Abstract
Multiple sclerosis (MS) is a progressive chronic autoimmune-mediated disease. Recently, long non-coding RNAs (lncRNAs) are characterized to participate in the adjustment of immune responses. Here, we evaluated the expression levels of GSTT1-AS1 and IFNG-AS1 lncRNAs and their targets (TNF and IFNG, respectively) in Iranian MS patients.
In this case-control study, 50 relapsing-remitting MS patients and 50 healthy subjects were recruited. Expressions of GSTT1-AS1 and IFNG-AS1 lncRNAs, as well as TNF and IFNG genes, were assessed in their peripheral blood samples by SYBR Green-based Real-time quantitative PCR.
Expression levels of GSTT1-AS1 and IFNG-AS1 lncRNAs were both significantly downregulated (p values 0.032 and 0.013, respectively). On the other hand, the expression of TNF and IFNG showed increased levels, however, did not reach statistical significance after our analysis (p > 0.05). Spearman correlation analysis showed that GSTT1-AS1 had a significant positive moderate correlation with IFNG-AS1 (r = 0.541, p < 0.0001), IFNG (r = 0.329, p = 0.001), and TNF (r = 0.204, p = 0.041). Also, IFNG-AS1 revealed the same correlation with IFNG (r = 0.475, p < 0.0001) as well as TNF (r = 0.399, p < 0.0001). Furthermore, GSTT1-AS1 (r = 0.313, p = 0.027) and (IFNG r = 0.478, p < 0.0001) demonstrated a significant positive correlation with age at onset.
Briefly, the current study provided for the first time dysregulation of GSTT1-AS1 and IFNG-AS lncRNAs network in MS, which highlights the significant role of epigenetic pathways in this autoimmune disorder. Larger sample size and further investigation assays could shed light on the underlying mechanisms in this area of science.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Multiple sclerosis (MS) (OMIM 126200) is a progressive chronic autoimmune-mediated disease, characterized by the demyelination, chronic inflammation, and neurodegeneration processes in the central nervous system (CNS). The underlying immunopathogenic mechanisms of this disorder is yet unknown. However, there is a large body of literature supporting the role of perivascular infiltration of autoreactive T cells which recognize and react against autoantigens [1, 2]. There are reports which demonstrate the significant increase of MS rates especially in Iranian population [3].
Newly, accumulated evidence has displayed that defects in CD4+ T helper cells may be involved in the MS process [4, 5]. CD4+ T helper cells can be grouped into Th1, Th2, and Th17 as well as follicular helper T cells, based on the production of cytokines. The Th1 cell lineage is regarded as a subset of inflammatory CD4+ T cells mainly producing interferon gamma (IFN-γ), an inflammation-related cytokine that participates protectively against autoimmune diseases, delayed-type hypersensitivity, and intracellular microbes [6, 7]. Likewise, tumor necrosis factor (TNF) gene encodes for a multifunctional proinflammatory cytokine, which has been characterized to affect excitatory synaptic transmission and mediates immunological and neural cytological roles [8,9,10].
Long non-coding RNAs (LncRNAs) constitute a distinct class of non-coding RNAs with more than 200 nucleotides in length. LncRNAs are substantially appreciated as key regulators of genome expression, and so far, only some of their functions have been recognized [11, 12]. In some well-studied instances, their active involvement in many aspects of neuronal diseases such as neural degeneration or neurogenesis in both of central and peripheral nervous systems has been described [13, 14]. In addition, dysregulation of some lncRNAs has been recently shown in peripheral blood mononuclear cells of MS patients, suggesting their possible role in MS development [14].
GSTT1-AS1 (lncRNA-CD244 in another words) is a lncRNA by which CD244 was found to mediate the inhibition of IFNG and TNF cytokine genes in tuberculosis pathogenesis [15]. It is speculated that GSTT1-AS1 represses the transcription of TNF and IFNG genes, by recruiting EZH2 enzyme complex to their promoters, to facilitate H3K27 methylation that causes a repressive transcriptional environment [16]. IFNG-AS1 (IFNG Antisense RNA 1), also named Tmevpg1 (Theiler’s murine encephalomyelitis virus persistence candidate gene 1), or NeST (nettoie Salmonella pas Theiler’s) is another lncRNA initially characterized as a candidate gene to control the Theiler’s virus persistence [17, 18]. IFNG-AS1 is located adjacent to IFN-γ-encoding gene, IFNG, in human. Currently, studies have elucidated that IFNG-AS1 is determined to be a pivotal checkpoint contributing IFNG expression [19, 20]. However, it is not yet known whether IFNG-AS1 is involved in the pathogenesis of MS. Besides, the relationship between GSTT1-AS1 and IFNG-AS1 lncRNAs as well as TNF and IFNG is, to the best of our knowledge, not studied in MS disorder.
