Abstract
In this study, Streptococcus thermophilus and Lactobacillus bulgaricus strains from traditional Turkish yoghurts were isolated, identified by 16S rRNA sequencing and genotypically 14 S. thermophilus and 6 L. bulgaricus strains were obtained as distinct strains by MLST analysis. Lactic acid production levels of the L. bulgaricus strains were higher than S. thermophilus strains. HPLC analysis showed that EPS monosaccharide composition of the strains mainly consisted of glucose and galactose. In general, all strains were found to be susceptible for antibiotics, except some strains were resistance to gentamicin and kanamycin. Apart from two strains of S. thermophilus, all strains displayed strong auto-aggregation level greater than 95% at 24 h incubation. S. thermophilus strains showed higher cell surface hydrophobicity than L. bulgaricus strains. This study demonstrated the isolation, identification, genotypic discrimination and techno-functional features of wild type yoghurt starter cultures which can potentially find place in industrial applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Yoghurt is a fermented dairy product widely consumed around the world, formed as a result of lactic acid fermentation produced by Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus (İspirli and Dertli, 2018). For production of yoghurt, two main approaches is used; first one is traditional production method (includes back-slopping) and second one is industrial production (used defined starter cultures) (Sert et al., 2017). Wild strains of S. thermophilus and L. bulgaricus can be provided from various origins like milk enviroments, animals, soil (Gezginc et al., 2015) and pine cone as different sources (Sert et al., 2017) to be used for production of traditional yoghurt. In the industrial production, using repeatedly same strains give rise to sensorial similarity and also, fermentation process could be failure due to the phage attacks (Sert et al., 2017). Starter cultures used in the yoghurt production are responsible for occurring different aroma compounds and affect techno-functional features like acidification level and viscosity (İspirli and Dertli, 2018). Due to the emphasized reasons, new wild type strain nominees should be identified from traditional yoghurt samples for industrial production.
Multiple locus sequence typing analysis (MLST) was suggested earlier in 1998 as a general approach to ensure exact and portable information for bacterial epidemiological studies (Maiden et al., 1998). MLST defines bacterial isolates on the principle of sequence polymorphism within of domestic fragments of housekeeping genes and is a strong method for population investigations (Maiden et al., 1998). Also, genetic relationship among the isolates could be determine by using MLST and it is becoming a gold standart technique with progress in sequencing fields (Miyoshi-Akiyama et al., 2013). Additionally, relatively decreasing prices on sequencing applications increases the attention to MLST as a discrimination technique (Behringer et al., 2011). MLST is, in general, preferred genotyping technique for many bacterial pathogens (Boonsilp et al., 2013), but in recent times it become a method of choice for genotyping distinction of lactic acid bacteria in L. bulgaricus (Cebeci and Gürakan, 2011; Ivanova et al., 2008; Ivanov et al., 2021), S. thermophilus (Delorme et al., 2017), L. plantarum (Xu et al., 2015), L. casei (Bao et al., 2016; Feng et al., 2018), L. brevis (Sharma et al., 2017) and Leu. mesenteroides (Sharma et al., 2018).
The techno-functional properties offered by lactic acid bacteria including yoghurt starter cultures affect both last quality characteristics and the sensory acceptability of the food products and offer positive effects on human health (Yilmaz et al., 2022a). These properties include hydrophobicity, low pH and bile salt resistance, auto-aggregation, EPS and lactic acid production (İspirli et al., 2017; Yilmaz et al., 2022b). Especially in terms of yoghurt production technology, EPS and lactic acid production are important due to the last quality characteristics of product, sensory acceptability and positive effects on health (Ruas-Madiedo et al., 2002). In this study, hydrophobicity, auto-aggregation, eps and lactic acid production were carried out from the mentioned above techno-functional properties.
This work was goaled to: (a) isolation of L. bulgaricus and S. thermophilus strains from traditionally produced yoghurt samples, (b) determination of starter culture candidates that can be used as starter culture in industrial production, (c) revealing the discrimination power of the MLST technique in determining strain differences, (d) determination of some technological and functional characterization of strains that found to be genotypically different according to MLST technique.
For this purpose, yoghurt samples produced traditionally from different provinces of Turkey were collected and wild type starter cultures were isolated and identified by PCR analysis. Genotypic differences of wild type starter cultures in comparison to a commercial starter culture were determined using the MLST technique. Distinct strains were selected based on their MLST profiles and subjected to technological and functional characterisation.
