Abstract
Raman spectroscopy has been proposed as a tool for diagnosis of human blood diseases aiming a quick and accurate diagnosis. Sickle cell disease arises in infancy and causes a severe anemia; thus, an early diagnosis may avoid pathological complications such as vasoocclusion, hemolytic anemia, retinopathy, cardiovascular disease, and infections. This work evaluated spectral differences between hemoglobin S (HbS) and hemoglobin A (HbA) to be used in a diagnostic model based on principal components analysis. Blood samples of patients with a previous diagnosis of sickle cell disease were hemolyzed with water, centrifuged, and the pellet was collected with a pipette. Near-infrared Raman spectra (830 nm, 200 mW) were obtained from these samples, and a model based on principal components analysis and Mahalanobis distance were used to discriminate HbA from HbS. Differences were found in the spectra of HbS and HbA, mainly in the 882 and 1,373 cm−1 (valine, HbA) and 1,547 and 1,622 cm−1 (glutamic acid, HbS). The spectral model could correctly discriminate 100 % of the samples in the correspondent groups. Raman spectroscopy was able to detect the subtle changes in the polypeptide chain (valine and glutamic acid substitution) due to the sickle cell disease and could be used to discriminate blood samples with HbS from HbA with minimum sample preparations (hemolysis with water and centrifugation).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sickle cell disease (SCD) affects millions of people throughout the world and is particularly common among those whose ancestors came from sub-Saharan Africa, Spanish-speaking regions in the Western Hemisphere (South America, the Caribbean, and Central America), Saudi Arabia, and India [1]. In the USA, the exact number of people living with SCD is unknown, but it is estimated to affect 90,000 to 100,000 Americans [2]. Many hemoglobin variants are associated with a distinct geographic distribution. For example, hemoglobin S (HbS) is typically associated with areas in both eastern and western Africa as well as areas in Saudi Arabia and India, and in localized pockets all around the Mediterranean Sea. Variations in the prevalence of HbS from within a country or geographic region are due to the presence of malaria, population isolates, altitude, and miscegenation [3]. In Brazil, SCD is being detected in approximately 3,500 newborns yearly [4], and recent estimates indicate that about 5.0 to 6.0 % of the population carries the HbS gene [5]. The ethnic origin of the Brazilian population is heterogeneous and unevenly distributed; around 2.5–40 million slaves were brought to Brazil from Africa through the slave trade in the 1500s to 1800s and were distributed in nearly all regions of the country [6–8].
Globin, the apoprotein of hemoglobin, shelters the iron-bearing porphyrin heme ring, where reversible oxygen binding occurs and permits the molecule to operate efficiently in oxygen transport; it is defined as hemoglobin A (HbA). Mutations can alter the primary amino acid sequence of globin polypeptides and sometimes result in clinically significant diseases called hemoglobinopathies [3, 7, 8]. HbS results from a point mutation in the sixth codon of the beta-globin gene, such that at the sixth position, glutamic acid is replaced by valine [9, 10].
HbS polymerizes when it is deoxygenated, and when a critical amount of HbS polymer accumulates within a sickle erythrocyte, cellular injury occurs and a sufficient number of damaged erythrocytes cause the phenotype of sickle cell disease recognized by hemolytic anemia and vasoocclusion [3, 7, 8, 11, 12]. The hemolytic anemia and vasoocclusive events are found in varying degrees in all disease genotypes, but some genotypes are clinically more severe than others. This is due in large part to variation in the cellular concentration of HbS and the propensity for polymer formation, which is highly dependent on HbS concentration [13–16]. Therefore, early detection and characterization of the hemoglobinopathies is essential so that appropriate counseling can be provided to couples and families who may be at risk of severe hematological consequences [17].
