Abstract
Halophilic bacteria such as the genus Halomonas are promising candidates in diverse industrial, agricultural and biomedical applications. Here, we successfully isolated a halophilic Halomonas alkaliphila strain XH26 from Xiaochaidan Salt Lake, and studied its osmoadaptation strategies using transcriptome and ectoine analysis. Divergent mechanisms were involved in osmoadaptation at different salinities in H. alkaliphila XH26. At moderate salinity (6% NaCl), increased transcriptions of ABC transporters related to iron (III), phosphate, phosphonate, monosaccharide and oligosaccharide import were observed. At high salinity (15% NaCl), transcriptions of flagellum assembly and cell motility were significantly inhibited. The transcriptional levels of ABC transporter genes related to iron (III) and iron3+-hydroxamate import, glycine betaine and putrescine uptake, and cytochrome biogenesis and assembly were significantly up-regulated. Ectoine synthesis and accumulation was significantly increased under salt stress, and the increased transcriptional expressions of ectoine synthesis genes ectB and ectC may play a key role in high salinity induced osmoadaptation. At extreme high salinity (18% NaCl), 5-hydroxyectoine and ectoine worked together to maintain cell survival. Together these results give valuable insights into the osmoadaptation mechanisms of H. alkaliphila XH26, and provide useful information for further engineering this specific strain for increased ectoine synthesis and related applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Halophiles are a group of microorganisms which can survive in various hypersaline environments such as saline lakes, salterns, saline soils and salted food (Almeida et al. 2019; Chen et al. 2020; Rathour et al. 2020; Soltan Dallal et al. 2017; Zhu et al. 2020). Due to their high osmoadaptation ability to salt stress, halophiles are potential precious hosts for diverse industrial, agricultural and biomedical applications such as sewage treatment (Huang et al. 2019; Torbaghan and Khalili Torghabeh 2019), saline soil improvement (Rezaei Somee et al. 2018; SadrAzodi et al. 2019), cultivation of salt-tolerant plants (Sagar et al. 2020; Saghafi et al. 2019; Vega et al. 2019), enzyme production (Martinez-Perez et al. 2020), anti-inflammatory product development (Brands et al. 2019; Kiefer et al. 2018) and skin care (Bujak et al. 2020; Cho et al. 2017). They adapt to high salinities mainly using two osmoadaptation strategies, the “salt-in” strategy and the accumulation and synthesis of compatible solutes (Brown 1976). Cells apply the “salt-in” strategy to mainly accumulate KCl in their cytoplasm, which is commonly found in extreme halophiles who need salt for growth (Oren 2013). Compatible solutes are small organic compounds which do not disturb cell metabolism even at high concentration, therefore halotolerant and halophilic bateria could either intake these compatible solutes from surrounding environment or synthesize by themselves to cope with high salinity growth conditions (Brown 1976; Kuhlmann and Bremer 2002; Yancey 2005). Common compatible solutes include ectoine, glycine betaine, trehalose and small amino acids. The compatible solute accumulation/synthesis strategy is adopted by a wider range of halophiles than “salt-in” strategy, among which some halophiles could combine both strategies to better adapt to the salt environment (Edbeib et al. 2016; Gunde-Cimerman et al. 2018; Peng et al. 2020).
With persist efforts on studying the osmoadaptation mechanisms of halophiles, researchers have found that within the same genus, different halophiles may secrete different type and level of compatible solutes to survive in a diverse range of salinity (Joghee and Jayaraman 2016). Halomonas belongs to the phylum Gammaproteobacteria class Oceanospirillales family Halomonadaceae, and is one of the largest halophilic genus so far with over 100 species and diverse salinity growth range. Halomonas elongata and Halomonas sp. NY-011 rely on the synthesis of ectoine to adapt to high-salt environments (Kraegeloh and Kunte, 2002; Wang et al. 2010), Halomonas sp. SBS 10 mainly accumulates betaine (Kushwaha et al. 2019) and Halomonas sp. AAD12 relies on proline (Ceylan et al. 2012). Among all these compatible solutes, ectoine is of special interest to researchers. It is the most widespread compatible solute among bacteria, and has wide applications as DNA, protein and cell protectant and stabilizer in extreme environmental conditions (Hahn et al. 2020; Nayak et al. 2020; Tsai et al. 2020), anti-inflammatory reagent (Bilstein et al. 2021; Moffa et al. 2019; Rieckmann et al. 2019), and a useful ingredient in new bio-cosmetics (Bujak et al. 2020; Hseu et al. 2020). Ectoine is biosynthesized by three enzymes encoded by ectA, ectB and ectC respectively within halophiles (Göller et al. 1998), and 5-hydroxyectoine usually coexists as its hydroxylated derivative synthesized by the hydroxylase enzyme encoded by the ectD gene (Prabhu et al. 2004; Garcia-Estepa et al. 2006; Bursy et al. 2007). Since ectoine is difficult to synthesize through chemical synthesis, many researchers have been committed to the discovery of high-yielding ectoine halophilic strains and the improvement of ectoine biosynthesis yield.
In this paper, we successfully isolated a halophilic bacteria strain Halomonas alkaliphila XH26 from Xiaochaidan Salt Lake located in Qaidam Basin, Qinghai province, China, which can survive in 0–18% NaCl and produce high yield of ectoine. We sequenced its complete genome and performed quantitative RNA-seq under different salt stress to study its mechanism of osmoadaptation and its relevant metabolic regulating pathways for ectoine production. This work may enlarge the knowledge of osmoadaptation mechanism of Halomonas and provide theoretical references for the future development and application of this specific Halomonas strain.
Materials and methods
Bacteria strain and culture conditions
Halomonas alkaliphila XH26 was isolated in our laboratory from Xiaochaidan Salt Lake (altitude 3171 m) located at the northeast part of Qaidam Basin in Qinghai Province, China. The basal culture medium contains 2 g/L yeast extract, 25 g/L MgSO4·7H2O, 55 g/L KCl, 0.2 g/L CaCl2, 6.5 g/L sodium glutamate, 3.0 g/L sodium citrate and 7.5 g/L casein enzymatic hydrolysate. For transcriptome analysis, cells of H. alkaliphila XH26 were cultured in basal medium with 0%, 6% and 15% NaCl respectively at pH of 8.0.
Reference genome sequencing, assembly and annotation
Halomonas alkaliphila XH26 complete genome was sequenced using PacBio Sequel platform and assembled using HGAP4 software and Canu (v1.6) software by Frasergen Co., Ltd. Glimmer (v3.02) was applied for CDS prediction. For functional annotation, Diamond was applied to annotate genes across NR (http://www.ncbi.nlm.nih.gov/protein), Swiss-Prot (https://web.expasy.org/docs/swiss-prot_guideline.html), eggNOG (http://eggnog.embl.de/) and KEGG (http://www.genome.jp/kegg/) databases, while hmmer3 and Blast2go were applied for Pfam (http://pfam.xfam.org/) and GO (http://www.geneontology.org/) annotations respectively.