Material and methods
Isolation of yoghurt starter cultures and their pre-identification
Traditional yoghurt samples produced from cow, sheep, goat and buffalo milks were collected for isolation and identification of yoghurt starter cultures, which are L. delbrueckii subsp. bulgaricus and S. thermophilus, from eleven different provinces of Turkey (Table1) and 57 isolates were obtained from these yoghurt samples. These isolates were tested by colony PCR for their identity to L. bulgaricus and S. thermophilus strains. For the isolation of starter cultures, 10 g of yoghurt sample was weighed, mixed with 90 mL of Ringer's solution and homogenized for 2 min. Serial dilutions were prepared and inoculated on Man, Rogosa, Sharpe (MRS Merck, Darmstadt, Germany), enriched with fructose 10 g/L, peptone from casein 8 g/L, Tween 80 2 g/L, cysteine 0.7 g/L,) and M17 (Merck, Darmstadt, Germany) agar plates for L. bulgaricus and S. thermophilus isolates, respectively and plates were incubated at 45 °C for 48 h. Different colonies were purified and stored at − 80 °C in glycerol (20% v/v) until it used.
A preliminary PCR evaluation was applied to test whether the colonies were belonging to L. bulgaricus and S. thermophilus strains. For this, colony PCR reactions were set with primer sets P1: 5′-CACTATGCTCAGAATACA-3′—P2: 5′-CGAACAGCATTGATGTTA-3′ and delF188:5′-CAACATGAGTCGCATGATTCAAG-3′—delR1042 5′-GGAACCACCTCTCTCTAGCTGTAG-3′ designed previously specific to S. thermophilus and L. bulgaricus strains, respectively (Giraffa et al., 2001; Lu et al., 2015). Details of PCR reactions were as below: 2 min at 95 ºC, 30 s at 95 ºC, 30 s at 50 ºC and 1 min at 72 ºC with 25 cycles and 5 min at 72 ºC for final elongation. Positive isolates of these PCR reactions were then subjected to identification studies.
DNA extraction of starter cultures and bacterial ıdentification by 16S rRNA gene sequencing
A commercial isolation kit (Thermofisher, Pure Link Genomic DNA Mini Kit, Turkey) was used according to manufacturer’s protocol for obtaining L. bulgaricus strains genomic DNAs and classical phenol:chloroform:isoamyl alcohol protocol was utilized to remove genomic DNAs of S. thermophilus strains. 16S rRNA gene of yoghurt starter cultures were amplified with primers AMP_F (GAGAGTTTGATYCTGGCTCAG) and AMP_R (AAGGAGGTGATCCARCCGCA) (İspirli et al., 2017). PCR mixtures and conditions for analysis of 16 s rRNA gene sequencing were performed as stated previously (İspirli and Dertli, 2018). Obtained amplicons were sequenced and strains were defined by using BLAST database with a resemblance criteria of 97–100%. Using MEGA X software phylogenetic trees were formed according to the neighbour-joining method (NJ).
Selection of different strains by MLST method
MLST methodology was applied to discriminate L. bulgaricus and S. thermophilus strains. The gene regions specific to two bacterial strains were amplified by PCR using the primer pairs given in Table 2. PCR reactions were prepared for MLST analyses containing 1 µL DNA template, 10 µL buffer, 5 µL dNTPs, 1 µL primer F, 1 µL primer R and 0.25 µL Taq polymerase (Promega) and up to 50 µL of deiyonized sterile water. Also, PCR conditions were applied using a thermal cycler (T100 Thermal Cycler; Bio-;Rad, North Carolina, USA) as determined follows: initial denaturation for 2 min at 95 ºC, 30 cycles at 95 ºC for 30 s, 48 ºC for 30 s, and 72 ºC for 1 min; and final elongation step of 72 ºC for 5 min., and then PCR products were sent to Medsantek (Istanbul) for sequencing. Database of https://pubmlst.org/ was used to get allele type of S. thermophilus strains since it has sufficient data for S. thermophilus strains. For discrimination of L. bulgaricus strains, UPGMA method of the MEGA-X software programme was used due to the insufficient data on the mentioned database and phylogenetic trees were created and then MLST profiles were determined.