Altogether, here we aimed to study the relative expression levels of GSTT1-AS1 and IFNG-AS1 lncRNAs in addition to their target coding genes (TNF and IFNG, respectively) in Iranian MS patients.
Materials and methods
Subjects
The subjects of this case-control study included 50 unrelated sporadic relapsing-remitting MS (RR-MS) patients (38 women and 12 men), as diagnosed and identified by MRI (magnetic resonance imaging) and 2017 McDonald criteria [21] by specialized neurologists. All patients were treated with Interferon (IFN)-β therapy for at least 2 years (intramuscular injection of CinnoVex 30 mcg [CinnaGen Co, Tehran, Iran] once a week) and were recognized as IFN-β responders [22, 23]. Moreover, 50 sex- and age-matched healthy controls (37 women and 13 men) were involved. The blood samples were gathered from MS Society of Iran and, in addition, some hospitals in Tehran. All MS patients were, prior to this study, HLA-typed. Therefore, HLA-DRB1*15 was ruled out due to its major effect in MS predisposition[24].
Blood sampling
The present study was carried out on 5-ml peripheral blood samples collected from the participants (MS patients and controls). Blood samples of MS patients were obtained during stable phase of disease. Samples were collected in K3-EDTA tubes. All individuals gave their informed consent to incorporate into this study. Then, a complete history of patients was acquired. Study design and the entire measurements were approved by the Ethics Committee of Shahid Beheshti University of Medical Sciences (SBUMS).
Quantitative real-time PCR assay
We extracted RNA from whole blood samples by using GeneAll Hybrid-R™ blood RNA extraction kit (cat No. 305-101). Then, 150 ng of RNA was used to synthesize the single strand cDNA through kit of Biosystems High-Capacity cDNA Reverse Transcription (PN 4375575), according to the manufacturer’s instruction. To design the specific primers, Allele ID6 for × 64 windows software (Premier Biosoft, Palo Alto, USA) was used (primer sequences and PCR product length are shown in Table 1). B2M was applied as a housekeeping gene in order to normalize the expression level for all samples including lncRNAs and target coding genes.
SYBR Green-based Real-time quantitative PCR assay was carried out, by Corbett Rotor Gene 6000 machine (Corbett Life Science), in duplicates for all samples comprising housekeeping gene, lncRNAs, and target genes in both groups of MS patients and healthy controls. Afterwards, a mean value was reported for each one. Routinely, the NTC (No Template Control) sample was considered for each primer in each run for method quality control and detection of contamination.
Statistical analysis
To examine the data obtained from participants, independent t tests and one-way ANOVA test were both conducted. As well, Pearson correlation coefficient was used to identify the correlation level between the variables. The significance level was determined as less than 5% (p ≤ 0.05). The analysis was accomplished via SPSS version 18 statistical package (Chicago, IL, USA). Spearman correlation test was performed to evaluate the correlations between relative expression levels of GSTT1-AS1, IFNG-AS1, TNF, and IFNG.
Results
Clinical information of all participants is given in Table 2. MS patients and healthy controls were evaluated in three separate ways: the results of total participants (regardless of their age and sex), sex-related results (male or female), and age-linked results (> 30, 30–40, and 40 ˂ years). The whole group of patients was then compared with healthy subjects and independently analyzed for age and sex.
Relative expression level of GSTT1-AS1 LncRNA
Statistical analysis revealed a significant downregulation in GSTT1-AS1 lncRNA expression in total MS patients (p = 0.032) and total female subgroup as well as the subgroup of females 40 ˂ years (p values 0.045 and 0.0005, respectively) versus healthy controls (relative expression ratios are demonstrated in Table 3).
Relative expression level of IFNG-AS1 LncRNA
Table 4 displays the results of total IFNG-AS1 expression level in RR-MS patients in comparison with the healthy group as well as those based on the age and sex of the participants. IFNG-AS1 lncRNA expression is significantly decreased in almost half of all categories, including total (p = 0.0.13), male (p = 0.03), female (p = 0.049), and women > 40 years (p = 0.037) MS patients compared to healthy subjects.
