign tumors, and 0.87 ± 0.05 for malignant tissue. We applied this idea to our results for I 1,446 and I 1,651 and found that \({\left( {{I_{{1,446}} } \mathord{\left/ {\vphantom {{I_{{1,446}} } {I_{{1,658}} }}} \right. \kern-\nulldelimiterspace} {I_{{1,658}} }} \right)} = 0.91 \pm 0.04\) for the control group and 0.83 ± 0.10 for the breast cancer patients. The statistical error of the mean of breast cancer patient is one order of magnitude greater than the control group, showing great variation in the band ratios. However, after analyzing the standard deviation and p value (p > 0.05), we concluded that this approach did not provide sufficient discrimination between the control and breast cancer groups, possibly because the samples were from patients at different stages of the disease. Hence, it may be necessary to increase the number of patients for each stage to discriminate between the control and breast cancer patients using this specific band ratio analysis. On the other hand, using fluorescence and Raman spectroscopy, Li and Bai [18] studied 1,022 serum blood samples from patients with different cancers (stomach, lung, liver, rectum, and esophagus). In the spectra of the cancerous serum, no apparent Raman peaks due to beta carotene were observed, but those peaks were observed in normal serum plasma. Using nonaqueous reversed-phase high-performance liquid chromatography, authors concluded that the concentration of beta carotene was lower in serum of cancer patients than in their control group. In our study, PCA analysis showed that two beta carotene-related peaks had differences between the control and breast cancer groups (1,155 and 1,523 cm−1). The mean value of the ratio of the beta carotene peaks for the breast cancer patients was lower (1.1 ± 0.3) than that of the controls (1.5 ± 0.4); however, this difference was not statistically significant and therefore, these bands could not be used to differentiate the spectra. The analysis of band ratios proposed by Alfano et al. and Xiaozhou et al. to discriminate between benign and malignant breast tissue was extrapolated to analyze and discriminate between RS of serum of control and breast cancer patients. The statistical results show that the bands proposed by these authors do not allow us to discriminate among the RS of serum samples. On the other hand, the PCA analysis allowed us to reduce the number of variables for later analysis with LDA, which was able to discriminate the RS of the control group from those of breast cancer patients.
PCA gave information on the position of the principal major differences between the RS of the control group and breast cancer patients. These bands were used to normalize the spectra and we found seven band ratios, which also allowed the discrimination of serum blood samples; the characteristic average values of band ratios for control group and breast cancer patients are described in Table 3. Also these bands were used to explain that associated comorbidities like atherosclerosis were not found in our group of patients. To prove, this conclusion we performed two different analyses, first grouping the controls by age and conducting PCA, which produced no group differences. Therefore, age was not a parameter that discriminated between groups. Our second analysis used the previous work of Nogueira and Silveira, who studied healthy and atherosclerotic human carotid arteries using Raman spectroscopy and PCA. They found three peaks that mark the principal group differences between the groups: 1,453, 1,439, and 1,663 cm−1. In our case, only one of these peaks was found by the loading vectors (1,450 cm−1). However, since the other peaks are not identified by the loadings and this single band does not appear among the band ratios of Table 3, we propose that atherosclerosis alone was not responsible for our findings. Our conclusions based on PCA results are in agreement with the clinical information that only one of our patients had a cardiovascular problem, the rest of the patients did not have any associated comorbidities, and none of the patients had atherosclerosis.