Acid producing ability of cultures
Acid producing ability of L. bulgaricus and S. thermophilus strains was determined according to pH changes in over time (ΔpH). Overnight cultures were inoculated to the Skim Milk Broth (peptone from casein 5 g/L, skim milk powder 28 g/L, yeast extract 2.5 g/L and glucose 1 g/L) at ratio 2% and incubated at 42 ºC for 10 h. pH degree of medium was measured with a pH meter (Metler Toledo, Seven Compact) at two hours of intervals and ΔpH values of the strains were calculated according to formulation below:
Titratable acidity
L. bulgaricus and S. thermopilus strains were twicely activated in MRS broth with 1% fructose and M17 broth with 1% lactose, respectively. Then strains were seperately inoculated in 100 mL Skim Milk Medium at 2% ratio and incubated at 42 ºC for 24 h. After the incubation, cultures were centrifuged (Universal 320 Centrifuge, Hettich) at 6000 × g for 10 min. and 3 mL of the supernatants were mixed with 6 mL of distilled water. 0.5 mL of phenolphthalein used as a indicator was added and titrated with 0.1N NaOH solution. By using expended NaOH amount (milliliter), acid produced ability of strains were calculated as titratable percent acidity in terms of lactic acid:
V (mL): used volume of NoaH in the titration.m (mL): used supernatants for titration
Determination of the EPS production characteristics of yoghurt starters
For EPS extraction, a method was adopted from a previously described methodology (İspirli and Dertli, 2018). Briefly, overnight cultures of L. bulgaricus and S. thermophilus strains propagated at MRS Broth containing 1% fructose and M17 Broth containing 1% lactose, respectively, were inoculated in 1% ratio in Brain Heart Infusion (BHI) Broth containing 2% lactose and incubated at 42 ºC for 48 h. After the incubation period, supernatants were get with centrifugation for 10 min at 4000 rpm and to this supernatants two-fold cold ethanol was added to obtain crude EPS and kept for 48 h at + 4 ºC to precipitate the crude EPS. After the centrifugation process for 15 min at 4000 rpm, supernatants were discarded and precipitated crude EPS was obtained. The crude EPSs were dissolved in 3 mL distilled water which were then freeze-dried and lyophilised EPSs were then stored at 4 ºC for further analysis. EPS production levels of yoghurt starters was defined by phenol–sulfuric acid procedure. Briefly, 200 µL dissolved crude EPS was placed into spectro cuvette and 600 uL of sulfuric acid (98%) was suffixed. Then 120 µL 5% phenol was added and waited for 5 min for color change. Absorbance values were measured at 490 nm and finally using glucose calibration curve EPS amounts were determined as glucose equivalent per mg/10 CFU. However, a HPLC (Shimadzu, Japan) methodology was applied for definition of the EPS compositions using an CARBOsep CHO-682 Pb Column and RID-10A detector (İspirli and Dertli, 2018). H2O was used as mobile phase with 0.7 mL/min flow rate and at 25 ºC column temperature.
Determination of antibiotic susceptibility
Susceptibility of selected yoghurt cultures for some antibiotics were detected with formerly summarized procedure (İspirli et al., 2017). Gentamicin (CN, 10 mcg), Chloramphenicol (C, 30 mcg), Erythromycin (E, 15 mcg), Tetracycline (TE, 30 mcg), Kanamycin (K, 30 mcg), Oxytetracycline (T, 30 mcg), Ampicillin (AM, 10 mcg), Streptomycin (S, 10 mcg) and Penicillin (P, 10 U) were selected to determination antibiotic susceptibility.
Auto-aggregation features
Auto-aggregation properties of selected yoghurt starter cultures were determined in accordance with (Gil-Rodríguez et al., 2015). Briefly, the pellet of overnight cultures were collected by centrifugation and twicely washed with a sterile salty solution (0.9% NaCI). The pellet was resuspended with same solution and absorbance values were measured by spectrophotometer (Schimadzu 150 UV-1800, Kyoto, Japan) for 0, 2, 4 and 24 h at OD600. Using following mathematical equation auto-aggregation percentage of cultures were determined:
where AT shows OD600 for 2, 4 and 24 h and A0 shows OD600 for 0 h.
Hydrophobicity characteristics
Adhere to hydrocarbon was used to determine their hydrophobicity capacity of yoghurt starter cultures and n-hexadecane selected as a hydrocarbon (Vinderola and Reinheimer, 2003). Overnight cultures were centrifuged for 10 min at 7000 rpm, washed two times in 0,05 M K2HPO4 and lastly resuspended in the same solution. All strains were adjusted to OD560 of 1.0 (A1). Then, 3 mL of the culture suspension was mixed with 0.6 mL n-hexadecane, mixed for 2 min and incubated for 20 min at 37 °C. The aqueous phase was meticulously removed and its absorbance value was measured at 560 nm (A2). Hydrophobicity characteristic of yoghurt starter cultures was determined using following mathematical equation:
Results and discussion
Identification and selection of different cultures
MLST is a more powerful appliance for differentiation of bacterial strains than 16S rRNA technique (Sharma et al., 2020). Thanks to its great discrimination power, MLST maintains clear results when comparing results get in other laboratories. However, it has disadvantages such as high cost and limited accessibility (Sharma et al., 2020). MLTS uses nucleotide sequences of several reference genes (housekeeping genes) for isolation characterization. These genes generally slow evolve and reveal the genetic relationship between bacterial isolates more precisely (Urwin and Maiden, 2003). For this reason, different gene regions defined as these housekeeping in both bacterial species were specifically sequenced by MLST method and a more detailed gene scan was performed. Thus, the species were differentiated at the strain level. For discrimination of S. thermophilus strains, there are sufficient data on database of https://pubmlst.org and allele types of S. thermophilus strains were obtained to determine different strains. However, for L. bulgaricus, there are insufficient data and therefore, UPGMA method of the MEGA-X software programme was used and phylogenetic trees were created and then MLST profiles were determined.