Relative expression level of TNF gene
TNF expression in MS patients in all categories was upregulated. Nonetheless, these discrepancies did not reach a significant level. The most considerable difference was for male patients > 40 years (sevenfold expression ratio compared with controls), while was not statistically significant. Table 5 indicates the TNF relative expression ratio in age- and sex-based subgroups.
Relative expression level of IFNG gene
Table 6 reveals the expression level of IFNG in patients and controls. All subgroups in relation to age- and sex-matched healthy subjects showed upregulated expression, but these changes were not statistically significant.
Correlation analysis between LncRNAs and target genes with expanded disability status scale (EDSS)
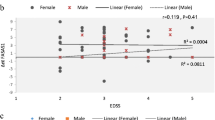
As seen in Fig. 1, TNF and IFNG expression levels both showed a significant and moderate correlation with EDSS (r = − 0.28, p = 0.049; and r = − 0.382, p = 0.006, respectively), while the correlations for GSTT1-AS1 and IFNG-AS1 with EDSS was not statistically significant (data not shown).
Correlation analysis between LncRNAs and target genes with disease duration
The correlation between all lncRNAs and target genes with disease duration did not reach a statistical significance (data not shown).
Correlation analysis between LncRNAs and target genes with age at onset
Correlation analysis results showed that there is a significant correlation between expression levels of GSTT1-AS1 (r = 0.313, p = 0.027) and IFNG (r = 0.478, p˂0.0001) with age at onset of MS disease (Fig. 2). The correlation of other lncRNAs and genes with age at onset was not significant (not shown).
Correlations between expression levels of LncRNAs and target genes
Spearman correlation analysis between the expression levels of lncRNAs and target genes is illustrated in Fig. 3, as described below:
For GSTT1-AS1
There was a significant and positive weak correlation with TNF (r = 0.204, p = 0.041) in patients.
For IFNG-AS1
Significant and positive correlations were obtained with GSTT1-AS1 (r = 0.541, p ˂ 0.0001) and TNF (r = 0.399, p˂ 0.0001).
For IFNG
There was a significant and positive moderate correlation with two studied lncRNAs, GSTT1-AS1 (r = 0.329, p ˂ 0.0001) and IFNG-AS1 (r = 0.475, p ˂ 0.0001). Moreover, the correlation with TNF was not significant (r = 0.19, p = 0.058) (not shown).
Discussion
In the current study, we presented downregulation of GSTT1-AS1 and IFNG-AS1 lncRNAs and upregulation of their coding targets TNF and IFNG in whole blood of MS patients. The importance of such differential expressions and dysregulations would be embedded in the pathogenesis of MS and emphasizes their role as potential biomarkers for different stages of disease course or treatment response [11]. In addition, finding the interaction between lncRNAs and their targets can provide us with potential new therapeutic avenues for future measurements.
IFN-β therapy has been extensively used over decades as a first-line treatment for RR-MS. Its beneficial effects in MS are, in part, owing to the inhibition of CD4+ T cell proliferation and modulation of proinflammatory cytokines expression, which are all necessary for the development of inflammation [25]. A large number of studies have examined the cytokines’ production in MS, but results have been extremely inconsistent and most studies, even those with a longitudinal study design, have utilized a cross-sectional approach to evaluate remission samples with samples collected during or shortly before relapse and controls [26,27,28,29,30,31]. Expression of IFNG and TNF is regulated through various epigenetic elements, including methylation, histone acetylation, and noncoding RNAs [32, 33]. It was previously showed that CD244 signaling during active tuberculosis infection can exploit GSTT1-AS1 lncRNA and histone-modifying enzymes to regulate effector functions of CD244+CD8+ T cells (Fig. 4) [15]. Indeed, CD244 provokes the expression of GSTT1-AS1 via creating a permissive transcriptional environment at the GSTT1-AS1 gene locus that results in loss of repressive histone mark, H3K27Me3. In turn, GSTT1-AS1 interestingly appears to physically interact with a polycomb protein, EZH2. This interaction consequently mediates the EZH2 recruitment to TNF and IFNG loci. Moreover, it is thought that this could trimethylate H3K27 at promoters of TNF and IFNG toward repressive chromatin status and suppression of TNF and IFNG expression [15, 34, 35]. Our findings support this hypothesis and suggest that GSTT1-AS1 might have interaction with chromatin and mediate the targeted recruitment of repressive histone-modifying elements in epigenetic control of transcriptional silencing in MS patients. Therefore, the inverse relationship between expression levels of GSTT1-AS1 lncRNA (downregulated) and the TNF and IFNG target genes (both upregulated) in MS patients of our study could be explained by this hypothesis, though for the two target genes, this increase was not significant after our analysis. Importantly, elevated expression levels of TNF and IFNG can contribute to the inflammatory responses and trigger cell death as well as tissue degeneration found in various autoimmune diseases [36, 37]. In harmony with our results, previous studies have shown higher IFN-γ production in stimulated cells of MS patients during or just prior to relapse [26, 29, 30, 38]. Particularly, in two of these studies, the difference in IFN-γ between MS patients and healthy controls did not reach statistical significance [29, 38]. These findings, however, are in contrast with Van Oosten work who showed no evidence of different stimulated production of IFN-γ in peripheral blood mononuclear cells (PBMCs) between relapse and remission samples [39]. Such differences might be, in part, due to the diversity of cross-sectional analysis method and sample size, which both may account for the failure to reach a significant difference.