Phylogenetic analysis
The total DNA of XH26 was extracted using Ezup Column Bacteria Genomic DNA Purification Kit (SK8255, Sangon Biotech) and characterized by 16S rRNA sequencing. The acquired 16S rRNA sequence was aligned in the EzBioCloud (http://www.ezbiocloud.net/identify) and NCBI GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) databases to obtain its taxonomic status. MEGA v7.0 (https://www.megasoftware.net/) was used to compare 16S rRNA sequences of XH26 and reference gene sequences of closely related species with high similarity downloaded from NCBI database. Escherichia coli K-12 (LT899983) was used as an outgroup. The neighbor-joining method with 1000 bootstrap replicates was adopted for phylogenetic analysis. The complete genome of XH26 was uploaded to the Type (Strain) Genome Server (TYGS), a free bioinformatics platform available under https://tygs.dsmz.de, for a whole genome-based taxonomic analysis (Meier-Kolthoff and Göker 2019; Meier-Kolthoff et al. 2022). A whole-genome sequence-based phylogenetic tree was constructed using the Genome BLAST Distance Phylogeny approach (GBDP) and the digital DNA-DNA hybridization (dDDH) values were calculated (Meier-Kolthoff et al. 2013).
RNA extraction, RNA library preparation and sequencing
Cells were harvested after 18 h of culture by centrifugation. Total RNA of each sample was extracted using TRIzol Reagent (Invitrogen)/RNeasy Mini Kit (Qiagen). Total RNA of each sample was quantified and qualified by Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), NanoDrop (Thermo Fisher Scientific Inc.) and 1% agarose gel. 1 μg total RNA with RIN value above 7 was used for following library preparation. Next generation sequencing library preparations were constructed according to the manufacturer’s protocol (NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina®). The rRNA was depleted from total RNA using Ribo-Zero rRNA Removal Kit (Bacteria) (Illumina). The ribosomal depleted mRNA was then fragmented and reverse-transcribed. First strand cDNA was synthesized using ProtoScript II Reverse Transcriptase with random primers and Actinomycin D. The second-strand cDNA was synthesized using Second Strand Synthesis Enzyme Mix (include dACGTP/dUTP). The purified double-stranded cDNA by AxyPrep Mag PCR Clean-up (Axygen) was then treated with End Prep Enzyme Mix to repair both ends and add a dA-tailing in one reaction, followed by a T-A ligation to add adaptors to both ends. Size selection of Adaptor-ligated DNA was then performed using AxyPrep Mag PCR Clean-up (Axygen), and fragments of ~360 bp (with the approximate insert size of 300 bp) were recovered. The dUTP-marked second strand was digested with Uracil-Specific Excision Reagent (USER) enzyme (New England Biolabs). Each sample was then amplified by PCR for 11 cycles using P5 and P7 primers, with both primers carrying sequences which can anneal with flow cell to perform bridge PCR and P7 primer carrying a six-base index allowing for multiplexing. The PCR products were cleaned up using AxyPrep Mag PCR Clean-up (Axygen), validated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), and quantified by Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). The RNA-seq was performed using HiSeq (2 × 150) paired-end (PE) configuration (Illumina).
RNA-seq data analysis
Clean RNA-seq data were aligned to the reference genome using Bowtie2 software (v2.1.0). Gene expression levels were estimated using HTSeq (v0.6.1p1), and differential expression analysis was performed using the DESeq Bioconductor package, and differential expressed genes (DEGs) were identified if |log2(fold change)|≥ 1, FDR < 0.05.
Swarming motility assay
Swarming motility assay in semisolid agar plates was conducted based on the procedure described by (Salvador et al. 2018) and modified for agar concentration. Swarming plates of basal medium containing 0%, 6% or 12% NaCl were prepared with an optimized agar concentration of 1.9%. Cells were grown in tubes with basal medium containing 0%, 6% or 12% NaCl, and incubated at 37 °C, 160 rpm until OD600 = 0.6. Aliquots of 3 µl of each culture were seeded at the centre of the respective swarming plates, and incubated at 37 °C in a humid environment. The migration halo was monitored during 96 h and the swarming diameters were measured. Three independent experiments were conducted to avoid artifacts caused by handling.
Quantitative real time polymerase chain reaction (qPCR) validation
The cDNA was synthesized from the extracted RNA using PrimeScript™ RT reagent Kit with gDNA Eraser (RR047A, Takara). The qPCR was performed using TB Green™ Premix Ex Taq™ II (RR820A, Takara) according to the manufacturer’s standard protocol. The internal reference gene was GADPH and the relative transcript quantification of each gene was calculated using the ΔΔCT method. The primers used in this study were synthesized by Nanjing GenScript Biotechnology Co., Ltd., and the primer sequences of the target genes can be found in Table S1.
Compatible solute extraction and accumulation detection
After 48 h of cultivation, cells were measured for their OD600 value and ectoine and 5-hydroxyectoine accumulation. Ectoine and 5-hydroxyectoine extraction was conducted as described by (Parwata et al. 2019). First, 1 mL of the bacterial culture was cold centrifuged at 10,625×g for 5 min, and the supernatant was discarded. Then an equal volume (1 mL) of ultrapure water was added to the tube and ground for 5 min with the 3rd Gen Variable Speed TGrinder (OSE-Y50, Tiangen Biotech (Beijing) Co., Ltd). After centrifugation at 10,625×g for 5 min, the water phase containing the compatible solutes was separated and filtered with a 0.22 µm filter membrane for high-performance liquid chromatography (HPLC) analysis. A standard curve was built using compatible solute standard product diluted in a gradient. Ectoine and 5-hydroxyectoine was identified using Agilent Technologies 1260 Infinity HPLC (USA) with a SeQuant ZIC- HILIC, 15.0 cm by 4.6 mm (5 μm) column (Sigma-Aldrich, USA). Detection conditions were as follows: mobile phase acetonitrile/pure water (v/v: 80/20), detection wavelength of 210 nm, flow rate of 1.0 mL/min, column pressure of 3.486–4.761 MPa, column temperature of 30 °C, and sample volume of 10 µl. The measured ectoine and 5-hydroxyectoine content was standardized by the cell dry weight (CDW).
Results
16S rRNA sequencing, phylogenetic analysis and genome sequencing of XH26
The 16S rRNA gene of XH26 was sequenced and undergone sequence alignment and phylogenetic analysis (Fig. 1). Results showed that XH26 belongs to the Halomonas genus, and its 16S rRNA gene is most closely related to Halomonas alkaliphila X3 (Yue et al. 2019) and Halomonas campaniensis LS21 (Yue et al. 2014) with 99.93% nucleotide identity. After that, the whole genome of XH26 was sequenced using PacBio Sequel platform. The XH26 circular genome was 4,112,053 bp in size, with 52.62% GC content and 3927 genes. We compared its whole genome with genomes of bacterial type strains using TYGS, and results confirmed that XH26 belongs to H. alkaliphila, and is most related to H. alkaliphila DSM16354 (dDDH > 80%, Fig. S1). The functional annotation details across databases could be found in Table S2.
Morphology and growth conditions of H. alkaliphila XH26
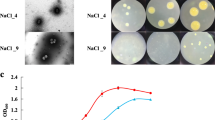
Halomonas alkaliphila XH26 could survive in a pH range of 6.0–10.0 with optimal pH of 8.0. To test XH26 tolerance to salinity, cells were cultured in a NaCl concentration range of 0–18% at optimal pH and measured for their OD600 values for 72 h (Fig. 2a). XH26 could survive in 0–18% NaCl, with fastest growth rate at 3% NaCl (Fig. 2b). After 48 h of culture, similar OD600 values could be measured in 3–9% NaCl. Morphologically, H. alkaliphila XH26 colonies were milky white, moist, opaque, small and round with smooth edges, and exhibited adhesion and swelling properties (Fig. 2c). Scanning electron and transmission electron micrographs revealed that H. alkaliphila XH26 was long rod-shaped with peritrichous flagella, and normally 3.0–5.0 μm in length and 0.5–0.75 μm in width (Fig. 2d, Fig. S2).
