Abstract
Background
A novel portable platform for nucleic acid amplification enables rapid detection of diarrhoea causing toxigenic Clostridium difficile directly from faeces, even in resource-limited settings. We evaluated the accuracy and precision of the new commercial molecular test system.
Methods
One thousand one hundred and sixty faecal samples from patients suspected of having Clostridium difficile infection (CDI) were analysed using the Orion GenRead C. difficile test system (Orion Diagnostica Oy, Espoo, Finland) and comparative methods in three teaching hospital laboratories in Finland and France. The precision of the Orion GenRead C. difficile test system was evaluated in a reproducibility study with a set of blind-coded samples. The test system is based on a new isothermal amplification technology (Strand Invasion Based Amplification, SIBA®) and detection of the tcdB gene of C. difficile. We calculated the sensitivity, specificity, and the overall agreement according to Clinical and Laboratory Standards Institute recommendations.
Findings
The overall agreement of the Orion GenRead C. difficile test when compared to the comparative methods in routine use in the participating laboratories was between 96.7% and 98.8%. In the reproducibility study; the total percent agreement between three laboratories was 99.8%.
Interpretation
The identification of toxigenic C. difficile from faeces with the light-weight portable Orion GenRead test system was highly sensitive and specific, and the results were reproducible in the participating laboratories. This platform could enable fast and accurate molecular pathogen detection even in resource-limited or point-of-care settings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During the last decade, nucleic acid testing has played an increasing role in clinical diagnosis and infection control. Through the development of novel molecular techniques, major improvements in the diagnostics of infectious diseases and emerging pathogens, as well as efficiency in laboratory analysis, have been attained [1, 2]. However, current nucleic acid tests often require high-end instrumentation, which confines them to a laboratory or other diagnostic settings. In most cases, specimens have to be transported to the laboratory for analysis, which increases the total turnaround time. Moreover, the high-end instrumentation makes nucleic acid tests impractical in many resource-limited settings with endemic diseases and insufficient laboratory capacity [3]. Therefore, new easy-to-use, small-scale nucleic acid tests that could perform as true point-of-care (POC) tests, bridging the gap between centralised laboratory diagnostics and peripheral healthcare units, are urgently needed.
A novel isothermal nucleic acid amplification based platform [4] now allows a small-scale and portable assay for rapid detection of toxigenic Clostridium difficile in faecal specimens. The Orion GenRead C. difficile assay (Orion Diagnostica Oy, Espoo, Finland) targets the tcdB gene of C. difficile. We compared the diagnostic accuracy and time to identification of this new molecular platform with methods used in routine C. difficile diagnostics at three teaching hospital laboratories.
Materials and methods
Clinical samples and setting
One thousand one hundred and sixty clinical samples in total were tested in this study between November and December 2014. The faecal samples were prospectively collected from patients suspected of having C. difficile infection (CDI) according to hospital routine practice in antibiotic-associated diarrhoea. The samples collected into conventional sample containers (n = 723) or into FecalSwabs™ (Copan, Italy) (n = 437) were analysed immediately after receipt into the laboratory.