Summary
Our study demonstrated that Raman spectroscopy and multivariate analysis can be used to discriminate between serum samples from breast cancer and healthy patients and that PC-DA gives better discrimination when a baseline correction was made (case A). However, the scatter plot of PC-DA did not show a clear discrimination between patients with and without metastases since the number of samples in each disease stage was small. To explore whether PC-DA can really discriminate between patients with and without metastases, the number of cases should be increased. On the other hand, the use of PCA loading vectors allowed us to detect differences between both groups based on several bands. These bands were analyzed by means of a pairwise comparison of mean intensity. The results show seven band ratios that were statistically accepted as markers for discrimination between the spectra of control and breast cancer patients. These bands correspond to proteins, polysaccharides, and phospholipids. Our results did not reveal whether the values of the band ratios are due to changes in concentration or morphology of these molecules nor did they determine exactly what molecules were responsible for the differences. Other techniques such as Fourier transform infrared spectroscopy or magnetic resonance could be used to acquire this information. Nevertheless, we obtained specific bands that did indeed indicate group differences and could therefore be used as potential screening markers for breast cancer.
References
Ernst MF, Roukema JA (2002) Diagnosis of non-palpable breast cancer: a review. Breast 11:13–22
Hans-Uldrich G, Yan B (2001) Infrared and Raman spectroscopy of biological materials. Marcel Dekker, New York
Parker FS (1983) Applications of infrared Raman, and resonance Raman spectroscopy in biochemistry. Plenum, New York
Das K, Stone N, Kendall C, Fowler C, Christie-Brown J (2006) Raman spectroscopy of parathyroid tissue pathology. Lasers Med Sci 21(4):192–197
Alfano RR, Liu CH et al (1991) Human breast tissue studied by IR Fourier transform Raman spectroscopy. Lasers in Life Sci 4:23–28
Hanlon EB, Manoharan R et al (2001) Prospects for in vivo Raman spectroscopy. Phys Med Biol 45:R1–R59
Mahadevan-Jansen A, Richards-Kortum R (1996) Raman spectroscopy for the detection of cancers and pre-cancers. J Biomed Opt 1:31–70
Shafer-Peltier KE, Haka AS (2002) Raman micro-spectroscopic model of human breast tissue: implications for breast cancer diagnosis in vivo. J Raman Spectrosc 33:552–563
Stone N, Kendall C et al (2002) Near-infrared Raman spectroscopy for the classification of epithelial pre-cancers and cancers. J Raman Spectrosc 33:564–573
Enejder AMK, Koo TW et al (2002) Blood analysis by Raman spectroscopy. Opt Lett 27:2004–2006
Berger AJ, Koo TW et al (1999) Multicomponent blood analysis by near-infrared Raman spectroscopy. Appl Opt 38:2916–2926
Berger AJ (1998) Measurement of analytes in human serum and whole blood samples by near infrared Raman spectroscopy. Ph.D. dissertation, Massachusetts Institute of Technology
Jollife IT (1986) Principal component analysis. Springer, New York
Brereton RG (2003) Chemometrics, data analysis for the laboratory and chemical plant. Wiley, New York
Chalmers JM, Griffiths PR (2002) Handbook of vibrational spectroscopy, vol. 5. Application in life, pharmaceutical and natural science. Wiley, New York
Frank CJ, McCreery RL, Redd DCB (1995) Raman spectroscopy of normal and diseased human breast tissue. Anal Chem 67:777–783
Nogueira VG, Silveira L (2005) Raman spectroscopy study of atherosclerosis in human carotid artery. J Biomed Opt 10:031117-1–031117-7
Li X, Bai J (2001) Study of serum fluorescence and Raman spectroscopy for diagnosis of cancer. Proc SPIE 4432:124–129
Acknowledgements
The authors wish to thank CONACYT and CONCyTEG for financial support under grant numbers 42891-F, C02-44058, 03-02-K118-039-A01, and 06-04-K117-90-Anexo1. We want to thank the editor and the referees for their valuable comments to improve this work. Also, we thank Q. F. B. Yolanda Pérez Valentín and Martin Olmos.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pichardo-Molina, J.L., Frausto-Reyes, C., Barbosa-García, O. et al. Raman spectroscopy and multivariate analysis of serum samples from breast cancer patients. Lasers Med Sci 22, 229–236 (2007). https://doi.org/10.1007/s10103-006-0432-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10103-006-0432-8