The most traditional and largely used methods of identifying and studying normal and mutant hemoglobin are the electrophoretic methods. The most used is cellulose acetate electrophoresis, also called alkaline electrophoresis, as it is performed at a pH of 8.6. At this pH, the hemoglobin molecule is negatively charged, and it will move toward the positive pole when placed in electric field. With hemoglobin variants, the amino acid substitution alters the overall charge of the molecule (due to a charge difference in the new amino acid side change), then it will have an electrophoretic mobility different from HbA as observed on cellulose acetate electrophoresis [18]. Despite the precise results in identifying the HbS, electrophoresis is a complex technical procedure, employing several reagents and time consuming. Optical techniques such as Raman spectroscopy could address this problem, by lowering costs and allowing screening examinations in a rapid way.
Raman spectroscopy has revealed to be an effective tool as a biomedical and bioanalytical technique, promising interesting application even for regular clinical use [19]. Raman spectroscopy can be employed to unveil the biochemical constitution of tissues with little sample preparation, in vivo and in real time [20]. The Raman scattering is a fundamental process in which energy is exchanged between light and matter [21]. When irradiated by a monochromatic laser beam, the molecule’s electronic cloud can be polarized. A change in the polarizability alters the energy of the scattered photon, thus causing a shift in the frequency of the scattered light depending on the molecular vibrational frequency. The scattered light is then collected by a spectrometer and displayed as the relative number of photons in each shifted frequency in a particular range [20].
Raman spectroscopy is particularly suitable for implementing the optical blood analysis techniques [20]. Campbell and Durek [22] provide detailed spectral information than the broad overtone bands of NIR absorption spectra. This is particularly important for complex biological systems such as blood [20]. Despite the relatively small cross-sections for Raman scattering, instrumentation advances have promoted rapidly increasing interest in, and have begun to reveal the benefits and capabilities of, Raman spectroscopy for blood analysis [20]. Biochemical assays based on Raman spectroscopy could be used for testing blood components and metabolites in the serum for clinical analysis as glucose, urea, total protein, albumin, cholesterol and triglycerides [20, 23, 24], doping control [25–27], detecting antibodies in cat’s sera [28], and even monitoring heparin levels in blood during surgeries [29].
More specifically, studies involving human hemoglobin investigated possible solution-phase conformational changes in HbC compared with HbA. Intrinsic fluorescence combined with UV resonance Raman data demonstrated a weakening of the Trpß15-Serß72 hydrogen bond that most likely leads to a displacement of the α-helix A away from the α-helix E [30]. Hirsch et al. [30] presented data based upon allosteric perturbation monitored by front-face fluorescence, ultraviolet resonance Raman spectroscopy, circular dichroism, and oxygen equilibrium studies that confirm and significantly expand previous findings suggesting solution-active structural differences in liganded forms of HbS and HbC distal to the site of mutation. The liganded forms of these hemoglobin are of significant interest because HbC crystallizes in the erythrocyte in the oxy form, and oxy HbS exhibits increased mechanical precipitability and a high propensity to oxidize showed that for hemoglobin the uncovering of solution-active structural differences at the tetrameric molecular level and, distal to the site of the ß6 mutations when compared with HbA. Their results suggests that point to A-helix alterations that may be the likely candidate in the primary mechanism driving oxy HbC to crystallize and causing destabilization of oxy HbS, which, under deoxy conditions, may give rise to the polymerization process [31]. Juszczak et al. [32] reported for the first time the specific conformational changes for a homogeneous population of ligand-bound adult deoxy human HbA generated by introducing CO into a sample of deoxy-HbA with the effector, inositol hexaphosphate, encapsulated in a porous sol-gel by UV resonance Raman [32].
Wood et al. [33] studied resonance Raman spectra of oxygenated and deoxygenated functional erythrocytes recorded using 785 nm laser excitation. They concluded that it can be used as a nondestructive molecular probe that has potential as an analytical and diagnostic tool for a variety of erythrocyte disorders. The technique may also have applications in monitoring heme perturbation in response to in vivo drug therapy within functional erythrocytes [33].
This work proposes the use of Raman spectroscopy for diagnosis of sickle cell disease in blood analysis. We intended to identify the spectral differences in the blood samples of a group of subjects with sickle cell anemia compared to a group of non-anemics (control) in order to detect spectral changes related to the HbS compared to the HbA related to the changes in the polypeptide chain induced in the β-globin, which has a crucial role in determining the clinical profile; this means that the highest concentration of this is a more serious disease. Then, a model based on principal components analysis (PCA) and Mahalanobis distance (MD) is presented to discriminate samples of the subjects with HbS from the ones with HbA, aiming a future diagnosis of hemoglobinopathies.