Morphology and growth conditions of H. alkaliphila XH26. a Growth curves of H. alkaliphila XH26 in medium containing 0%, 3%, 6%, 9%, 12%, 15% and 18% NaCl respectively. b The growth rates at each time point at different salinities. c XH26 morphology under light microscope. d Scanning electron microscopy micrograph of XH26. Scale bar: 1 μm
Transcript analysis under different salt stress
To address the key mechanisms of salt tolerance for H. alkaliphila XH26, cells were cultured in basal medium with 0%, 6% and 15% NaCl respectively for 18 h before collected for RNA-seq. Three biological replicates were obtained for each treatment condition. The number of raw reads obtained ranged from 13,407,768 to 21,569,442, with Q30 no lower than 92.56% (Table S3). After filtering out the contaminated and low-quality sequences, the clean reads were mapped to its own genome sequence. The total mapped rate of all samples ranged from 93.2% to 99.6%, with unique mapped reads ratio ranged from 91.16% to 97.21%.
Transcriptionally expressed gene analysis
Gene transcriptional expression levels of each sample were analyzed using Transcripts per Million (TPM). Similar number of genes could be found for all groups, ranging from 3656 in 15% NaCl group to 3704 in 0% NaCl group. Strong correlations existed within each treatment group (≥0.82), with highest correlation scores in 15% NaCl group (≥0.97) (Fig. S3a). The results were consistent with principle component analysis (PCA) (Fig. S3b). Samples in 15% NaCl group showed more distinct expression level patterns compared with 6% and 0% NaCl groups and had lowest intra-group variation.
Differentially expressed genes (DEG) analysis on the transcriptional level
Significant changes of gene transcriptional expressions were observed in 15%/0% and 15%/6% comparison groups (Fig. S4, Table 1). There were 786 down-regulated genes and 521 up-regulated genes at 15% NaCl when compared to 0% NaCl (the DEG15%/0% set) (FDR < 0.05, |log2FC|≥ 1). A slightly less number of DEGs were found at 15% NaCl when compared to 6% NaCl (the DEG15%/6% set), with 734 down-regulated genes and 492 up-regulated genes, respectively. More genes were transcriptionally down-regulated than up-regulated in 15% NaCl culture. The gene transcription levels were quite similar for H. alkaliphila XH26 in 0% and 6% NaCl, with only 116 down-regulated genes and 182 up-regulated genes at 6% NaCl when compared to 0% NaCl (the DEG6%/0% set). All three DEG sets were annotated across eggNOG (http://eggnog.embl.de/), GO (http://www.geneontology.org/), KEGG (http://www.genome.jp/kegg/), Swiss-Prot (https://web.expasy.org/docs/swiss-prot_guideline.html), Pfam (http://pfam.xfam.org/) and NR (http://www.ncbi.nlm.nih.gov/protein) databases and the respective statistical results are shown in Table 1.
Global transcriptional response to osmotic stress
We first analyzed the Cluster of Orthologous Groups (COG) functional categories of each DEG set (Fig. 3a). COG Type S had the largest number of genes among all three DEG sets which were poorly characterized and therefore excluded from the following analysis. In DEG6%/0% set, functional categories related to inorganic ion transport and metabolism (P), amino acid transport and metabolism (E), carbohydrate transport and metabolism (G), energy production and conversion (C) and transcription (K) were mostly represented (> 5% of DEG6%/0% no.). Functional categories related to posttranslational modification, protein turnover, chaperones (O), signal transduction mechanisms (T), replication, recombination and repair (L) and coenzyme transport and metabolism (H) also increased their representations to a lesser extent (4–4.88% of DEG6%/0% no.). In 15% NaCl condition, besides the functional categories E, C, P, K, O, L mentioned above, category related to cell wall/membrane/envelope biogenesis (M) was also mostly represented compared to both 0% NaCl and 6% NaCl condition (4.15% of DEG15%/0% no. and 4.84% of DEG15%/6% no. respectively), indicating different mechanisms involved in high salinity-induced osmoadaptation.
Functional categories of DEGs in H. alkaliphila XH26. a COG annotation. B: Chromatin structure and dynamics; C: Energy production and conversion; D: Cell cycle control, cell division, chromosome partitioning; E: Amino acid transport and metabolism; F: Nucleotide transport and metabolism; G: Carbohydrate transport and metabolism; H: enzyme transport and metabolism; I: Lipid transport and metabolism; J: Translation, ribosomal structure and biogenesis; K: Transcription; L: Replication, recombination and repair; M: Cell wall/membrane/envelope biogenesis; N: Cell motility; O: Posttranslational modification, protein turnover, chaperones; P: Inorganic ion transport and metabolism; Q: Secondary metabolites biosynthesis, transport and catabolism; S: Function unknown; T: Signal transduction mechanisms; U: Intracellular trafficking, secretion, and vesicular transport; V: Defense mechanisms. b GO annotation. c KEGG annotation
For GO annotation, the top ten GO groups comprising most number of DEGs for all three DEG groups were unanimous, with four GO groups belonging to biological process category (cellular process, metabolic process, localization and biological regulation), three belonging to cellular component category (cell part, membrane part and membrane) and three belonging to molecular function category (catalytic activity, binding and transporter activity) (Fig. 3b).
For KEGG annotation, top represented KEGG pathways were quite similar among all three DEG groups except for cell motility pathway, which was among the top ten most represented pathways in DEG15%/0% and DEG15%/6% sets but not in the DEG6%/0% set, indicating possible role of cell motility regulation in high salinity-induced osmoadaptation (Fig. 3c).
GO and KEGG enrichment analysis for three DEG sets
To figure out top GO groups and KEGG signaling pathways involved in different salinity-induced osmoadaptation, GO and KEGG enrichment analysis were performed for all three DEG sets (Figs. 4, 5, 6). Total 62 GO groups were significantly represented in the DEG6%/0% set, with 14 annotated to biological process category, 4 annotated to cellular component category and 44 annotated to molecular function category (FDR < 0.05). The top 20 GO groups were most related to transmembrane transporter activity (10 GO groups containing 30 DEGs) and fatty acid catabolism (4 GO groups containing 3 DEGs), the latter of which were down-regulated (Fig. 4a). In KEGG enrichment analysis, the genes were annotated to 95 signaling pathways and ABC transporters (map02010) was the only significantly mapped reference canonical pathway with 21 up-regulated DEGs and 11 down-regulated DEGs (Fig. 4b).
For high-salinity induced osmoadaptation, there were 117 GO groups significantly enriched in the DEG15%/6% set and 51 in the DEG15%/0% set (FDR < 0.05), the top 20 of which were shown in Fig. 5a and Fig. 6a, respectively. For KEGG enrichment analysis, there were 161 and 160 annotated signaling pathways for the DEG15%/6% set and the DEG15%/0% set respectively. Significantly enriched signaling pathways were flagellar assembly (map02040) and phenylalanine metabolism (map00360) in both DEG15%/6% and DEG15%/0% sets, and valine, leucine and isoleucine degradation (map00280), fatty acid degradation (map00071) and propanoate metabolism (map00640) in the DEG15%/0% set (Fig. 5b, Fig. 6b).