Schematic model illustrating mechanism of action of GSTT1-AS1 lncRNA [15, 16]. Orientation of transcription is indicated by green arrows bellow. GSTT1-AS1 associates with EZH2 cargo and both connect to the target mRNAs. Locations of H3K27Me epigenetic marks which suppress the transcription of target genes are showed
IFNG-AS1 is characterized to cooperate with T-bet to stimulate the transcription of IFNG by effector Th1 cells [16]. This lncRNA associates with the WDR5 (Fig. 5), a component of histone H3K4 methyl transferase complex, and recruits this complex to create H3K4-methylation marks at the IFNG promoter as well as intronic regions to facilitate transcription [40]. While recently researchers have established that lncRNA IFNG-AS1 could change the epigenetic marking of IFNG encoding chromatin and induce IFNG transcription [19, 20, 40], other researchers have obtained the contrary results [18]. We observed a significant decrease in IFNG-AS1 expression in MS patients, but, to our surprise, the expression of IFNG was upregulated. In accordance with our data, a study performed by Li et al. [41] reported that IFNG-AS1 expression in PBMCs of primary immune thrombocytopenia (ITP), as an autoimmune disease, was lower than in healthy controls. They speculated that IFNG-AS1 can promote IFNG transcription, and that IFNG over-expression negatively regulates the expression of IFNG-AS1. It seems that the same applies to our data and we may suggest that significant decrease in IFNG-AS1 and insignificant increase of IFNG could be mechanisms for avoiding excessive IFNG expression [41]. Again, in Li study, the upregulation of whole blood IFNG concentration in active ITP patients did not reach the statistical significance. It may be that findings of our work and other studies, related to lncRNAs and targets, can be explained, in part, by their sample size limitation and heterogeneity of autoimmune disorders such as MS and ITP.
Schematic model illustrating mechanism of action of IFNG-AS1 lncRNA [16][16][16][16][16]. Orientation of transcription is indicated by green arrows. IFNG-AS1 associates with WDR5 cargo and both connect to the target mRNA. Locations of H3K4Me epigenetic marks which favor transcriptional activation are displayed. By IFNG over-expression, IFNG can negatively feedback and suppress the expression of IFNG-AS1 lncRNA
Of note, we assessed the expression levels of the abovementioned lncRNAs and target genes in IFN-β responder MS patients to exclude the effects of numerous drugs and to assimilate the group of our patients and to unify the results; however, a limitation of our study can be that we did not include non-responder MS patients.
Briefly, the current study provided for the first time dysregulation of GSTT1-AS1 and IFNG-AS lncRNAs network in MS, which highlights the significant role of epigenetic pathways. Larger sample size and further investigation assays could shed light on the underlying mechanisms in this area of science.