Material and methods
This study was approved by the Research Ethics Committee from UNICASTELO (protocol no. 64133). About 5 mL of blood from 22 subjects (both gender, average age of 21.1 ± 14.9) were collected in test tubes containing EDTA 10 %, which were divided as follows: 19 subjects with sickle cell disease (HbS) and 3 normal subjects (HbA). The blood samples were centrifuged at 1,500 rpm for 5 min, and then were resuspended in saline, three times until translucent. Then, hemolysates-washed red blood cells were obtained by adding deionized water; debris were separated by centrifugation. The samples were placed in cryogenic tubes and stored at 8 °C for spectral analysis.
For Raman spectroscopy, the samples were passively warmed up to room temperature and placed in an aluminum sample holder with a vessel of 5 mm diameter and about 0.1 mL volume. Spectra were taken with the laser directed to the vessel, perpendicular to the holder’s surface, through a Raman probe connected to a Raman spectrometer. The spectrometer (model Dimension P-1 Raman, Lambda Solutions, Inc., MA, USA) is composed by a semiconductor laser (830 nm, 350 mW, adjustable power) coupled to the Raman probe (model 830 Vector Probe, Lambda Solutions, Inc.) that is used to illuminate sample and collect the scattered light. The probe is connected to an imaging spectrograph with diffraction grating (1,200 lines/mm) and Peltier-cooled, deep depleted/back illuminated CCD camera (−75 °C), which collects high-resolution Raman spectrum from the sample in the fingerprint region (400–1,800 cm−1). The laser power was adjusted to 300 mW, and the integration time to collect the Raman signal was set to 3 s X 10 accumulations for all samples.
Triplicate spectra were obtained from each sample, which were averaged after preprocessing. Spectra were preprocessed to remove the undesired background fluorescence using a seventh-order polynomial fitted over the spectral range of 400–1,800 cm−1 and subtracted from the gross spectrum, as described elsewhere [34], and normalized to the intensity of the Raman peak at 1,640 cm−1 (weak Raman band of water), averaged and plotted with the aim to identify spectral differences between HbA and HbS.
After preprocessing, the spectra dataset was submitted to PCA, which is a multivariate statistical technique that transforms a set of original, correlated variables (spectra) to a set of uncorrelated variables called principal components (the principal components vectors and scores) based on the maximum variance [35]. PCA can be used to group spectra according to their similar variances, since each principal component is orthogonal to each other, so a unique spectral characteristic is present in each component. The first principal components vectors bring the most relevant spectral features presented in the dataset, whereas the principal components scores (PCs) bring the intensity of each principal component vector in each spectrum of the dataset [35]. Then, one could use the scores to discriminate spectra from individuals of a population according to variation of the spectral characteristics (using a suitable discriminant analysis) or use them to correlate the spectral information of the sample (using a regression line) [36]. Principal components were calculated using MATLAB 7.0 (The MathWorks Inc, MA, USA).
The term discriminant analysis refers to several different types of analyses; it is a classification method. It assumes that different classes generate data based on different Gaussian distributions (normally distributed) [35]. Classificatory discriminant analysis is used to classify observations into two or more known groups on the basis of one or more quantitative variables. Classification can be done by either a parametric method or a nonparametric method in the discrimination procedure. A parametric method is appropriate only for approximately normal within-class distributions. The method generates either a linear discriminant function (the within-class covariance matrices are assumed to be equal) or a quadratic discriminant function (the within-class covariance matrices are assumed to be unequal). It has used the “classify.m” function from MATLAB’S Statistics package, where the independent variables were the principal component’s scores of the two clinical groups, being the most relevant scores identified by calculating the t test (5 % significance level); the ones with lower significance was chosen. Mahalanobis discriminant function was used to classify discrimination capability using the PC scores [36].