Role of transmembrane transport in osmoadaptation
Both GO and KEGG enrichment analysis of the DEG6%/0% set suggested that transmembrane transport may play a major role in moderate salinity-induced osmotic adaptation (Fig. 4). For high-salinity induced osmoadaptation, a relatively large number of genes related to transmembrane transport were also differentially expressed at the transcription level in 15% NaCl compared to 0% and 6% NaCl. There were 32, 66 and 56 DEGs in the DEG6%/0%, DEG15%/6% and DEG15%/0% sets annotated to ABC transporters (map02010) respectively, comprising a gene set of 92 (Table S4). These genes were mostly related to mineral and organic ion transport, phosphate and amino acid transport, as well as transport of monosaccharides, oligosaccharides and lipids.
As for mineral ion transport, iron transport seemed important in both moderate and high salinity-induced osmoadaptation. In moderate salinity (6% NaCl), the transcription levels of ABC transporter genes related to iron (III) transport including fbpB1, fbpB2, idiA, cysA2, potA, HI_0301 and NGR_a03680 were significantly up-regulated. In high salinity (15% NaCl), the transcriptions of most of these genes stayed up-regulated compared to 0% NaCl, but somehow stayed no changes or even down-regulated compared to those in 6% NaCl. Instead, iron import in high salinity was implemented by both single ion transport and iron complex import in the form of iron3+-hydroxamate indicated by significantly up-regulation of fhuB, fhuC and fhuD transcriptions. At the same time, transcriptions of cytochrome biogenesis and assembly-related transporter genes ccmA, ccmB, ccmC, cydC and cydD were significantly up-regulated, indicating possible role of cytochrome in high salinity-induced osmoadaptation in H. alkaliphila XH26. In addition, sulfate import-related ABC transporter genes were transcriptionally down-regulated under both moderate and high salt stress, tungstate transporter genes were down-regulated in high salinity-induced osmoadaptation while manganese transporter genes were down-regulated in moderate salinity condition. Molybdate transporter genes were transcriptionally up-regulated under high salt stress while stayed unchanged under moderate salt stress.
In high salinity condition, transcriptions of ABC transporter genes related to glycine betaine (GB) and putrescine uptake were significantly up-regulated. Trehalose, maltose, ribose and sn-Glycerol 3-phosphate transporter genes were up-regulated under salt stress, but stayed no change or even down-regulated in 15% NaCl compared to 6% NaCl. Amino acid transport seemed most complicated, with mixed changes of ups and downs for different amino acid transporter transcription levels, which needed to be further analyzed with changes of amino acid metabolism pathways. Together these results indicated that transmembrane transport and the transcription levels of ABC transporter genes might play an important role in salt-induced osmoadaptation in H. alkaliphila XH26, and divergent mechanisms were involved in moderate and high salinity-induced osmoadaptation.
In addition, we checked the existence and the transcriptional changes of the Tripartite ATP-independent periplasmic transporter (TRAP-T) and the betaine-choline-carnitine-transporter (BCCT) families for compatible solute transport in XH26 (Table S5). H. alkaliphila XH26 has the TRAP-T system for ectoine transport, and the teaABCD gene cluster corresponds to orf03787-orf03784 on the XH26 genome respectively. TeaABC is an osmoregulated ectoine transporter with TeaA as the substrate-binding protein (Tetsch and Kunte 2002; Grammann et al. 2002), and the universal stress protein TeaD negatively regulates the transport of ectoine and hydroxyectoine (Schweikhard et al. 2010). Under salt stress, the transcriptions of teaB and teaC were not significantly changed. The transcription of teaA was significantly up-regulated in 6% NaCl compared to 0% NaCl, and the transcription of teaD was significantly down-regulated in 15% NaCl compared to both 0% and 6% NaCl. As for the BCCT system, H. alkaliphila XH26 doesn’t have the sodium-coupled BCCT gene betP for betaine transport (Krämer and Morbach 2004). Instead, it has the BCCT genes ectT, betS, betT, VP1456 (bccT1), VP1723 (bccT2) and VPA0356. Under salt stress, the transcriptional levels of all these BCCT genes were not significantly changed except VP1456, which encodes VP1456 for GB, proline, choline, and ectoine uptake (Ongagna-Yhombi et al. 2015). In 15% NaCl, the transcription of VP1456 was significantly up-regulated compared to 0% NaCl.
Reduced flagellum assembly and cell motility in high salinity-induced osmoadaptation
For high-salinity induced osmoadaptation, there were 48 DEGs in DEG15%/6% group annotated to 16 significantly enriched GO groups related to cell motility (FDR < 0.005, Table S6), which were all transcriptionally down-regulated, showing reduced flagellum assembly and cell motility in high salinity condition. Similarly, 12 significantly enriched GO groups related to cell motility (FDR < 0.005) were down-regulated in the DEG15%/0% group, which contained 43 DEGs mostly included in the DEG15%/6% set except two (orf00232 and orf00240). Checking the transcription levels of all these 50 genes in DEG6%/0% group, we found that only two of them were significantly up-regulated (orf00123 and orf02416) while others were not significantly changed (Table S6). This was consistent with previous KEGG annotation result showing a significant larger number of cell motility genes represented in the 15% NaCl condition (Fig. 3c). In addition, KEGG enrichment analysis showed that flagellar assembly (map02040) was the top significantly enriched signaling pathway in 15% NaCl group compared with both 6% and 0% NaCl groups, which was down-regulated (Fig. 5b and Fig. 6b). Together these results indicated that in 15% NaCl which was less optimal for cell growth (Fig. 2a), reduced flagellum assembly and cell motility might be maintained compared to 6% and 0% NaCl culture conditions.
To verify the role of cell motility in high salinity-induced osmoadaptation, swarming motility assay was performed to test cells’ migration ability under various salt stress (Fig. 7). Results showed that cells moved at a slightly slower speed on agar in 6% NaCl compared to those in 0% NaCl, and migrated much slower in 12% NaCl compared to those in both 0% and 6% NaCl. The swarming motility test results confirmed the significant decrease of cell motility in high salt induction.
