Abstract
Psychrophilic fungi are a critical biotic component in cold deserts that serves a central role in nutrient recycling and biogeochemical cycles. Despite their ecological significance, culture-independent studies on psychrophilic mycobiome are limited. In the present study, the fungal diversity patterns across the Drass, an Indian cold desert in the Himalaya, were indexed by targeted amplicon pyrosequencing (ITS). In the Drass dataset, Ascomycota was represented by 92 genera, while 22 genera represented Basidiomycota. The most abundant genus was Conocybe (20.46%). Most of the identified genera were reported in the literature to be prolific extracellular hydrolytic enzyme producers. To identify whether the Drass fungal assemblages share similarities to other cold deserts, these were further compared to Antarctic and Arctic cold deserts. Comparative analysis across the three cold deserts indicated the dominance of Dikarya (Ascomycota and Basidiomycota). The observed alpha diversity, Shannon index as well as Pielou's evenness was highest in the Antarctic followed by Drass and Arctic datasets. The genera Malassezia, Preussia, Pseudogymnoascus, Cadophora, Geopora, Monodictys, Tetracladium, Titaea, Mortierella, and Cladosporium were common to all the cold deserts. Furthermore, Conocybe was represented predominantly in Drass. Interestingly, the genus Conocybe has not been previously reported from any other studies on Antarctic or Arctic biomes. To the best of our knowledge, this is the first fungal metagenome study in Drass soil. Our analysis shows that despite the similarities of low temperature among the cold deserts, a significant differential abundance of fungal communities prevails in the global cold deserts.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The mycobiome represents the fungal community within a biome (Schnecker et al. 2014) and these biotic communities participate in crucial ecological processes operating in the ecosystems. The literature holds ample reports on the fungal community structure of contrasting ecosystems (Zimmerman and Vitousek 2012; Moll et al. 2016; Durán et al. 2019). However, psychrophilic and psychrotolerant microbes, inhabiting ice-covered regions of the cold desert have gained much attention in recent decades as they play a significant role in decomposing organic matters, nutrient recycling, and biogeochemical cycles in intensely cold environments (Margesin and Miteva 2011; Gesheva and Vasileva-Tonkova 2012). Besides, they are a significant producer of cold-tolerant enzymes and secondary metabolites with industrial and pharmaceutical applications such as cold-adapted lipase in detergent Lipoclean® (Duncan et al. 2008; Krishnan et al. 2011; Wang et al. 2013; Sarmiento et al. 2015; Duarte et al. 2018). Moreover, the temperature has an essential role in microbial ecology (Pietikäinen et al. 2005). Interestingly, about 85% of the earth experiences below 5 °C permanently or seasonally (Hassan et al. 2016). Psychrotolerant microbes can withstand temperatures close to the freezing point as well as mild temperatures (Ahmad et al. 2010). The studies on cold-adapted fungi in Antarctic, Arctic, Finland, and Siberian tundra have reported the dominance by Ascomycetes and Basidiomycetes (Frank-Fahle et al. 2014; Heino et al. 2014; Schnecker et al. 2014; Wang et al. 2017).
Drass, a cold desert in the Himalaya, world's highest mountain range, is located at an estimated elevation of 3280 m above sea level (34.45 °N latitude, 75.77 °E longitude). It is the coldest human inhabited place in India and the second coldest in the world after Siberia. The great Himalayan range acts as an obstruction, blocking most of the monsoons in Ladakh, converting it into cold arid desert. There are reports pertaining to the bacterial diversity in the Himalayan habitats (Gangwar et al. 2009; Shivaji et al. 2011, 2013; Srinivas et al. 2011; Gupta et al. 2015; Gupta and Vakhlu 2015; Yadav et al. 2015). However, to the best of our knowledge, psychrophilic and psychrotolerant fungal diversity in Drass cold desert remains unexplored. Hence, in this study, we have implemented a pyrosequencing approach to explore and map the fungal community in Drass soil. Furthermore, microbial communities including mycobiome are known to be influenced by geography, elevation, receding glacier (Siles and Margesin 2016; Dresch et al. 2019; Řezáčová et al. 2019). Hence, to identify the similarities and dissimilarities between Drass mycobiome and other cold deserts, we compared the Drass mycobiome to the Antarctic and Arctic cold deserts. In this study, we addressed three main questions (i) What are the dominant fungal OTUs that inhabit in the soil of Drass (ii) To identify the differentially abundant fungal taxa with different ecotypes (Drass, Antarctic and Arctic) (iii) What are the similarities between fungal communities in different cold deserts across the globe.