In total 49 of the isolates were obtained for 16 s rRNA gene sequencing which resulted in the identification of 17 L. bulgarcius and 32 S. thermophilus strains from traditional yoghurt samples. The distribution of the identified yoghurt cultures by yoghurt type and provinces were shown in the Table 1.
For the discrimination of 17 L. bulgarcius and 32 S. thermophilus strains by MLST analysis pyrG, recA and rpoB and ß-pyrG, ß-recA and ß-rpoB genes were amplified from genomic DNAs of S. thermophilus and L. bulgaricus strains, respectively. Sequence data were uploaded to MLST database (https://pubmlst.org) and allele type of S. thermophilus strains were determined (Table 2). For pyrG, recA and rpoB primers six, two and four different gene regions had emerged, respectively. Strains with different allele types according to the gene regions of the primers were determined as different strains. For example, T1 strain has 2, 6 and 10 allele types for pyrG, recA and rpoB primers, respectively, while T4 strain has 2, 6 and 8 allele types for the same primers, respectively, and these two strains were identified as different strains. However, 6, 6 and 8 allelic types for the same primers were determined for 8 strains and only one of these strains was randomly selected as a different strain. Consequently, it was determined that 14 of 32 S. thermophilus strains identified according to 16 s RNA gene region were different strains as a result of MLST analyses. The allelic types of the isolated strains were compared with the commercial S. thermophilus DGCC7710 strain (Yu et al., 2015) and there was no similarity between the strains within the three reference gene regions with the tested commercial strain.
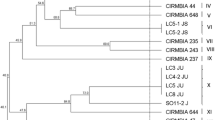
For L. bulgaricus strains, phylogenetic trees were created for the MLST analyses using ß-pyrG, ß recA and ß-rpoB gene regions and with the gene regions tested 11, 12 and 12 different groups were formed, respectively with the phylogenetic tests (Fig. 1). Base sequences of the L. bulgaricus strains were loaded into the BLAST (http://goo.gl/lohXcq) system and compared to the commercial cultures (Cebeci and Gürakan, 2011) and no similarity was observed when three genes were tested together. However, strains of B12 and B6 had the same branching point as the commercial culture in β-pyrG, β-recA and β-rpobG primer pairs, respevtively and they could be a close relative with commercial culture. Different strains for L. bulgaricus were determined according to the relationship in the phylogenetic trees created with primer pairs of ß-pyrG, ß-recA and ß-rpoB. B1, B2, B3, B4, B5, B9, B10, B11, B13 and B14 strains of L. bulgaricus do not have at least one consanguinity in one of three primer pairs and are candidates as different strain. B1, B9 and B14 strains that were not related at branching points for at least 2 primer pairs were selected as different strains. B2, B3, B4, B5, B10, B11 and B13 strains are not consanguineous in only one primer pair and if there was a consanguinity in one of the other two primer pairs, it was selected as a different strain. For example, the B4 strain is not related only to the ß-recA primer, and the B4 strain was chosen differently because it is related to the B9 strain in the pyrG primer and the B2 strain in the rpoB primer. Finally, B1, B4, B7, B9, B14 and B15 strains of L. bulgaricus were selected as different strains and were used in the following analyzes. It has been stated in various studies that MLST analysis can be successfully applied to distinguish between different bacterial species (De Las Rivas et al., 2004, Yu et al., 2015) and similarly, in this study, we obtained distinct strains of S. thermophilus and L. bulgaricus strains by MLST. Identification of distinct strains from traditional yoghurts by MLST technique can be also important for further starter culture applications of these strains. In a recent study, 35 L. bulgaricus strains were subjected to both phenotypic characterisation as well as MLST discrimination based on the gens mainly related with the tested phenotypic features and results of this study demonstrated the correlation of the MLST data with the alterations of the phenotypic features suggesting the importance of the MLST discrimination for further applicability of these strains for technological purposes (Liu et al., 2016). In another study, worldwide MLST scheme of S. thermophilus strains was obtained by testing 178 S. thermophilus strains originated from different regions. In terms of MLST application, this study demonstrated the agreement of the results of the MLST with the whole-genome sequencing application suggesting the capacity of the MLST technique. Importantly, the new MLST scheme of S. thermophilus strains revealed the domination of the commercial starter cultures around the world which was suggested to be a negative factor for the traditional starter cultures and natural biodiversity (Delorme et al., 2017). These findings reveal the importance of the identification of traditional starter cultures and thanks to MLST application, novel strains can be identified in a cost-effective way. Another importance of the identification of the traditional strains is their protection in terms of biodiversity. Also, technologically promising strains can be transferred to commercial applications and nowadays there is an increasing trend for this as consumers expect the traditional sensorial aspects of dairy products, which can be provided by the utilisation of this traditional starter cultures. From this perspective, MLST discriminated S. thermophilus and L. bulgaricus strains were further subjected to the technological and functional characterisation tests to reveal their potential for commercial applications.