Ectoine synthesis for osmoadaptation
The synthesis and accumulation of compatible solute ectoine has been found to be important in many halophilic bacteria including Halomonas. The typical ectoine and hydroxyectoine biosynthesis and degradation pathways were shown in Fig. 8a. Those genes all exist in H. alkaliphila XH26 genome. As in H. elongata DSM 2581 (Schwibbert et al. 2011), the position of the ectD gene (3,346,765–3,347,748) is apart from the ectABC cluster (19,453,512–1,945,966), and the ectoine catabolic genes also form a doeABCD gene cluster which is located at 3,091,719–3,104,268 in the genome (Table S7). In 15% NaCl condition, the transcriptions of ectoine and 5-hydroxyectoine synthesis genes including lysC, ectB, ectC and ectD were all significantly up-regulated compared to those in 0% NaCl while the degradation genes including doeC and doeD were significantly down-regulated (Fig. 8b, Table S7). In 6% NaCl condition, the transcription levels of most genes were not significantly different from those in 0% NaCl, except for a significant increase in the transcription of the 5-hydroxyectoine synthesis gene ectD. The fold change of ectC was 2.836. However, due to large intra-group variations, the increase was considered not significant. From 6 to 15% NaCl, significant increase of ectB transcription and decrease of doeC and doeD transcriptions were observed. We further performed qPCR to measure the mRNA expression levels to confirm the transcription changes of these ectoine and hydroxyectoine synthesis and degradation genes found in RNA-seq. Results showed consistent trends for all of the ten genes tested with the RNA-seq data, and a significant increase of ectC and asd mRNA expressions in the 15%/6% comparison group (p < 0.05, |log2FC|≥ 1) as well as an increased lysC and doeC mRNA expressions in the 6%/0% comparison group were observed (Fig. 8b). Although decreases of doeC and doeD transcriptions were shown in the 15%/0% comparison group as in the RNA-seq data, the decreases were not significant. Together these data indicated that ectoine and 5-hydroxyectoine synthesis might be an important metabolic pathway for osmoadaptation in H. alkaliphila XH26. When exposed to medium salinity stress (6% NaCl), the transcriptions of ectoine and 5-hydroxyectoine synthesis genes lysC and ectD were increased. When exposed to high salinity stress (15% NaCl), besides lysC and ectD, the increased transcriptions of ectB and ectC played a key role. The transcription changes of asd and ectA seemed not significant in ectoine and 5-hydroxyectoine synthesis for osmoadaptation in H. alkaliphila XH26.
Effect of ectoine biosynthesis in osmoadaptation. a Ectoine biosynthesis and degradation pathways. b RNA-seq and qPCR results showing the expression changes of ectoine synthesis and degradation genes under salt stress. For qPCR, data are presented as the mean ± standard error (n > 3). *, Significant. c The effect of NaCl concentration on the growth of H. alkaliphila XH26 and the accumulation of ectoine and 5-hydroxyectoine
To validate the important role of ectoine synthesis in osmoadaptation, we measured the intracellular ectoine and 5-hydroxyectoine contents and respective OD600 values under different NaCl concentrations after 48 h of culture (Fig. 8c). High salinity such as 18% NaCl can significantly inhibit cell growth. The accumulation of ectoine was quite small at 0% NaCl (32.53 mg/g CDW), and increased with increasing salinity, reaching the maximum value of 219.35 mg/g at 9% NaCl; then started to decrease with the increase of NaCl concentration. At 15% NaCl, the ectoine content dropped to 76.74 mg/g, which was similar to that at 3% NaCl (65.93 mg/g), and further dropped to 20.97 mg/g at 18% NaCl. The 5-hydroxyectoine was not synthesized at 0% NaCl. From 3% NaCl, the synthesis of 5-hydroxyectoine started to slowly increase with increasing salinity, reaching 21.29 mg/g at 15% NaCl. At 18% NaCl, the 5-hydroxyectoine production was 10.22 mg/g, which was a little less than the ectoine production at the same salinity. Together with transcriptome analysis results, these findings confirmed that H. alkaliphila XH26 accumulated ectoine as one of the main compatible solutes for salt-induced osmoadaptation. In extreme high salinity condition (18% NaCl) when cell growth was significantly inhibited, 5-hydroxyectoine and ectoine contributed together to maintain cell survival.
Discussion
Osmoadaptation strategies in H. alkaliphila XH26
Halophiles usually adopt two strategies to adapt to salt stress, which are the “salt-in” strategy and the accumulation and synthesis of compatible solutes (Brown 1976; Kindzierski et al. 2017; Chen et al. 2017). Our study showed that salt-induced osmoadaptation could be quite complicated in H. alkaliphila XH26 and divergent mechanisms were involved in osmoadaptation to moderate and high salinity conditions. In moderate salinity condition (6% NaCl), transmembrane transport and ectoine synthesis played a major role for osmoadaptation. Increased transcriptions of iron (III), phosphate and phosphonate transporters and decreased transcriptions of sulfate and manganese transporters were observed. Transcriptions of monosaccharide and oligosaccharide import genes including trehalose, maltose, ribose and sn-Glycerol 3-phosphate were up-regulated, and ectoine synthesis and accumulation was significantly increased. In high salinity condition (15% NaCl), besides transmembrane transport and ectoine and 5-hydroxyectoine synthesis which were crucial for osmoadaptation, flagellum assembly and cell motility were significantly inhibited compared to both 0% NaCl and 6% NaCl. This was different to Chromohalobacter salexigens which showed up-regulated transcriptions of flagellum synthesis related genes and increased cell motility at 15% NaCl (Salvador et al. 2018). In Bacillus subtilis, similar findings to our H. alkaliphila XH26 were observed when cultured in 7.2% NaCl, which exhibited a repressed transcription of chemotaxis and motility genes and severe impairment of the swarming ability (Steil et al. 2003). However, the growth salinity range of this sigB mutant strain was not reported in the article.
Role of amino acid and glucose metabolism and citrate cycle in osmoadaptation and ectoine synthesis
Compatible solute ectoine is a cyclic amino acid derivative with hydrophilic and zwitterionic characteristics which can protect cells from extreme environments such as high osmotic pressure and high temperature (Bursy et al. 2008; Galinski and Trüper 1994; Garcia-Estepa et al. 2006; Malin and Lapidot 1996). Under salt-induced conditions, halophilic bacteria of different genera can biosynthesize ectoine, such as Chromohalobacter (Galinski and Trüper 1994; Salvador et al. 2018), Marinococcus (Galinski and Trüper 1994; Louis and Galinski 1997; Wei et al. 2011), Bacillus (Anburajan et al. 2019; Galinski and Trüper 1994; Kuhlmann and Bremer 2002), Stenotrophomonas (Sajjad et al. 2018), and Halomonas (Chen et al. 2018; Galinski and Trüper 1994; Kraegeloh and Kunte, 2002; Zhao et al. 2019). In our study, ectoine was synthesized and accumulated under salt stress from l-aspartate in H. alkaliphila XH26. At high salinity (15% NaCl), the transcriptional levels of lysC, ectB and ectC were significantly up-regulated compared to 0% NaCl which were verified by qPCR analysis while the transcriptions of asd and ectA stayed unchanged (Fig. 8b). This result was not exactly the same as that found in H. elongata DSM 2581 (Kindzierski et al. 2017). In H. elongata DSM 2581, the transcriptions of ectABC were all significantly elevated at 12% NaCl compared to 0.6% NaCl, and the corresponding enzymes EctA, EctB and EctC were up-regulated at 6% NaCl compared to cells from 0.6% NaCl whereas the enzymes LysC and Asd were not significantly changed under salt stress (6% and 12% NaCl). While not consistent with H. elongata DSM 2581, the significant increase of lysC transcription in H. alkaliphila XH26 was also found in Vibrio parahaemolyticus under salt stress (Yang et al. 2010), and an increased lysC expression could lead to a 9% increase of ectoine production in E. coli (Ning et al. 2016). Since the gene transcriptional levels do not represent the actual protein expression levels, further validation studies need to be performed to confirm our findings in H. alkaliphila XH26. Besides, the decrease of doeD in transcription at high salinity (15% NaCl) was consistent with H. elongata DSM 2581, which showed a significant decrease of doeD transcription and DoeD protein level under salt stress (Kindzierski et al. 2017).