Results
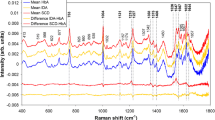
Figure 1 presents the mean Raman spectra of hemoglobin blood samples from normal (HbA) and with sickle cell disease (HbS) subjects and EDTA that was used as anticoagulant. The main spectral differences of HbA compared to HbS samples can be found in the shift positions of 882 and 1,373 cm−1, which are assigned to valine (Val) that present greater intensity in HbS spectra, and in the positions of 1,547 and 1,622 cm−1, which are assigned to glutamic acid (Glu), with prominent bands in the spectra of hemoglobin A. The Raman spectra of EDTA were not seen in the spectra of hemoglobin.
In order to identify and discriminate the spectral differences among HbA and HbS, the discrimination based on PCA technique and Mahalanobis distance was used. The 66 spectra were divided into two groups: HbA (normal) and HbS (sickle cell disease). PCA identified the first three principal components with higher relevance for discrimination, which exhibited more than 95 % of the spectral variance. ANOVA (5 % of significance) was applied to the scores of these PCs to verify which one could introduce greater power of HbA and HbS discrimination. Figure 2 (left) presents the scores of these first three PCs, and, as seen by the statistical analysis, the PC1 and PC2 were effective in discriminating HbS and HbA (p < 0.05). PC3 was not effective in discriminating these Hb variations (p > 0.05). In addition, the principal components loading vectors of PC1 and PC2 (Fig. 2, right) showed features that are in accordance with the structural modification of hemoglobin, featured bands on PC1 (882 and 1,373 cm−1 bands) are assigned to valine with greater intensity in patients with sickle cell disease. However, the spectral features in the PC2 (1,547 and 1,622 cm−1 bands) are assigned to glutamic acid, which are present in greater intensity in normal patients. The differences in the Raman bands of valine (at 882 and 1,373 cm−1) and glutamic acid (at 1,547 and 1,622 cm−1) were clearly seen in PC1 and PC2, and were correlated to the known changes in the polypeptide chain. PC3 presented a spectral feature that is not relevant for discriminating among HbA and HbS.
Left Plot of the mean and standard deviation of the first two principal components scores (PC1, PC2, and PC3) for HbS and HbA groups. Asterisks represent statistically significance difference among specific groups (t test) *p < 0.05. Right Plot of the first three principal components vectors, with spectral features of PC1 related to valine and glutamic acid; PC2 related to glutamic acid, as indicated by arrows. PC3 did not present spectral feature significant for discriminating among HbA and HbS
Figure 3 presents binary scatter plot of PC1 and PC2, which were found to be relevant for the discrimination between HbA and HbS groups. The diagnosis line is based on the Mahalanobis distance between the data in the groups formed by the covariance ellipse of the two groups, which could distinguish the groups of sickle cell disease and normal subjects. PCA/DA was able to discriminate the HbS from HbA with 100 % accuracy.
Discussion
The present work aimed to evaluate whether Raman spectroscopy was able to discriminate the presence of mutant hemoglobin (HbS) or normal hemoglobin (HbA) in samples of patients with facilforme disease. Raman spectroscopy is an optical technique with advantages over traditional biochemical techniques; it has been used as a tool for the spectroscopic study of the most different organic and inorganic compounds demonstrating being a high-tech tool to be used in the diagnosis of diseases that affect humans [37]. In order to identify the changes of various biomolecules that are found in biological fluids like blood and urine, Raman spectroscopy has great sensitivity in detecting chemical compounds present in these biological samples [38, 39].
Hirsch et al. [30] presented a comparison of the UV resonance Raman spectra of the liganded (CO) forms of HbA compared with HbC. All differences can be attributed to the Trp (W3)α14/β15 band since no other spectral differences are observed [30]. Others studies involving human hemoglobin and Raman spectroscopy aim to identify the molecular structure because the function is directly related to it is molecular conformation [31–33].