Acid producing ability
Acid producing ability of yoghurt starter cultures is a principal parameter for the selection of starter cultures, so acid producing abilities of 14 S. thermophilus and 6 L. bulgaricus strains selected according to MLST analysis were determined and as expected, pH values of all strains of S. thermophilus and L. bulgaricus decreased with the increase in incubation time (Fig. 2). In general, after 6 h of incubation, a rapid decrease in pH value of S. thermophilus and L. bulgaricus strains was observed and it was defined that the pH values of these strains were close to about 4. However, a similar decrease was not observed in T3, T5 and T28 strains and pH values of this three strains were found to be close to 6. The increase in the count of S. thermophilus and L. bulgaricus during the incubation period resulted more lactic acid production and thus a rapid decrease of the pH value. However, a slight decrease in pH values after 4 h of incubation may be related to the adaptation process of the strains to the medium. At last of 10 h of incubation period, it was defined that L. bulgaricus strains lowered the pH value more than S. thermophilus strains and the average pH values were 3.68 and 4,08 for L. bulgaricus and S. thermophilus strains, respectively. The data obtained in this study are compatible with the literature and it has been stated that L. bulgaricus decreases the pH value more than S. thermophilus (Lourens-Hattingh and Viljoen, 2001).
Titratable acidity
S. thermophilus and L. bulgaricus alter lactose to lactic acid during fermentation, which emerges unique acidic taste of yoghurt (Sert et al., 2017). The lactic acid production levels of S. thermophilus and L. bulgaricus strains changed between as 0.42, 0.62 (%) and 0.65, 0.98 (%), respectively (Table 3). L. bulgaricus strains were greater lactic acid producer than S. thermophilus strains, so the highest and lowest lactic acid production level determined for L. bulgaricus and S. thermophilus strains, respectively. Also, these results were compatible with pH changes. Similarly reported that L. bulgaricus strains have ability to produce more lactic acid than S. thermophilus strains (Gezginc et al., 2015). However, they stated that lactic acid production level could be change depending on the species, strain and growth conditions.
EPS production
EPS production is an important properties for cultures in order to give desired rheological properties to yoghurt (İspirli and Dertli, 2018), in this respect, EPS production characteristics and EPS composition of the selected isolates were defined. EPS production levels of S. thermophilus and L. bulgaricus strains ranged between as 38.03 ± 1.8, 196.56 ± 1.6 and 60.86 ± 0.5, 197.33 ± 0.0 mg EPS/cfu and these values were determined for T17, T4 and B14, B4 strains for yoghurt cultures, respectively (Table 3). According to the HPLC analysis, EPS composition of S. thermophilus and L. bulgaricus strains consists of monomeric sugars glucose and galactose. EPS compositions of the strains differ from each other in terms of total glucose and galactose (mg/mL). As can be seen in the Table 3, maximum and minimum total EPS production level of the strains were observed for strain T26 (0.814 mg/mL) and strain B7 (0.305 mg/mL), respectively. These results showed that the amount of EPS may vary depending on the strain. These findings are in consistent with previous studies which are reported that EPS compositions of S. thermophilus strains (Pachekrepapol et al., 2017; Xu et al., 2018) and L. bulgaricus strains (Schiraldi et al., 2006) are composed of glucose and galactose.
Antibiotic susceptibility
Most strains of the bacteria defined as probiotics are either unable to resist or to tolerate antibiotics (Amorim et al., 2018). Our findings revealed that yoghurt starter cultures are generally susceptibility to tested antibiotics (Table 3). It was defined that the growth of L. bulgaricus and S. thermophilus strains were inhibited at the same degree from antibiotics like chloramphenicol, erythromycin, tetracycline, oxytetracycline, ampicillin and penicillin. Three strains of S. thermophilus, T3, T6 and T10, were found to be resistant to kanamycin, but all strains were found to be susceptible to other antibiotics. It has been determined that some strains of L. bulgaricus are resistant to gentamicin (strains ofB1, B4, B7 and B9) and kanamycin (strains of B1, B4, B7, B9 and B14), yet sensitive to other antibiotics. The resistance of yoghurt starter cultures to certain antibiotics such as kanamycin and gentamicin may be a natural feature of bacterial species and even genera (Szutowska and Gwiazdowska, 2021). These findings are in consistent with previous studies (Akpinar et al., 2011; Iyer et al., 2010; Yerlikaya et al., 2020) and antibiotic resistance may vary from strain to strain. In addition, different results in the literature may be related to strains diversity, methods applied or culture media used.