The ectoine synthesis could be regulated by general amino acid metabolism and citrate cycle (TCA cycle) within the cell. The amino acid metabolism (E) was among the top 3 represented KEGG pathways, whose transcription was up-regulated under salt stress (Fig. 3c). A summary of KEGG enrichment analysis for top enriched amino acid metabolic pathways is shown in Table S8 (FDR < 1). Both valine, leucine and isoleucine degradation (map00280) and phenylalanine metabolism (map00360) were significantly down-regulated under high salt stress. Alanine, aspartate and glutamate metabolism (map00250) can be related to ectoine synthesis pathways through oxaloacetate within the citrate cycle (map00020), and L-glutamate can serve as the carbon source for ectoine synthesis (Fig. 9). The transcriptions of relevant genes along this signaling pathway mostly stayed unchanged under moderate salinity condition (6% NaCl), and significantly increased at high salinity (15% NaCl), including glutamate synthase genes gltB and gltD, glutaminase gene glsA, 4-aminobutyrate transaminase gene davT, succinate semialdehyde dehydrogenase genes sad and gabD, succinate dehydrogenase genes sdhA and sdhD, and aconitate hydratase gene acnA. L-glutamate also serves as the nitrogen source for ectoine synthesis by generating L-2,4-diaminobutanoate through the transamination reaction catalyzed by EctB (Peters et al. 1990; Ono et al. 1999; Richter et al. 2019). In addition, glucose metabolism can serve as the carbon source for ectoine synthesis as well. Both Entner-Doudoroff (ED) and Embden-Meyerhof-Parnas (EMP) pathways were significantly up-regulated in 15% NaCl, and the generation of oxaloacetate from pyruvate was up-regulated under salt stress (Fig. 9).
Possible role of betaine synthesis and uptake in osmoadaptation
Besides ectoine, glycine betaine has also been shown to be an important compatible solute in halophile osmoadaptation, which could be synthesized through either choline oxidation or glycine methylation (Gunde-Cimerman et al. 2018; Nyyssola et al. 2000). In our study, H. alkaliphila XH26 only possesses choline oxidation genes betA, betB and betI2, which is consistent with existing studies (Canovas et al. 2000; Gadda and McAllister-Wilkins 2003; Gu et al. 2008). The transcriptions of these three genes were significantly up-regulated in 15% NaCl condition compared to both 0% NaCl and 6% NaCl conditions, while no significant changes were found in 6% NaCl condition compared to 0% NaCl condition (Table S9). Together with the ABC transporter analysis results which showed significantly increased transcriptional expressions of glycine betaine uptake genes at 15% NaCl (Table S4), these data indicate that glycine betaine uptake and synthesis may also play a role in high salt stress-induced osmoadaptation in H. alkaliphila XH26. Interestingly, glycine betaine might be also involved in ectoine synthesis. Studies have shown that inhibition of betaine synthesis may increase ectoine production under low salt stress (Kushwaha et al. 2019), and whether it is the case in H. alkaliphila XH26 need to be further studied and verified.
Conclusions
In this work, we have investigated the osmoadaptation mechanism of H. alkaliphila XH26 under differential salt stress conditions. To adapt to low salt stress, H. alkaliphila XH26 increased ectoine synthesis and the transcriptions of the transmembrane import of iron (III), phosphate, phosphonate, monosaccharide and oligosaccharide. When exposed to high salinity stress, H. alkaliphila XH26 tried to survive by significantly decreased transcriptions of flagellum assembly and cell motility genes together with reduced cell motility, increased ectoine and 5-hydroxyectoine synthesis, and increased transcriptions of ABC transporters related to iron (III) and iron3+-hydroxamate import, glycine betaine and putrescine uptake, and cytochrome biogenesis and assembly. The synthesis and accumulation of ectoine and 5-hydroxyectoine under salt stress could be preferentially regulated by the increased transcriptions of lysC, ectB, ectC and ectD genes. The transcription of TRAP-T gene teaA was increased while the transcription of teaD was decreased under salt stress, indicating a potential increase in ectoine import. Besides, other compatible solutes such as glycine betaine may also play a role in the high salinity-induced osmoadaptation of H. alkaliphila XH26.
Data availability
The H. alkaliphila XH26 16S rRNA gene sequence has been submitted to GenBank database under accession number MK696282 (Bioproject number PRJNA527735). The complete genome sequence has been deposited at GenBank under accession number CP071889 (Bioproject number PRJNA705502, Biosample number SAMN18316568). The raw reads of RNA-seq data are available in the NCBI Sequence Read Archive (SRA) under accession numbers SRR13808843-SRR13808851 (Bioproject number PRJNA705502, Biosample numbers SAMN18090744-18090752).
References
Almeida E, Dias TV, Ferraz G, Carvalho MF, Lage OM (2019) Culturable bacteria from two Portuguese salterns: diversity and bioactive potential. Antonie Van Leeuwenhoek 113:459–475
Anburajan L, Meena B, Sreelatha T, Vinithkumar NV, Kirubagaran R, Dharani G (2019) Ectoine biosynthesis genes from the deep sea halophilic eubacteria, Bacillus clausii NIOT-DSB04: its molecular and biochemical characterization. Microb Pathog 136:103693
Bilstein A, Heinrich A, Rybachuk A, Mosges R (2021) Ectoine in the treatment of irritations and inflammations of the eye surface. Biomed Res Int 2021:8885032
Brands S, Schein P, Castro-Ochoa KF, Galinski EA (2019) Hydroxyl radical scavenging of the compatible solute ectoine generates two N-acetimides. Arch Biochem Biophys 674:108097
Brown AD (1976) Microbial water stress. Bacteriol Rev 40:803–846
Bujak T, Zagorska Dziok M, Niziol-Lukaszewska Z (2020) Complexes of ectoine with the anionic surfactants as active ingredients of cleansing cosmetics with reduced irritating potential. Molecules 25:1433
Bursy J, Kuhlmann AU, Pittelkow M, Hartmann H, Jebbar M, Pierik AJ, Bremer E (2008) Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl Environ Microbiol 74:7286–7296
Bursy J, Pierik AJ, Pica N, Bremer E (2007) Osmotically induced synthesis of the compatible solute hydroxyectoine is mediated by an evolutionarily conserved ectoine hydroxylase. J Biol Chem 282:31147–31155
Canovas D, Vargas C, Kneip S, Moron MA, Ventosa A, Bremer E, Nieto JNJ (2000) Genes for the synthesis of the osmoprotectant glycine betaine from choline in the moderately halophilic bacterium Halomonas elongata DSM 3043, USA. Microbiology (reading) 146:455–463
Ceylan S, Yilan G, Akbulut BS, Poli A, Kazan D (2012) Interplay of adaptive capabilities of Halomonas sp. AAD12 under salt stress. J Biosci Bioeng 114:45–52