The participating laboratories were Nordlab Oulu (Oulu, Finland), Fimlab Laboratories (Tampere, Finland), and Grenoble University Hospital Centre (CHU) (Grenoble, France). Intended sample size was planned to contain a minimum of 150 samples per laboratory. The samples were tested with the Orion GenRead C. difficile test, and the result was compared to the result of a test routinely used in the laboratory for C. difficile diagnostics. The routine (or reference) methods were as follows; in Nordlab, illumigene® C. difficile (Meridian Bioscience Inc, USA); in Fimlab, IMDx™ C. difficile for Abbott m2000 (Abbott Laboratories, USA) and toxigenic culture on CCFA (cycloserine cefoxitin fructose agar, Oxoid Limited, UK); in Grenoble CHU, a two-step algorithm was used, where faecal samples were first screened with the C. DIFF QUIK CHEK® test (TECHLAB, USA), after which confirmatory toxigenic C. difficile testing was conducted with the Xpert® C. difficile assay (Cepheid, USA) for all GDH-positive samples (Fig. 1). In Grenoble, in case of a discrepant result between C. DIFF QUIK CHEK and Orion GenRead C. difficile, the sample was analysed with the Xpert C. difficile assay. The sample was defined as true positive or negative based on the result with the Xpert assay. In Nordlab, in case of discrepant result between illumigene and Orion GenRead, the sample was analysed with the Xpert C. difficile test. The sample was defined as true positive or negative according to the Xpert assay. In Fimlab, the sample was defined as true positive when toxigenic culture or both nucleic-acid tests (GenRead and IMDx) were positive and true negative when toxigenic culture and one of the nucleic-acid tests were negative.
Reproducibility samples and setting
The reproducibility testing was conducted using a panel of five samples that included three samples with various concentrations of C. difficile ATCC 43255 cells and two C. difficile negative samples. The amounts of C. difficile cells that were spiked into the artificial faecal matrix were 5 × 105 CFU/ml, 8 × 104 CFU/ml, and 4 × 104 CFU/ml in the high, medium, and low positive sample respectively. One C. difficile negative sample contained C. sordellii cells of the amount of 2 × 105 CFU/ml. The other negative sample was composed of artificial faecal matrix only containing DMEM (Dulbecco’s modified Eagle’s medium, Sigma–Aldrich), 5% fetal bovine serum (Sigma–Aldrich), 1.3% Ultra Dawn (Procter & Gamble) and 1% (w/v) FECLONE™- BFPS4 (SiliClone Studio, USA). The samples were prepared at Orion Diagnostica (Espoo, Finland) and divided into three identical panels that were distributed to the three laboratories (Nordlab, Fimlab, and the R&D laboratory at Orion Diagnostica), where they were tested blindly.
Orion GenRead C. difficile
The Orion GenRead C. difficile test is a qualitative test based on a proprietary isothermal nucleic acid amplification technology, Strand Invasion Based Amplification (SIBA®) [4]. The test specifically amplifies and detects a conserved region in the C. difficile toxin B gene (tcdB).
The test was performed as follows; a sample was taken with a flocked swab and transferred into a filtration vial. Three to five drops of the lysis buffer was squeezed from the vial to an empty micro-tube and the tube was heated in a heating block at 95 °C for 5 minutes. The heat-treated sample was mixed with reaction buffer, and 40 μl of the mixture was pipetted into a reaction tube containing freeze-dried reagents for the C. difficile assay and the internal control reaction, which controls for the integrity of the reagents and successful amplification.
The reaction tubes were run in the Orion GenRead instrument for 50 minutes at a constant temperature of 41 °C. The results were reported as positive, negative, or invalid by the Orion GenRead instrument that includes embedded automated result reporting software. The positive reactions were reported by the instrument as soon as the reaction reached the level of positivity as determined by software algorithm. In cases where an invalid result was reported, a new sample preparation was conducted as per the manufacturer’s protocol, and the run was repeated in all participating centers.
illumigene C. difficile
The illumigene C. difficile test is a qualitative test based on isothermal loop-mediated amplification technology (LAMP) [5]. The target of the test is a toxin A gene (tcdA) [6].
The test was performed according to manufacturer’s instructions. Briefly, stool samples were collected on sample brush and diluted in a collection apparatus with sample diluent. The sample was vortexed, and five to ten drops were placed into an illumigene extraction tube. The tube was heated for 10 minutes at 95 °C and vortexed. Fifty microliters of the extracted sample was added to the reaction buffer tube. After vortexing the buffer tube, 50 μl was transferred to the test chamber and control chamber of the Illumipro-10 instrument which reports a positive, negative or invalid test results. In case of an invalid result, the sample was re-run with less sample material.