In HbA, glutamic acid is substituted by valine HbS [40]. Spectral differences of HbS samples were compared to HbA (Fig. 1), are related with the bands 882 and 1,373 cm−1, and are assigned to Val that are present in greater intensity in HbS average spectra. However, predominant bands in the spectra of HbA (1,547 and 1,622 cm−1) are assigned to Glu. These data are in line with the expected, since sickle cell disease is characterized by the substitution of adenine for thymine (GAG > GTG), encoding valine instead of glutamic acid in position 6 of globin chain, with production of HbS [9].
In order to identify and discriminate the spectral differences among hemoglobin A and HbS, PCA technique was used and PC1 and PC2 presented statistically significant difference for discrimination of HbS and HbA. Indeed, PC1 and PC2 spectral vectors presented features according to the structural modification of hemoglobin, PC1 are related to bands at 882 and 1,373 cm−1, and PC2 are related to bands at 1,547 and 1,622 cm−1.
The diagnostic line based on PCA/Mahalanobis could correctly discriminate 100 % of the samples in the correspondent groups. On the basis of this information and the development of new studies, it is possible in the future that Raman spectroscopy can be used in clinical practice as a useful tool in the diagnosis of sickle disease. It is critical that future research using Raman spectroscopy with biological material may be performed and compared with the conventional techniques, such electrophoresis that have already been adopted for the diagnosis of hemoglobinopathies.
The main disadvantages of electrophoresis are complex technical procedures that include electrophoresis apparatus and support (gel), and voltage source; requires several reagents (buffer); and it is a time-consuming process that takes about 30 min (electrophoretic running), besides the fact that it has a relatively high cost and requires a well-experienced laboratory staff [41] with costs ranging from about US$ 10.00 to US$ 20.00 [42, 43] per running. By using dispersive spectrometer in the near infrared (785 or 830 nm), a Raman spectrum of biological sample could be collected and processed in real time with total time exposure as low as 2 s, with signal-to-noise ratio suitable for disease diagnosis [44, 45]. The possibility of performing real-time diagnosis without the use of reagents or extensive preprocessing could bring advantages to the Raman technique terms of both rapidness and cost compared to electrophoresis, allowing screening examinations in a rapid way.
In conclusion, this work showed that near-infrared Raman spectroscopy was capable of identifying the subtle changes in hemoglobin in blood of subjects with sickle cell anemia, as the exchange of amino acids (glutamic acid and valine) in protein structure globin chain is a constituent of hemoglobin due to this genetic disorder. The differences in the Raman bands of valine (at 882 and 1,373 cm−1) and glutamic acid (at 1,547 and 1,622 cm−1) were clearly seen and were correlated to the known changes in the polypeptide chain. PCA/DA was able to discriminate the HbS from HbA with 100 % accuracy. It has been shown that Raman spectroscopy could be developed to the intent of the identification of predominant HbA in normal individuals and HbS from patients with sickle cell disease.
References
Center of Disease Control (CDC). http://www.cdc.gov/ncbddd/sicklecell/data.html#references Dec 2013
US Department of Health and Human Services, National Institutes of Health, National Heart, Lung, and Blood Institute (2009) Disease and conditions index. Sickle cell anemia: who is at risk? Bethesda, MD: US Department of Health and Human Services, National Institutes of Health, National Heart, Lung, and Blood Institute. http://www.nhlbi.nih.gov/health/dci/Diseases/Sca/SCA_WhoIsAtRisk.html. Dec 2013