Auto-aggregation
Probiotic strains must present auto-aggregation feature that is related to the microbial adherence to the gastrointestinal tract (GIT) (Gil-Rodríguez et al., 2015). After 24 h of incubation period, all yoghurt cultures, except two strains of S. thermophilus (T6 and T13), had auto-aggregation percentage over 95% (Fig. 3). The strains showed different auto-aggregation profiles after 2 h of incubation period, which may be due to the fact that the early incubation period affects the adaptation of the strains to the medium. It was determined that all strains showed an aggregation profile over 50% after 4 h incubation. Different results can be obtained for distinct strains as (Aslim et al., 2007) previously reported that at the end of the 4 h of incubation period, 4 different L. bulgaricus strains showed an aggregation profile between 45 and 93%. Finally, these results indicated that L. bulgaricus strains exhibited better aggregation ability than S. thermophilus strains.
Hydrophobicity
One of the desired probiotic properties for microorganisms is hydrophobicity and this characteristic can be linked to adhesion to the GIT (Gut et al., 2019). As seen in the Fig. 3, the hydrophobic properties of L. bulgaricus strains (B1 and B15 strains) varied between 2 and 11%, respectively. It was determined that the hydrophobicity of S. thermophilus strains increased up to 26% (T15 strain) and one strain (T26) decreased below 1%. T14 and T15 from S. thermophilus strains and B15 from L. bulgaricus strains were observed with their high hydrophobic properties. (Vinderola and Reinheimer, 2003) et al. reported that the hydrophobicity features of 8 different S. thermophilus strains and L. bulgaricus strains varied between 12 and 26%, 5% and 27%, respectively. Also, (Iyer et al., 2010) reported that the hydrophobicity features of 2 different S. thermophilus strains were 18% and 19%. On the other hand, (Flint et al., 1997) reported that the hydrophobicity properties of S. thermophilus strains ranged from 24 to 98%. Hydrophobicity characteristic depends on the origin of strains, surface features, solvent type and environmental conditions (Kaushik et al., 2009) and this study revealed that cell surface hidrofobicity of the yoghurt starter cultures may vary depending on the strain.
In conclusion, in this study, potential starter culture candidates were identified for industrial yoghurt production and distinct strains were successfully discriminated by MLST analysis. All selected strains were EPS producers and the monomeric composition of cultures consisted of glucose and galactose as the sugar units and EPS production levels altered depending on strain specific conditions. Similarly, results showed that technological (such as titratable acidity and lactic acid production) and functional properties (such as hydrophobicity, auto-aggregation profile) of strains can be changed depending on the strain specific conditions. This study demonstrated the importance of the isolation, discrimination by MLST and characterisation of traditional yoghurt cultures, which can be further transferred to industrial applications.
Data availability
The data that assist the results of this work provides from the corresponding writer upon acceptable demand.
References
Akpinar A, Yerlikaya O, Kiliccedil S. Antimicrobial activity and antibiotic resistance of Lactobacillus delbrueckii ssp. bulgaricus and Streptococcus thermophilus strains isolated from Turkish homemade yoghurts. African Journal of Microbiology Research. 5: 675–682 (2011)
Amorim JC, Piccoli RH, Duarte WF. Probiotic potential of yeasts isolated from pineapple and their use in the elaboration of potentially functional fermented beverages. Food Research International. 107: 518-527 (2018)
Aslim B, Onal D, Beyatli Y. Factors influencing autoaggregation and aggregation of Lactobacillus delbrueckii subsp. bulgaricus isolated from handmade yogurt. Journal of Food Protection. 70: 223–227 (2007)
Bao Q, Song Y, Xu H, Yu J, Zhang W, Menghe B, Zhang H, Sun Z. Multilocus sequence typing of Lactobacillus casei isolates from naturally fermented foods in China and Mongolia. Journal of Dairy Science. 99: 5202-5213 (2016)
Behringer M, Miller WG, Oyarzabal OA. Typing of Campylobacter jejuni and Campylobacter coli isolated from live broilers and retail broiler meat by flaA-RFLP, MLST, PFGE and REP-PCR. Journal of Microbiological Methods. 84: 194-201 (2011)