References
PILLI D, ZOU A, TEA F, DALE RC, BRILOT F (2017) Expanding role of T cells in human autoimmune diseases of the central nervous system. Front Immunol 8:652
RIEDHAMMER C, WEISSERT R (2015) Antigen presentation, autoantigens, and immune regulation in multiple sclerosis and other autoimmune diseases. Front Immunol 6
SAHRAIAN MA, SAHEBKAR M, DEHGHANI R, DERAKHSHAN-JAZARI M, KAZAMI-MOGHADDAM V, KOUCHAKI E (2017) Multiple sclerosis-a disease on a dramatically rising trend in Iran: review of possible reasons. Iran J Neurolt 16:34
DENDROU CA, FUGGER L, FRIESE MA (2015) Immunopathology of multiple sclerosis. Nat Rev Immunol 15:545–558
JONES A, KERMODE A, LUCAS R, CARROLL W, NOLAN D, HART P (2017) Circulating immune cells in multiple sclerosis. Clin Exp Immunol 187:193–203
COSMI L, MAGGI L, SANTARLASCI V, LIOTTA F, ANNUNZIATO F (2014) T helper cells plasticity in inflammation. Cytometry Part A 85:36–42
RAPHAEL I, NALAWADE S, EAGAR TN, FORSTHUBER TG (2015) T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine 74:5–17
MARQUES-ROCHA JL, SAMBLAS M, MILAGRO FI, BRESSAN J, MARTÍNEZ JA, MARTI A (2015) Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J 29:3595–3611
OLMOS G, LLADÓ J (2014) Tumor necrosis factor alpha: a link between neuroinflammation and excitotoxicity. Mediat Inflamm 2014:1–12
TAHERI M, NEMATI S, MOVAFAGH A, SABERI M, MIRFAKHRAIE R, EFTEKHARIAN MM, ARSANG-JANG S, REZAGHOLIZADEH A, SAYAD A (2016) TRAIL gene expression analysis in multiple sclerosis patients. Hum Antibodies 24:33–38
EFTEKHARIAN MM, GHAFOURI-FARD S, SOUDYAB M, OMRANI MD, RAHIMI M, SAYAD A, KOMAKI A, MAZDEH M, TAHERI M (2017) Expression analysis of long non-coding RNAs in the blood of multiple sclerosis patients. J Mol Neurosci 63:333–341
SANTORO M, NOCITI V, LUCCHINI M, DE FINO C, LOSAVIO FA, MIRABELLA M (2016) Expression profile of long non-coding RNAs in serum of patients with multiple sclerosis. J Mol Neurosci 59:18–23
WANG A, WANG J, LIU Y, ZHOU Y (2017) Mechanisms of long non-coding RNAs in the assembly and plasticity of neural circuitry. Front Neural Circuits 11
ZHANG F, GAO C, MA XF, PENG XL, ZHANG RX, KONG DX, SIMARD AR, HAO JW (2016) Expression profile of long noncoding RNAs in peripheral blood mononuclear cells from multiple sclerosis patients. CNS Neurosci Ther 22:298–305
WANG Y, ZHONG H, XIE X, CHEN CY, HUANG D, SHEN L, ZHANG H, CHEN ZW, ZENG G (2015b) Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci 112:E3883–E3892
AUNE TM, CROOKE PS, SPURLOCK CF (2016) Long noncoding RNAs in T lymphocytes. J Leukoc Biol 99:31–44
PENG H, LIU Y, TIAN J, MA J, TANG X, RUI K, TIAN X, MAO C, LU L, XU H (2015) The long noncoding RNA IFNG-AS1 promotes T helper type 1 cells response in patients with Hashimoto’s thyroiditis. Sci Rep 5
VIGNEAU S, ROHRLICH P-S, BRAHIC M, BUREAU J-F (2003) Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J Virol 77:5632–5638
COLLIER SP, COLLINS PL, WILLIAMS CL, BOOTHBY MR, AUNE TM (2012) Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol 189:2084–2088
COLLIER SP, HENDERSON MA, TOSSBERG JT, AUNE TM (2014) Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J Immunol 193:3959–3965
CSEPANY T (2018) Diagnosis of multiple sclerosis: a review of the 2017 revisions of the McDonald criteria. Ideggyogy Sz 71:321–329
SAYAD A, GHAFOURI-FARD S, OMRANI MD, NOROOZI R, TAHERI M (2017) Myxovirus resistance protein A (MxA) polymorphism is associated with IFNβ response in Iranian multiple sclerosis patients. Neurol Sci 38:1093–1099
TAHERI M, GHAFOURI-FARD S, SOLGI G, SAYAD A, MAZDEH M, OMRANI MD (2017) Determination of cytokine levels in multiple sclerosis patients and their relevance with patients’ response to Cinnovex. Cytokine 96:138–143