IMDx™ C. difficile for Abbott m2000
The IMDx C. difficile assay for the Abbott m2000 is an in-vitro PCR assay for the qualitative detection of tcdA and tcdB genes [7]. The test was performed according to the manufacturer’s instructions. Shortly, samples were transferred to 2.5 ml Tris-EDTA buffer tubes, after which automated sample lysing and target amplification and detection were performed on the m2000 Specimen Processor and RealTime Systems. The results were reported as positive, negative or invalid. In case of an invalid result, the sample was re-run and cultured to C. difficile selective CCFA medium to observe C. difficile growth.
C. DIFF QUIK CHEK
The C. DIFF QUIK CHEK® test is a rapid membrane enzyme immunoassay used as a screening test to detect glutamate dehydrogenase (GDH) [8]. GDH is an antigen that indicates the presence of C. difficile in faecal samples. The test does not distinguish toxigenic from non-toxigenic strains of C. difficile. The C. DIFF QUIK CHEK® was used according to the manufacturer’s instructions.
Xpert C. difficile
The Xpert C. difficile assay is a qualitative in-vitro diagnostic test for the identification and differentiation of tcdB, and cdt (binary toxin) genes, as well as tcdCΔ117 from stool samples [9]. The test utilizes automated real-time PCR performed on the Cepheid GeneXpert® Systems. It was performed as instructed by the manufacturer. Briefly, a stool sample was taken with a swab from the sample container and the sample was transferred into the sample reagent vial. The vial was vortexed for 10 seconds and the solution was transferred into the test cartridge, which was placed into the GeneXpert instrument. The test was then performed using the C. difficile assay program.
Controls
An additional negative and a positive control were included in each Orion GenRead C. difficile test series in all of the study sites. A clean swab without a sample was used as a negative control and a pure culture of a toxigenic C. difficile strain ATCC 9689 (at Fimlab and Nordlab) or a clinical strain isolated from a faecal sample of a patient, and confirmed as toxigenic C. difficile with Xpert C. difficile test (at Grenoble CHU), was used as a positive control.
Statistical analysis
The sensitivity, specificity, overall agreement, and confidence intervals values for the Orion GenRead test system were calculated according to the FDA’s Statistical Guidance on Reporting Results from Studies Evaluating Diagnostic Tests (Document issued on: March 13, 2007) and NCCLS document EP12-A — User Protocol for Evaluation of Qualitative Test Performance; Approved Guideline (ISBN 1-56238-468-6).
Results
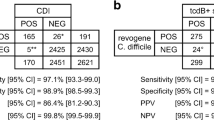
A total of 1160 faecal samples were analysed with the Orion GenRead C. difficile test and the comparative methods (Fig. 1). One hundred and eighty-four samples (15.9%) were considered as true positives and 949 samples (81.8%) as true negatives for toxigenic C. difficile. A total of 11 false positives (0.9%) and 16 false negatives (1.4%) were reported with the Orion GenRead C. difficile. The sensitivities, specificities, and overall agreement values of the Orion GenRead test system relative to the comparative methods are summarised in Table 1.
Method comparison against illumigene C. difficile
Three hundred and ninety-seven faecal samples in conventional sample containers were analysed with both the Orion GenRead C. difficile test and the illumigene C. difficile. Of these samples, 79/397 (19.9%) were defined as true positives and 305/397 (76.8%) as true negatives. Two samples were re-classified according to the discrepancy resolution conducted with the Xpert C. difficile test. One sample reported as negative with illumigene C. difficile, but as positive with the Orion GenRead C. difficile test, was also positive with the Xpert® C. difficile test. The other sample reported as positive with illumigene® C. difficile but as negative with the Orion GenRead C. difficile test, was also negative with the Xpert C. difficile test. The false negativity and positivity rates were 1.7% (7/397) and 1.5% (6/397), with the Orion GenRead C. difficile. The sensitivity of the Orion GenRead test system when compared with the illumigene test was 91.9%, the specificity 98.1%, and the overall agreement 96.7%.