Chen S, Xu Y, Helfant L (2020) Geographical isolation, buried depth, and physicochemical traits drive the variation of species diversity and prokaryotic community in three typical hypersaline environments. Microorganisms 8:120
Chen WC, Hsu CC, Lan JC, Chang YK, Wang LF, Wei YH (2018) Production and characterization of ectoine using a moderately halophilic strain Halomonas salina BCRC17875. J Biosci Bioeng 125:578–584
Chen YH, Lu CW, Shyu YT, Lin SS (2017) Revealing the saline adaptation strategies of the halophilic bacterium Halomonas beimenensis through high-throughput omics and transposon mutagenesis approaches. Sci Rep 7:13037
Cho BR, Jun HJ, Thach TT, Wu C, Lee SJ (2017) Betaine reduces cellular melanin content via suppression of microphthalmia-associated transcription factor in B16–F1 murine melanocytes. Food Sci Biotechnol 26:1391–1397
Edbeib MF, Wahab RA, Huyop F (2016) Halophiles: biology, adaptation, and their role in decontamination of hypersaline environments. World J Microbiol Biotechnol 32:135
Gadda G, McAllister-Wilkins EE (2003) Cloning, expression, and purification of choline dehydrogenase from the moderate halophile Halomonas elongata. Appl Environ Microbiol 69:2126–2132
Galinski EA, Trüper HG (1994) Microbial behaviour in salt-stressed ecosystems. FEMS Microbiol Rev 15:95–108
Garcia-Estepa R, Argandona M, Reina-Bueno M, Capote N, Iglesias-Guerra F, Nieto JJ, Vargas C (2006) The ectD gene, which is involved in the synthesis of the compatible solute hydroxyectoine, is essential for thermoprotection of the halophilic bacterium Chromohalobacter salexigens. J Bacteriol 188:3774–3784
Göller K, Ofer A, Galinski EA (1998) Construction and characterization of an NaCl-sensitive mutant of Halomonas elongata impaired in ectoine biosynthesis. FEMS Microbiol Lett 161:293–300
Grammann K, Volke A, Kunte HJ (2002) New type of osmoregulated solute transporter identified in halophilic members of the bacteria domain: TRAP transporter TeaABC mediates uptake of ectoine and hydroxyectoine in Halomonas elongata DSM 2581(T). J Bacteriol 184(11):3078–3085
Gu ZJ, Wang L, Le Rudulier D, Zhang B, Yang SS (2008) Characterization of the glycine betaine biosynthetic genes in the moderately halophilic bacterium Halobacillus dabanensis D-8(T). Curr Microbiol 57:306–311
Gunde-Cimerman N, Plemenitas A, Oren A (2018) Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol Rev 42:353–375
Hahn MB, Smales GJ, Seitz H, Solomun T, Sturm H (2020) Ectoine interaction with DNA: influence on ultraviolet radiation damage. Phys Chem Chem Phys 22:6984–6992
Hseu YC, Chen XZ, Vudhya Gowrisankar Y, Yen HR, Chuang JY, Yang HL (2020) The skin-whitening effects of ectoine via the suppression of α-MSH-stimulated melanogenesis and the activation of antioxidant Nrf2 pathways in UVA-irradiated keratinocytes. Antioxidants (basel) 9:63
Huang JL, Wang HH, Alam F, Cui YW (2019) Granulation of halophilic sludge inoculated with estuarine sediments for saline wastewater treatment. Sci Total Environ 682:532–540
Joghee NN, Jayaraman G (2016) Biochemical changes induced by salt stress in halotolerant bacterial isolates are media dependent as well as species specific. Prep Biochem Biotechnol 46:8–14
Louis P, Galinski EA (1997) Characterization of genes for the biosynthesis of the compatible solute ectoine from Marinococcus halophilus and osmoregulated expression in Escherichia coli. Microbiology (reading, England) 143:1141–1149
Kiefer J, Harati K, Muller Seubert W, Fischer S, Ziegler B, Behr B, Gille J, Kneser U, Lehnhardt M, Daigeler A, Dragu A (2018) Efficacy of a gel containing polihexanide and betaine in deep partial and full thickness burns requiring split-thickness skin grafts: a noncomparative clinical study. J Burn Care Res 39:685–693
Kindzierski V, Raschke S, Knabe N, Siedler F, Scheffer B, Pflüger-Grau K, Pfeiffer F, Oesterhelt D, Marin Sanguino A, Kunte HJ (2017) Osmoregulation in the halophilic bacterium Halomonas elongata: a case study for integrative systems biology. PLoS ONE 12:e0168818
Kraegeloh A, Kunte H (2002) Novel insights into the role of potassium for osmoregulation in Halomonas elongata. Extremophiles 6:453–462
Krämer R, Morbach S (2004) BetP of Corynebacterium glutamicum, a transporter with three different functions: betaine transport, osmosensing, and osmoregulation. Biochem Biophys Acta 1658(1–2):31–36
Kuhlmann AU, Bremer E (2002) Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl Environ Microbiol 68:772–783
Kushwaha B, Jadhav I, Verma HN, Geethadevi A, Parashar D, Jadhav K (2019) Betaine accumulation suppresses the de-novo synthesis of ectoine at a low osmotic concentration in Halomonas sp SBS 10, a bacterium with broad salinity tolerance. Mol Biol Rep 46:4779–4786
Malin G, Lapidot A (1996) Induction of synthesis of tetrahydropyrimidine derivatives in Streptomyces strains and their effect on Escherichia coli in response to osmotic and heat stress. J Bacteriol 178:385–395
Martinez-Perez RB, Rodriguez JA, Cira-Chavez LA, Dendooven L, Viniegra-Gonzalez G, Estrada-Alvarado I (2020) Exoenzyme-producing halophilic bacteria from the former lake texcoco: identification and production of n-butyl oleate and bioactive peptides. Folia Microbiol (praha) 65:835–847
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10(1):2182
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M (2022) TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res 50:D801–D807
Moffa A, Carbone S, Costantino A, Fiore V, Rinaldi V, Baptista P, Cassano M, Casale M (2019) Potential role of topical ectoine for prevention of pediatric upper respiratory tract infection: a preliminary observational study. J Biol Regul Homeost Agents 33:1935–1940
Nayak PK, Goode M, Chang DP, Rajagopal K (2020) Ectoine and hydroxyectoine stabilize antibodies in spray-dried formulations at elevated temperature and during a freeze/thaw process. Mol Pharm 17:3291–3297
Ning Y, Wu X, Zhang C, Xu Q, Chen N, Xie X (2016) Pathway construction and metabolic engineering for fermentative production of ectoine in Escherichia coli. Metab Eng 36:10–18
Nyyssola A, Kerovuo J, Kaukinen P, von Weymarn N, Reinikainen T (2000) Extreme halophiles synthesize betaine from glycine by methylation. J Biol Chem 275:22196–22201
Ongagna-Yhombi SY, McDonald ND, Boyd EF (2015) Deciphering the role of multiple betaine-carnitine-choline transporters in the Halophile Vibrio parahaemolyticus. Appl Environ Microbiol 81(1):351–363
Ono H, Sawada K, Khunajakr N, Tao T, Yamamoto M, Hiramoto M, Shinmyo A, Takano M, Murooka Y (1999) Characterization of biosynthetic enzymes for ectoine as a compatible solute in a moderately halophilic Eubacterium, Halomonas elongata. J Bacteriol 181(1):91–99
Oren A (2013) Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes. Front Microbiol 4:315
Parwata IP, Wahyuningrum D, Suhandono S, Hertadi R (2019) Heterologous ectoine production in Escherichia coli: optimization using response surface methodology. Int J Microbiol 2019:5475361