Steinberg MH (2009) Genetic etiologies for phenotypic diversity in sickle cell anemia. Sci World J 9:46–67
Silva-Pinto AC, Angulo IL, Brunetta DM, Neves FIR, Bassi SC, De Santis GC, Covas DT (2013) Clinical and hematological effects of hydroxyurea therapy in sickle cell patients: a single-center experience in Brazil. Sao Paulo Med J 131:238–243
Salzano FM (1985) Incidence, effects and management of sickle cell disease in Brazil. Am J Pediatr Hematol Oncol 7:240–244
Gonçalves MS, Bomfim GC, Maciel E, Cerqueira I, Lyra I, Zanette A, Bomfim G et al (2003) BetaS-haplotypes in sickle cell anemia patients from Salvador, Bahia, Northeastern Brazil. Braz J Med Biol Res 36:1283–1288
Wainscoat JS, Bell JI, Thein SL, Higgs DR, Sarjeant GR, Peto TE, Weatherall DJ (1983) Multiple origin of the sickle mutations: evidence from β S-globin cluster polymorphisms. Mol Biol Med 1:191–197
Steinberg MH (2008) Sickle cell anemia, the first molecular disease: overview of molecular etiology, pathophysiology, and therapeutic approaches. Sci World J 8:1295–1324. doi:10.1100/tsw.2008.157
Pauling L, Itano H, Singer SJ, Wells IC (1949) Sickle cell anemia: a molecular disease. Science 110:543–548. doi:10.1126/science.110.2865.543
Ingram VM (1956) A specific chemical difference between the globins of normal human and sickle-cell anaemia haemoglobin. Nature 178:792–794
Eaton JW, Hebbel RP (1981) Pathogenesis of sickle cell disease. Pathobiol Annu 11:31–52
Hebbel RP, Eaton JW, Steinberg MH, White JG (1981) Erythrocyte/endothelial interactions and the vasocclusive severity of sickle cell disease. Prog Clin Biol Res 55:145–162
Eaton WA, Hofrichter J (1990) Sickle cell hemoglobin polymerization. Adv Protein Chem 40:63–280
Ferrone FA, Hofrichter J, Eaton WA (1985) Kinetics of sickle hemoglobin polymerization. II. A double nucleation mechanism. J Mol Biol 183:611–631
Mozzarelli A, Hofrichter J, Eaton WA (1987) Delay time of HbS gelation prevents most cells from sickling in vivo. Science 237(4814):500–506
Coletta M, Hofrichter J, Ferrone FA, Eaton WA (1982) Kinetics of sickle haemoglobin polymerization in single red cells. Nature 300(5888):194–197
Trent RJ (2006) Diagnosis of the haemoglobinopathies. Clin Biochem Rev 27(1):27–38
Hoyer MD (2011) 79 Hemoglobinopathies: the how, why, and what. Division of Hematopathology Mayo Clinic Rochester, 2011 ASCP Annual Meeting, Las Vegas
Rusciano G, De Luca AC, Pesce G, Sasso A (2008) Review: Raman tweezers as a diagnostic tool of hemoglobin-related blood disorders. Sensors 8:7818–7832
Hanlon EB, Manoharan R, Koo TW, Shafer KE, Motz JT, Fitzmaurice M, Kramer JR, Itzkan I, Dasari RR, Feld MS (2000) Prospects for in vivo Raman spectroscopy. Phys Med Biol 45:R1–R59
Diem M (1993) Introduction to modern vibrational spectroscopy. Wiley, New York
Campbell ID, Durek RA (1984) Biological spectroscopy. Benjamin Cummings, New York
Berger AJ (1998) Measurement of analytes in human serum and whole blood samples by near-infrared Raman spectroscopy. PhD dissertation. Massachusetts Institute of Technology, Massachusetts
Villanueva LAE, Castro RJ, Vazquez SM, Flores GA, Ortiz LCM, Delgado AJA (2012) Raman spectroscopy of blood in-vitro. Proc. SPIE 8229, Optical diagnostics and sensing XII: toward point-of-care diagnostics and design and performance validation of phantoms used in conjunction with optical measurement of tissue IV, 82291D. doi:10.1117/12.908689
Guimarães AE, Pacheco MTT, Silveira L, Barsottini DJ, Villaverde AB, Zângaro RA (2006) Near infrared Raman spectrocopy (NIRS): a technique for doping control. Spectrosc Int J 20(4):185–194
Ryder AG, O’Connor GM, Glynn TJ (2000) Quantitative analysis of cocaine in solid mixtures using Raman spectroscopy and chemometric methods. J Raman Spectrosc 31(3):221–227