Method comparison against IMDx C. difficile
The method comparison between IMDx C. difficile and Orion GenRead C. difficile was conducted using two different types of sample material, namely faecal sample collected into a conventional sample container and into a FecalSwab containing Cary–Blair medium. Of the 170 samples in conventional sample containers, 23 (13.5%) were true positives and 145 (85.3%) true negatives. The false negativity and positivity rates were 1.2% (2/170) and 0% (0/170) with the Orion GenRead C. difficile respectively. The sensitivity and specificity were 92.0% and 100%. Of the 437 samples in FecalSwabs, 68 (15.6%) were true positives and 362 (82.8%) true negatives. Six (1.4%) false negatives and one (0.2%) false positive were obtained with the Orion GenRead C. difficile. When the samples collected into FecalSwabs were analysed, the sensitivity and specificity were 91.9% and 99.7% respectively. The overall agreement between the tested methods was 98.8% when conventional sample containers were used for sample collection, and 98.4% with samples collected into FecalSwabs.
Method comparison against the two-step algorithm with C. DIFF QUIK CHEK and Xpert C. difficile
The overall agreement between the assay combination of a GDH-based C. DIFF QUIK CHEK and the Xpert C. difficile assay and the Orion GenRead C. difficile test was 96.8%, when 156 faecal patient specimens collected in dry containers were analysed: 14/156 (8.9%) samples were true positives and 137/156 (87.8%) true negatives. One (0.6%) false negative and four (2.6%) false positives were obtained with the Orion GenRead C. difficile. The sensitivity and specificity values were 93.3% and 97.2% respectively.
The reproducibility study
The precision of the Orion GenRead C. difficile test system was evaluated by a reproducibility study, where the same sets of blind-coded samples were tested at the three participating laboratories. During a 5-day testing protocol, five samples containing various amounts of C. difficile or C. sordellii cells (in three replicates) were analysed twice per day by two laboratory technicians. A total of 449/450 samples were correctly identified (Table 2). Only one sample was reported as false positive due to a probable contamination caused by a user. The total agreement between the three laboratories was 99.8%.
Invalid results during the study
A total of 45 invalid results (3.9%) were obtained with the Orion GenRead test system during the study (Fig. 1). False amount of or error in sample processing gave rise to an invalid result and thus, practise was deemed necessary for the sampling of the correct amount of the variable faecal sample material. The reagent container that was noticed to be leaking was replaced during the study. In case of an invalid result, the result was excluded from the performance characteristics calculations and the sample was re-run. The percentage of invalid results for illumigene, IMDx, and Xpert were 11.8%, 1.0%, and 0% respectively.
Discussion
In recent years, alongside centralisation of molecular diagnostic with automated high-throughput instruments, movement towards nucleic-acid-based POC testing is being pursued through simplification and miniaturisation of the testing process and test platforms [10–12]. As a part of this trend, we evaluated the performance and usability of a new small-scale portable nucleic acid amplification assay, the Orion GenRead C. difficile. The Orion GenRead instrument is self-contained, comprising slots for 12 samples, a touch screen, and an integrated barcode reader. It is capable of operating with batteries for up to 8 hours or six consecutive assay runs. With battery-powered technology, the problem regarding inconsistent or non-existent local electrical sources, particularly in resource-limited settings, is minimised. The instrument is lightweight (3.55 kg) and small [15 cm (h) × 22.5 cm (L) × 23 cm (d)] and can easily be carried, for example, from a centralised laboratory to near bedside and back. The instrument is simple to use, and the results are given rapidly in an easy to interpret form as positive or negative answer, shown on the instrument screen; therefore, the interpretation of the results does not require highly skilled laboratory personnel. These features allow the diagnostic laboratory to redesign part of their analytical processes, creating mobile nucleic acid amplification test (NAAT) units. Mobile NAAT units enable sample analysis to be initiated directly after specimen collection without any delay in specimen transportation. Furthermore, since the Orion GenRead reports assay results automatically on the instrument touch screen within 15 minutes as its best, in a clear and unambiguous form as positive, negative, or invalid, it provides rapid assistance for patient management decision. It has recently been discussed by Peeling and Mabey that POC tests for infectious agents could save many lives, especially in developing countries, by increasing access to proper diagnosis and treatment [13]. Similarly, Buchan and Ledeboer stated in their review, that rapid and fully automated on-demand or single-test formats have potential to affect health-care decisions at its earliest stage [10]. They also state that the total cost of a patient’s care can be reduced by rapid diagnostic test results, even though the automated or single-test format are often more expensive than batched testing, if the price is calculated per one single test. The Orion GenRead test system, being based on isothermal technology that enables construction of cheaper instruments that do not require stringent control of the temperature and its rapid changes like PCR, is a step towards more cost-efficient POC technologies that could be more affordable for the developing economies.