MAZDEH M, TAHERI M, SAYAD A, BAHRAM S, OMRANI MD, MOVAFAGH A, INOKO H, AKBARI MT, NOROOZI R, HAJILOOI M, SOLGI G (2016) HLA genes as modifiers of response to IFN-beta-1a therapy in relapsing-remitting multiple sclerosis. Pharmacogenomics 17:489–498
WANG Y, ZHONG H, XIE X, CHEN CY, HUANG D, SHEN L, ZHANG H, CHEN ZW, ZENG G (2015a) Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci U S A 112:E3883–E3892
BECK J, RONDOT P, CATINOT L, FALCOFF E, KIRCHNER H, WIETZERBIN J (1988) Increased production of interferon gamma and tumor necrosis factor precedes clinical manifestation in multiple sclerosis: do cytokines trigger off exacerbations? Acta Neurol Scand 78:318–323
CHOFFLON M, JUILLARD C, JUILLARD P, GAUTHIER G, GRAU GE (1992) Tumor necrosis factor alpha production as a possible predictor of relapse in patients with multiple sclerosis. Eur Cytokine Netw 3:523–531
CHOFFLON M, ROTH S, JUILLARD C, PAUNIER A, JUILLARD P, DEGROOTE D, GRAU G (1997) Tumor necrosis factor production capacity as a potentially useful parameter to monitor disease activity in multiple sclerosis. Eur Cytokine Netw 8:253–257
DEBRUYNE J, PHILIPPÉ J, DEREUCK J, WILLEMS A, LEROUX-ROELS G (1998) Relapse markers in multiple sclerosis: are in vitro cytokine production changes reflected by circulatory T-cell phenotype alterations? Mult Scler J 4:193–197
HOLLIFIELD RD, HARBIGE LS, PHAM-DINH D, SHARIEF MK (2003) Evidence for cytokine dysregulation in multiple sclerosis: peripheral blood mononuclear cell production of pro-inflammatory and anti-inflammatory cytokines during relapse and remission. Autoimmunity 36:133–141
SIMPSON S, STEWART N, VAN DER MEI I, OTAHAL P, CHARLESWORTH J, PONSONBY A-L, BLIZZARD L, DWYER T, PITTAS F, GIES P (2015) Stimulated PBMC-produced IFN-γ and TNF-α are associated with altered relapse risk in multiple sclerosis: results from a prospective cohort study. J Neurol Neurosurg Psychiatry 86:200–207
KÜÇÜKALI Cİ, KÜRTÜNCÜ M, ÇOBAN A, ÇEBI M, TÜZÜN E (2015) Epigenetics of multiple sclerosis: an updated review. NeuroMolecular Med 17:83–96
KULAR L, CASTELO-BRANCO G, JAGODIC M (2017) Epigenetics and multiple sclerosis. In: Neuropsychiatric disorders and epigenetics. Elsevier
GUIL S, ESTELLER M (2012) Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol 19:1068–1075
NAGANO T, FRASER P (2011) No-nonsense functions for long noncoding RNAs. Cell 145:178–181
BECHER B, SPATH S, GOVERMAN J (2017) Cytokine networks in neuroinflammation. Nat Rev Immunol 17:49–59
GOVERMAN J (2009) Autoimmune T cell responses in the central nervous system. Nat Rev Immunol 9:393–407
PHILIPPÉ J, DEBRUYNE J, LEROUX-ROELS G, WILLEMS A, DEREUCK J (1996) In vitro TNF-α, IL-2 and IFN-γ production as markers of relapses in multiple sclerosis. Clin Neurol Neurosurg 98:286–290
VAN OOSTEN BW, BARKHOF F, SCHOLTEN PE, VON BLOMBERG BME, ADÈR HJ, POLMAN CH (1998) Increased production of tumor necrosis factor α, and not of interferon γ, preceding disease activity in patients with multiple sclerosis. Arch Neurol 55:793–798
GOMEZ JA, WAPINSKI OL, YANG YW, BUREAU J-F, GOPINATH S, MONACK DM, CHANG HY, BRAHIC M, KIRKEGAARD K (2013) The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell 152:743–754
LI H, HAO Y, ZHANG D, FU R, LIU W, ZHANG X, XUE F, YANG R (2016) Aberrant expression of long noncoding RNA TMEVPG1 in patients with primary immune thrombocytopenia. Autoimmunity 49:496–502
Funding
The present article is financially supported by “Research Department of the School of Medicine Shahid Beheshti University of Medical Sciences.”
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ganji, M., Sayad, A., Omrani, M.D. et al. Expression analysis of long non-coding RNAs and their target genes in multiple sclerosis patients. Neurol Sci 40, 801–811 (2019). https://doi.org/10.1007/s10072-019-3720-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-019-3720-3