Boonsilp S, Thaipadungpanit J, Amornchai P, Wuthiekanun V, Bailey MS, Holden MT, Zhang C, Jiang X, Koizumi N, Taylor K. A single multilocus sequence typing (MLST) scheme for seven pathogenic Leptospira species. PLoS Neglected Tropical Diseases. 7: e1954 (2013)
Cebeci A, Gürakan GC. Comparative typing of L. delbrueckii subsp. bulgaricus strains using multilocus sequence typing and RAPD–PCR. European Food Research and Technology. 233: 377–385 (2011)
De Las Rivas B, Marcobal A, Munoz R. Allelic diversity and population structure in Oenococcus oeni as determined from sequence analysis of housekeeping genes. Applied and Environmental Microbiology. 70: 7210-7219 (2004)
Delorme C, Legravet N, Jamet E, Hoarau C, Alexandre B, El-Sharoud WM, Darwish MS, Renault P. Study of Streptococcus thermophilus population on a world-wide and historical collection by a new MLST scheme. International Journal of Food Microbiology. 242: 70-81 (2017)
Feng J, Jiang Y, Li M, Zhao S, Zhang Y, Li X, Wang H, Lin G, Wang H, Li T. Diversity and evolution of Lactobacillus casei group isolated from fermented dairy products in Tibet. Archives of Microbiology. 200: 1111-1121 (2018)
Flint S, Brooks J, Bremer P. The influence of cell surface properties of thermophilic streptococci on attachment to stainlesssteel. Journal of Applied Microbiology. 83: 508-517 (1997)
Gezginc Y, Topcal F, Comertpay S, Akyol I. Quantitative analysis of the lactic acid and acetaldehyde produced by Streptococcus thermophilus and Lactobacillus bulgaricus strains isolated from traditional Turkish yogurts using HPLC. Journal of Dairy Science. 98: 1426–1434 v
Gil-Rodríguez AM, Carrascosa AV, Requena T. Yeasts in foods and beverages: In vitro characterisation of probiotic traits. LWT-Food Science and Technology. 64: 1156-1162 (2015)
Giraffa G, Paris A, Valcavi L, Gatti M, Neviani E. Genotypic and phenotypic heterogeneity of Streptococcus thermophilus strains isolated from dairy products. Journal of Applied Microbiology. 91: 937-943 (2001)
Gut AM, Vasiljevic T, Yeager T, Donkor ON. Characterization of yeasts isolated from traditional kefir grains for potential probiotic properties. Journal of Functional Foods. 58: 56-66 (2019)
İspirli H, Demirbaş F, Dertli E. Characterization of functional properties of Enterococcus spp. isolated from Turkish white cheese. LWT-Food Science and Technology. 75: 358–365 (2017)
İspirli H, Dertli E. Isolation and identification of exopolysaccharide producer lactic acid bacteria from Turkish yogurt. Journal of Food Processing and Preservation. 42: e13351 (2018)
Ivanov I, Petrov K, Lozanov V, Hristov I, Wu Z, Liu Z, Petrova P. Bioactive Compounds Produced by the Accompanying Microflora in Bulgarian Yoghurt. Processes. 9: 114 (2021)
Ivanova P, Peykov S, Dimitrova A, Dimov S. Molecular typing by genus-specific PCR and RAPD profiling of diverse Lactobacillus delbrueckii strains isolated from cow, sheep and buffalo yoghurts. Biotechnology & Biotechnological Equipment. 22: 748-753 (2008)
Iyer R, Tomar S, Kapila S, Mani J, Singh R. Probiotic properties of folate producing Streptococcus thermophilus strains. Food Research International. 43: 103-110 (2010)
Kaushik JK, Kumar A, Duary RK, Mohanty AK, Grover S, Batish VK. Functional and probiotic attributes of an indigenous isolate of Lactobacillus plantarum. PloS One. 4: e8099 (2009)
Liu W, Yu J, Sun Z, Song Y, Wang X, Wang H, Wuren T, Zha M, Menghe B, Heping Z. Relationships between functional genes in Lactobacillus delbrueckii ssp. bulgaricus isolates and phenotypic characteristics associated with fermentation time and flavor production in yogurt elucidated using multilocus sequence typing. Journal of Dairy Science. 99: 89-103 (2016)
Lourens-Hattingh A, Viljoen BC. Yogurt as probiotic carrier food. International Dairy Journal. 11: 1-17 (2001)
Lu W, Kong W, Yang P, Kong J. A one-step PCR-based method for specific identification of 10 common lactic acid bacteria and Bifidobacterium in fermented milk. International Dairy Journal. 41: 7-12 (2015)
Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proceedings of the National Academy of Sciences. 95: 3140-3145 (1998)
Miyoshi-Akiyama T, Hayakawa K, Ohmagari N, Shimojima M, Kirikae T. Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PloS One. 8: e66358 (2013)
Pachekrepapol U, Lucey J, Gong Y, Naran R, Azadi P. Characterization of the chemical structures and physical properties of exopolysaccharides produced by various Streptococcus thermophilus strains. Journal of Dairy Science. 100: 3424-3435 (2017)