The sample preparation and result interpretation of the Orion GenRead system proved to be simple, and thus, it may enable diagnostic aid even in settings where little decision support is available. However, even the simplest test can give the wrong answer, if no quality control is included in the test, due, for example, to inhibitory substances, which are known to have a notable effect on the diagnostic performance of the NAATs [14]. In the Orion GenRead C. difficile assay, every test includes an internal amplification control with an artificial DNA target to control successful amplification and the integrity of the assay reagents. In this study, inhibitory samples were observed as presupposed, but the level of inhibition with the Orion GenRead was less than 4%. By eliminating the problems due to inhibitors, for example through sample dilution, NAATs have proven to be superior to the conventional test methods, such as culture or antigen testing [15]. It should be borne in mind, though, that in addition to internal controls, a systematic external quality assurance is also needed to ensure the continuous performance of NAATs.
A limitation of this study is that a uniform reference method, i.e., “gold standard”, was not used but three methods instead. In comparison with the three routine methods, used here as the reference, the Orion GenRead C. difficile assay provided a slightly lower sensitivity and specificity (overall agreement varying from 96.7 to 98.8%). However, there is a balance to be found in the sensitiveness of the test for CDI; it should be sensitive enough, but not overly so, as to detect the asymptomatic carriage [16], unless detection of the carriage is wanted for reasons to control the spread of CDI [17–22]. It has been speculated that highly sensitive NAATs may have a role in infection control, if asymptomatic patients contribute to the spread of C. difficile in health care facilities [21, 22]. Furthermore, well-performing NAATs have a high negative predictive value and thus, potential to decrease the need for repeat testing and empirical treatment [23, 24]. With a good analytical sensitivity and short total turnaround time (25 to 55 minutes), the Orion GenRead system is suitable also for two-step testing of toxigenic C. difficile as a screening test, followed by a test for toxin production to confirm possible active infection.
Interestingly, apart from POC single-test analysis, the Orion GenRead system is adapted also for use in high-throughput screening in a more centralized laboratory. The instrument enabled four batches of 12 sample runs (48 samples) in less than 3.5 hours and five batches (72 samples) in less than 4.5 hours, with minimal space and energy requirement. This proved to be as fast as with the IMDx C. difficile assay, integrated on the fully automated high-throughput Abbott m2000 system. The total turnaround time for Abbott system is 3–4 hours for the analysis of 1–48 samples and 4–4.5 hours for the analysis of 49–96 samples.
In conclusion, the Orion GenRead C. difficile assay proved to be a well-performing test providing access to state of the art diagnostic support even in settings where health care infrastructure is minimal. The proprietary isothermal SIBA® technology used on Orion GenRead enables the system to be small-scale, and most interestingly portable, making it suitable for regions were distances are long between nursing units and for resource-limited settings.