Peng S, Kai M, Yang X, Luo Y, Bai L (2020) Study on the osmoregulation of “Halomonas socia” NY-011 and the degradation of organic pollutants in the saline environment. Extremophiles 24:843–861
Peters P, Galinski EA, Trüper HG (1990) The biosynthesis of ectoine. FEMS Microbiol Lett 71(1):157–162
Prabhu J, Schauwecker F, Grammel N, Keller U, Bernhard M (2004) Functional expression of the ectoine hydroxylase gene (thpD) from Streptomyces chrysomallus in Halomonas elongata. Appl Environ Microbiol 70:3130–3132
Rathour R, Gupta J, Mishra A, Rajeev A, Dupont CL, Thakur IS (2020) A comparative metagenomic study reveals microbial diversity and their role in the biogeochemical cycling of Pangong lake. Sci Total Environ 731:139074
Rezaei Somee M, Shavandi M, Dastgheib SMM, Amoozegar MA (2018) Bioremediation of oil-based drill cuttings by a halophilic consortium isolated from oil-contaminated saline soil. Biotech 3:229
Richter AA, Mais CN, Czech L, Geyer K, Hoeppner A, Smits S, Erb TJ, Bange G, Bremer E (2019) Biosynthesis of the stress-protectant and chemical Chaperon Ectoine: biochemistry of the transaminase EctB. Front Microbiol 10:2811
Rieckmann T, Gatzemeier F, Christiansen S, Rothkamm K, Munscher A (2019) The inflammation-reducing compatible solute ectoine does not impair the cytotoxic effect of ionizing radiation on head and neck cancer cells. Sci Rep 9:6594
SadrAzodi SM, Shavandi M, Amoozegar MA, Mehrnia MR (2019) Biodegradation of long chain alkanes in halophilic conditions by Alcanivorax sp. strain Est-02 isolated from saline soil. Biotechnology 3:141
Sagar A, Sayyed RZ, Ramteke PW, Sharma S, Marraiki N, Elgorban AM, Syed A (2020) ACC deaminase and antioxidant enzymes producing halophilic Enterobacter sp. PR14 promotes the growth of rice and millets under salinity stress. Physiol Mol Biol Plants 26:1847–1854
Saghafi D, Delangiz N, Lajayer BA, Ghorbanpour M (2019) An overview on improvement of crop productivity in saline soils by halotolerant and halophilic PGPRs. Biotech 3:261
Sajjad W, Qadir S, Ahmad M, Rafiq M, Hasan F, Tehan R, McPhail KL, Shah AA (2018) Ectoine: a compatible solute in radio-halophilic Stenotrophomonas sp. WMA-LM19 strain to prevent ultraviolet-induced protein damage. J Appl Microbiol 125:457–467
Salvador M, Argandona M, Naranjo E, Piubeli F, Nieto JJ, Csonka LN, Vargas C (2018) Quantitative RNA-seq analysis unveils osmotic and thermal adaptation mechanisms relevant for ectoine production in Chromohalobacter salexigens. Front Microbiol 9:1845
Schweikhard ES, Kuhlmann SI, Kunte HJ, Grammann K, Ziegler CM (2010) Structure and function of the universal stress protein TeaD and its role in regulating the ectoine transporter TeaABC of Halomonas elongata DSM 2581(T). Biochemistry 49(10):2194–2204
Schwibbert K, Marin-Sanguino A, Bagyan I, Heidrich G, Lentzen G, Seitz H, Rampp M, Schuster SC, Klenk HP, Pfeiffer F, Oesterhelt D, Kunte HJ (2011) A blueprint of ectoine metabolism from the genome of the industrial producer Halomonas elongata DSM 2581T. Env Microbiol 13:1973–1994
Soltan Dallal MM, Zamaniahari S, Davoodabadi A, Hosseini M, Rajabi Z (2017) Identification and characterization of probiotic lactic acid bacteria isolated from traditional Persian pickled vegetables. GMS Hyg Infect Control 12:15
Steil L, Hoffmann T, Budde I, Völker U, Bremer E (2003) Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J Bacteriol 185:6358–6370
Tetsch L, Kunte HJ (2002) The substrate-binding protein TeaA of the osmoregulated ectoine transporter TeaABC from Halomonas elongata: purification and characterization of recombinant TeaA. FEMS Microbiol Lett 211(2):213–218
Torbaghan ME, Khalili Torghabeh GH (2019) Biological removal of iron and sulfate from synthetic wastewater of cotton delinting factory by using halophilic sulfate-reducing bacteria. Heliyon 5:e02948
Tsai T, Mueller-Buehl AM, Satgunarajah Y, Kuehn S, Dick HB, Joachim SC (2020) Protective effect of the extremolytes ectoine and hydroxyectoine in a porcine organ culture. Graefes Arch Clin Exp Ophthalmol 258:2185–2203
Vega C, Rodriguez M, Llamas I, Bejar V, Sampedro I (2019) Silencing of phytopathogen communication by the halotolerant PGPR Staphylococcus equorum strain EN21. Microorganisms 8:42
Wang K, Zhang S, Li S, Sang X, Bai L (2010) Osmotolerance features and mechanisms of a moderately halophilic bacterium Halomonas sp. NY-011*. Chin J Appl Environ Biol 16:256–260
Wei YH, Yuan FW, Chen WC, Chen SY (2011) Production and characterization of ectoine by Marinococcus sp. ECT1 isolated from a high-salinity environment. J Biosci Bioeng 111:336–342
Yancey PH (2005) Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol 208:2819–2830
Yang L, Zhan L, Han H, Gao H, Guo Z, Qin C, Yang R, Liu X, Zhou D (2010) The low-salt stimulon in Vibrio parahaemolyticus. Int J Food Microbiol 137(1):49–54
Yue HT, Ling C, Yang T, Chen XB, Chen YL, Deng HT, Wu Q, Chen JC, Chen GQ (2014) A seawater-based open and continuous process for polyhydroxyalkanoates production by recombinant Halomonas campaniensis LS21 grown in mixed substrates. Biotechnol Biofuels 7:1–12
Yue W, Yan Z, Qiufen L (2019) Function verification and protein structure prediction of gene cluster narGYJV encoding dissimilatory nitrate reductase in Halomonas alkaliphila X3. Period Ocean Univ China 49:033–040
Zhao Q, Li S, Lv P, Sun S, Ma C, Xu P, Su H, Yang C (2019) High ectoine production by an engineered Halomonas hydrothermalis Y2 in a reduced salinity medium. Microb Cell Fact 18:184
Zhu D, Han R, Long Q, Gao X, Xing J, Shen G, Li Y, Wang R (2020) An evaluation of the core bacterial communities associated with hypersaline environments in the Qaidam Basin, China. Arch Microbiol 202:2093–2103
Acknowledgements
This work was supported by the National Natural Science Foundation Project of China (31860030, 21967018), Natural Science Foundation Project of Qinghai, China (2018-ZJ-930Q), Key Research Foundation of Development and Transformation of Qinghai Province (2019-SF-121) and Youth Science and Technology Project Fund of Medical College of Qinghai University (2017-KYZ-02).
Author information
Authors and Affiliations
Contributions
DZ and JX conceived and designed the experiments; XZ, TZ, NY and YL performed the experiments; TZ, XZ, LQ, YL, ZM, JX and DZ analyzed and interpreted the data; JX, XZ and TZ wrote the manuscript. JX and DZ ensured the accuracy or integrity of the work. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by L. Huang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, T., Zhang, X., Li, Y. et al. Study of osmoadaptation mechanisms of halophilic Halomonas alkaliphila XH26 under salt stress by transcriptome and ectoine analysis. Extremophiles 26, 14 (2022). https://doi.org/10.1007/s00792-022-01256-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00792-022-01256-1