Ruas-Madiedo P, Hugenholtz J, Zoon P. An overview of the functionality of exopolysaccharides produced by lactic acid bacteria. International Dairy Journal. 12: 163-171 (2002)
Schiraldi C, Valli V, Molinaro A, Cartenì M, De Rosa M. Exopolysaccharides production in Lactobacillus bulgaricus and Lactobacillus casei exploiting microfiltration. Journal of Industrial Microbiology and Biotechnology. 33: 384-390 (2006)
Sert D, Mercan E, Dertli E. Characterization of lactic acid bacteria from yogurt-like product fermented with pine cone and determination of their role on physicochemical, textural and microbiological properties of product. LWT-Food Science and Technology. 78: 70-76 (2017)
Sharma A, Kaur J, Lee S, Park Y-S. Molecular discrimination of Lactobacillus brevis strains isolated from food products in South Korea using multilocus sequence typing. LWT-Food Science and Technology. 86: 337-343 (2017)
Sharma A, Kaur J, Lee S, Park Y-S. Genetic diversity analysis of Leuconostoc mesenteroides from Korean vegetables and food products by multilocus sequence typing. Applied Microbiology and Biotechnology. 102: 4853-4861 (2018)
Sharma A, Lee S, Park Y-S. Molecular typing tools for identifying and characterizing lactic acid bacteria: a review. Food Science and Biotechnology. 29: 1301-1318 (2020)
Song Y, Sun Z, Guo C, Wu Y, Liu W, Yu J, . . . Zhang, H. Genetic diversity and population structure of Lactobacillus delbrueckii subspecies bulgaricus isolated from naturally fermented dairy foods. Scientific Report. 6(1): 1-8 (2016)
Szutowska J, Gwiazdowska D. Probiotic potential of lactic acid bacteria obtained from fermented curly kale juice. Archives of Microbiology. 203: 975-988 (2021)
Urwin R, Maiden MC. Multi-locus sequence typing: a tool for global epidemiology. Trends in microbiology. 11:479-87 (2003)
Vinderola CG, Reinheimer JA. Lactic acid starter and probiotic bacteria: a comparative “in vitro” study of probiotic characteristics and biological barrier resistance. Food Research International. 36: 895-904 (2003)
Xu H, Liu W, Zhang W, Yu J, Song Y, Menhe B, Zhang H, Sun Z. Use of multilocus sequence typing to infer genetic diversity and population structure of Lactobacillus plantarum isolates from different sources. BMC Microbiology. 15: 1-10 (2015)
Xu Z, Guo Q, Zhang H, Wu Y, Hang X, Ai L. Exopolysaccharide produced by Streptococcus thermophiles S-3: molecular, partial structural and rheological properties. Carbohydrate Polymers. 194: 132-138 (2018)
Yerlikaya O, Saygili D, Akpinar A. Evaluation of antimicrobial activity and antibiotic susceptibility profiles of Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus strains isolated from commercial yoghurt starter cultures. Food Science and Technology. 41: 418-425 (2020)
Yilmaz MT, İspirli H, Taylan O, Bilgrami AL, Dertli E. Structural and bioactive characteristics of a dextran produced by Lactobacillus kunkeei AK1. International Journal of Biological Macromolecules. 200: 293-302 (2022a)
Yilmaz MT, İspirli H, Taylan O, Taşdemir V, Sagdic O, Dertli E. Characterisation and functional roles of a highly branched dextran produced by a bee pollen isolate Leuconostoc mesenteroides BI-20. Food Bioscience. 45: 101330 (2022b)
Yu J, Sun Z, Liu W, Xi X, Song Y, Xu H, Lv Q, Bao Q, Menghe B, Sun T. Multilocus sequence typing of Streptococcus thermophilus from naturally fermented dairy foods in China and Mongolia. BMC Microbiology. 15: 1-13 (2015)
Acknowledgements
This study was financially supported by TUBİTAK (Turkey) with the grand number 116G024. Hilal Dikmen was supported by TUBİTAK.
Author information
Authors and Affiliations
Contributions
HD: writing-original draft. HG: supervision. FD: methodology. SK: Investigation. HI: methodology. MA: project administration, MT: project administration, OS: project administration. ED: supervision.
Corresponding author
Ethics declarations
Conflict of interest
The writers inform no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dikmen, H., Goktas, H., Demirbas, F. et al. Multilocus sequence typing of L. bulgaricus and S. thermophilus strains from Turkish traditional yoghurts and characterisation of their techno-functional roles. Food Sci Biotechnol 33, 625–635 (2024). https://doi.org/10.1007/s10068-023-01366-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-023-01366-2