References
van Belkum A, Durand G, Peyret M, Chatellier S, Zambardi G, Schrenzel J, Shortridge D, Engelhardt A, Dunne WM Jr (2013) Rapid clinical bacteriology and its future impact. Ann Lab Med 33:14–27. doi:10.3343/alm.2013.33.1.14
Procop GW (2007) Molecular diagnostics for the detection and characterization of microbial pathogens. Clin Infect Dis 1:S99–S111
Huggett J, Green C, Zumla A (2009) Nucleic acid detection and quantification in the developing world. Biochem Soc Trans 37:419–423. doi:10.1042/BST0370419
Hoser MJ, Mansukoski HK, Morrical SW, Eboiqbodin KE (2014) Strand Invasion Based Amplification (SIBA®): a novel isothermal DNA amplification technology demonstrating high specificity and sensitivity for a single molecule of target analyte.. PLoS One 9:e112656. doi:10.1371/journal.pone.0112656
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:12, e63
Norén T, Alriksson I, Andersson J, Akerlund T, Unemo M (2011) Rapid and sensitive loop-mediated isothermal amplification test for Clostridium difficile detection challenges cytotoxin B cell test and culture as gold standard. J Clin Microbiol 49:710–711. doi:10.1128/JCM.01824-10
Stellrecht KA, Espino AA, Maceira VP, Nattanmai SM, Butt SA, Wroblewski D, Hannett GE, Musser KA (2014) Premarket evaluations of the IMDx C. difficile for Abbott m2000 Assay and the BD Max Cdiff Assay. J Clin Microbiol 52:1423–1428. doi:10.1128/JCM.03293-13
Eastwood K, Else P, Charlett A, Wilcox M (2009) Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol 47:3211–3217. doi:10.1128/JCM.01082-09
Pancholi P, Kelly C, Raczkowski M, Balada-Llasat JM (2012) Detection of toxigenic Clostridium difficile: comparison of the cell culture neutralization, Xpert C. difficile, Xpert C. difficile/Epi, and Illumigene C. difficile assays. J Clin Microbiol 50:1331–1335. doi:10.1128/JCM.06597-11
Buchan BW, Ledeboer NA (2014) Emerging technologies for the clinical microbiology laboratory. Clin Microbiol Rev 27:783–822. doi:10.1128/CMR.00003-14
de Paz HD, Brotons P, Muñoz-Almagro C (2014) Molecular isothermal techniques for combating infectious diseases: towards low-cost point-of-care diagnostics. Expert Rev Mol Diagn 14:827–843. doi:10.1586/14737159.2014.940319
Peeling RW, McNerney R (2014) Emerging technologies in point-of-care molecular diagnostics for resource-limited settings. Expert Rev Mol Diagn 14:525–534. doi:10.1586/14737159.2014.915748
Peeling RW, Mabey D (2010) Point-of-care tests for diagnosing infections in the developing world. Clin Microbiol Infect 16:1062–1069. doi:10.1111/j.1469-0691.2010.03279.x
Pasternack R, Vuorinen P, Kuukankorpi A, Pitkäjärvi T, Miettinen A (1996) Detection of Chlamydia trachomatis infections in women by Amplicor PCR: comparison of diagnostic performance with urine and cervical specimens. J Clin Microbiol 34:995–998
Hirvonen JJ, Kaukoranta SS (2015) Comparison of BD Max Cdiff and GenomEra C. difficile molecular assays for detection of toxigenic Clostridium difficile from stools in conventional sample containers and in FecalSwabs. Eur J Clin Microbiol Infect Dis 34:1005–1009. doi:10.1007/s10096-015-2320-2
Polage CR, Gyorke CE, Kennedy MA, Leslie JL, Chin DL, Wang S, Nguyen HH, Huang B, Tang YW, Lee LW, Kim K, Taylor S, Romano PS, Panacek EA, Goodell PB, Solnick JV, Cohen SH (2015) Overdiagnosis of Clostridium difficile Infection in the Molecular Test Era. JAMA Intern Med 8:1–10. doi:10.1001/jamainternmed.2015.4114
Khanna S, Pardi DS, Rosenblatt JE, Patel R, Kammer PP, Baddour LM (2012) An evaluation of repeat stool testing for Clostridium difficile infection by polymerase chain reaction. J Clin Gastroenterol 46:846–849. doi:10.1097/MCG.0b013e3182432273