Virkler K, Lednev IK (2010) Raman spectroscopic signature of blood and its potential application to forensic body fluid identification. Anal Bioanal Chem 396:525–534
Duarte J, Pacheco MTT, Villaverde AB, Machado RZ, Zângaro RA, Silveira L (2010) Near-infrared Raman spectroscopy to detect anti-Toxoplasma gondii antibody in blood sera of domestic cats: quantitative analysis based on partial least-squares multivariate statistics. J Biomed Opt 15(4):047002
Monfared AMT, Tiwari VS, Tripathi MM, Anis H (2013) Raman spectroscopy for clinical-level detection of heparin in serum by partial least-squares analysis. J Biomed Opt 18(2):027010
Hirsch RE, Lin MJ, Vidugirus GVA, Huangi S, Friedmani JM, Nagel RL (1996) Conformational changes in oxyhemoglobin C (Glub6 3 Lys) detected by spectroscopic probing. Med J Biol Chem 271(5):372–375
Hirsch RE, Juszchq LJ, Fataliev NA, Friedman JM, Ronald L (1999) Solution-active structural alterations in ligand hemoglobins C (β6 Glu→Lys) and (β6 Glu→Val). J Biol Chem 274(20):13777–13782
Juszczak LJ, Hirsch RE, Nagel RL, Friedman JM (1998) Conformational differences in CO derivatives of HbA, HbC (Eβ6K) and HbS (Eβ6V) in the presence and absence of inositol hexaphosphate detected using ultraviolet resonance Raman spectroscopy. J Raman Spectrosc 29:963–968
Wood BR, Caspers P, Puppels GJ, Pandiancherri S, McNaughton D (2007) Resonance Raman spectroscopy of red blood cells using near-infrared laser excitation. Anal Bioanal Chem 387:1691–1703
Jolliffe IT (1995) Principal components analysis. Springer, New York, pp 1–27
The MathWorks Documentation Center. Discriminant analysis. http://www.mathworks.com/help/stats/discriminant-analysis.html#brah8i2. Feb 2013
Background: discriminant procedures. http://support.sas.com/documentation/cdl/en/statug/63347/HTML/default/viewer.htm#statug_introdisc_sect002.htm. Feb 2013
Chaiken J, Deng B, Goodisman J, Shaheen G, Bussjager RJ (2011) Analyzing near-infrared scattering from human skin to monitor change in hematocrit. J Biomed Opt 16(9):097005
Stone N, Hart Prieto MC, Crow P, Uff J, Ritchie AW (2007) The use of Raman spectroscopy to provide an estimation of the gross biochemistry associated with urological pathologies. Anal Bioanal Chem 387(5):1657–1688
Bispo JA, de Sousa Vieira EE, Silveira L, Fernandes AB (2013) Correlating the amount of urea, creatinine, and glucose in urine from patients with diabetes mellitus and hypertension with the risk of developing renal lesions by means of Raman spectroscopy and principal component analysis. J Biomed Opt 18(8):87004
Sigma Aldrich Catalog. http://www.sigmaaldrech/brazil.html. Jun 2013
Wajcman H, Moradkhani K (2011) Abnormal haemoglobins: detection and characterization. Indian J Med Res 134(4):538–546
Clinical Center Ltda (2008) Tabela de preços exames laboratoriais. http://www.sinescontabil.com.br/convenios/exames.htm. May 2014
NM Medical (2012) Price calculator. http://www.nmmedical.com/price-calculator-profile.htm. May 2014
Motz JT, Gandhi SJ, Scepanovic OR, Haka AS, Kramer JR, Dasari RR, Feld MS (2005) Real-time Raman system for in vivo disease diagnosis. J Biomed Opt 10(3):031113
Zhao J, Lui H, McLean DI, Zeng H (2008) Integrated real-time Raman system for clinical in vivo skin analysis. Skin Res Technol 14:484–492
Acknowledgments
L. Silveira Jr. acknowledges FAPESP (São Paulo Research Foundation, Brazil) for the partial financial support (Proc. no. 2009/018788-5 and 2012/20666-0).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Filho, A.C.B., Silveira, L., Yanai, A.L.S. et al. Raman spectroscopy for a rapid diagnosis of sickle cell disease in human blood samples: a preliminary study. Lasers Med Sci 30, 247–253 (2015). https://doi.org/10.1007/s10103-014-1635-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10103-014-1635-z