Sydnor ER, Lenhart A, Trollinger B, Avdic E, Maragakis LL, Carroll KC, Cosgrove SE (2011) Antimicrobial prescribing practices in response to different Clostridium difficile diagnostic methodologies. Infect Control Hosp Epidemiol 32:1133–1136. doi:10.1086/662381
Kyne L, Warny M, Qamar A, Kelly CP (2000) Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N Engl J Med 342:390–397
Samore MH, DeGirolami PC, Tlucko A, Lichtenberg DA, Melvin ZA, Karchmer AW (1994) Clostridium difficile colonization and diarrhea at a tertiary care hospital. Clin Infect Dis 18:181–187
Leekha S, Aronhalt KC, Sloan LM, Patel R, Orenstein R (2013) Asymptomatic Clostridium difficile colonization in a tertiary care hospital: admission prevalence and risk factors. Am J Infect Control 41:390–393. doi:10.1016/j.ajic.2012.09.023
Alasmari F, Seiler SM, Hink T, Burnham CA, Dubberke ER (2014) Prevalence and risk factors for asymptomatic Clostridium difficile carriage. Clin Infect Dis 59:216–222. doi:10.1093/cid/ciu258
Polage CR, Solnick JV, Cohen SH (2012) Nosocomial diarrhea: evaluation and treatment of causes other than Clostridium difficile. Clin Infect Dis 55:982–989. doi:10.1093/cid/cis551
Guerrero DM, Chou C, Jury LA, Nerandzic MM, Cadnum JC, Donskey CJ (2011) Clinical and infection control implications of Clostridium difficile infection with negative enzyme immunoassay for toxin. Clin Infect Dis 53:287–290. doi:10.1093/cid/cir361
Acknowledgements
We are grateful to all Nordlab Oulu, Fimlab Laboratories, and CHU clinical and laboratory staff who helped conducting the study; especially, we thank Liisa Venho, Marika Huttu, Sari Ansaranta, and Tiina Malila for excellent technical assistance in conducting the analyses of the clinical specimens. We also thank Orion Diagnostica for providing the instrument and reagents for the Orion GenRead test system.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
This study was not funded by any external party. The study was performed using the clinical microbiology services of Nordlab Oulu, Fimlab Laboratories, and Grenoble CHU.
Conflicts of interest
Pirjo Matero has been employed by Orion Diagnostica since June 2010. At present, Pirjo Matero is on a leave of absence from Orion Diagnostica to conduct PhD studies at the University of Helsinki. The other authors have no conflict of interest to declare.
Ethical approval
Neither human participants nor animals were involved in this study.
Informed consent
All authors have seen and approved the content and have contributed significantly to the work. Jari J. Hirvonen and Pirjo Matero have contributed equally to the designing and supervising the study, analyzing and interpreting the data, and writing and editing of the manuscript. Risto Vuento, Hanna Tuokko, and Jaana Kauppila have contributed in supervising the study, analyzing and interpreting the data, and critically reviewing the manuscript. Claire Siebert took part in analysing and interpratation of the data. Sandrine Boisset took part in designing and supervising the study, analyzing, and interpreting the data, and writing and critically reviewing the manuscript.
Rights and permissions
About this article
Cite this article
Hirvonen, J.J., Matero, P., Siebert, C. et al. Novel portable platform for molecular detection of toxigenic Clostridium difficile in faeces: a diagnostic accuracy study. Eur J Clin Microbiol Infect Dis 36, 783–789 (2017). https://doi.org/10.1007/s10096-016-2860-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2860-0