Materials and methods
Sampling sites and ITS metagenomic pyrosequencing
Soil samples from different locations (n = 10) in Drass (J&K, India) (34.45 N, 75.77 E) were collected. The soil temperature at the time of sampling was recorded to be between 10 °C and 15 °C. Soil samples (~ 1 cm below the surface) were collected in sterile containers with the help of a sterile spatula and transported to the laboratory at 4 °C. Subsequently, a composite soil sample was obtained by pooling the soil samples and further sieved through 2 mm sieve which was eventually stored at – 20 °C for future analysis. Metagenomic DNA was extracted based on sodium dodecyl sulfate (SDS) and cetyl trimethyl ammonium bromide (CTAB) lysis followed by phenol:chloroform:isoamyl alcohol (25:24:1) purification and ethanol precipitation as described in Gupta et al. (2015). NanoDrop 1000 Spectrophotometer (Thermo Scientific, US) confirmed the purity and concentration of DNA. Extracted metagenomic DNA was pooled, diluted to a final concentration of 50 ng/μl, and used as the template for PCR amplification. The inter transcribed spacer (ITS) region was amplified using ITS1F (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4R (5′- TCCTCCGCTTATTGATATGC-3′) (White et al. 1990). The amplification was carried out with initial denaturation at 95 °C for 5 min followed by 35 cycles of denaturation at 95 °C for 45 s, annealing at 55 °C for the 30 s, extension at 72 °C for 60 s, and a final extension at 72 °C for 8 min in Eppendorf master cycler Gradient. The length of the PCR amplicons was verified using the Low Mass DNA Ladder (Invitrogen™, US) by agarose gel electrophoresis (1% w/v). The amplicons were purified by gel elution (Qiagen, India) and outsourced for pyrosequencing.
Public metagenomic datasets of cold desert
Besides the Drass metagenome dataset, the Antarctic and Arctic cold desert metagenome datasets available in NCBI and MG-RAST were included in the present study. Two metagenomic datasets of the Antarctic cold desert were included. The first Antarctic dataset originated from the McMurdo Dry Valleys (Dreesens et al. 2014). As the metagenomic data was not available in NCBI-SRA, the sequences were acquired through electronic mail on request. The second Antarctic dataset originated from the Browning Peninsula situated in the Windmill Islands, Eastern Antarctica (Pudasaini et al. 2017). The data was obtained from the Australian Antarctic Data Centre, Australia (Siciliano et al. 2014). Two metagenomic datasets of Arctic cold deserts were also included in the present study. The first dataset originated from the Midtre Lovénbreen Glacier, Svalbard (Dong et al. 2016). The data was obtained from the MG-RAST server (Project ID mgp15403). The second dataset originated from the Ny-Ålesund Region, Svalbard (Zhang et al. 2016). The data was obtained from NCBI (Accession No. SRX1481175). The datasets had different sequencing depths that could lead to biased results on comparative analysis. Hence, the datasets were normalized by rarefying as recommended elsewhere (Weiss et al. 2017).
Pyrosequencing and analysis
Tag-encoded FLX amplicon pyrosequencing (TEFAP) was done at the research and testing laboratory (Lubbock, TX, USA). Raw metagenomic ITS sequences from this study were deposited in NCBI SRA under the project title “Cold desert Metagenome” with accession and experiment number PRJNA260660S and RX700597 respectively. The raw data from this study, as well as the public datasets, were annotated and analyzed with the same pipeline as described in Gupta et al. (2015). In brief, sequences below 200 bp or with homopolymers of more than 8 bp were discarded during the initial quality filtering steps. Sequences that passed the initial quality check were subsequently subjected to denoising by flow gram clustering to omit sequencing errors (Gontcharova 2010). Chimeric sequences were removed using the Uchime tool (Edgar et al. 2011). The cleaned sequences were annotated in Mothur (Schloss et al. 2009) against the UNITE fungal database (Nilsson et al. 2019) with the Wang et al. 2007 classification method of 8 kmer length and 80% bootstrap confidence threshold. Diversity indices (Chao1 richness estimator, Shannon's H-indices, and Pielou's evenness index) and rarefaction curves were estimated with microbiome v1.8.0, ranacapa v0.1.0, and, phyloseq v1.30.0 libraries in R (McMurdie and Holmes 2013; Lahti and Shetty 2017; R Team Core 2017; Kandlikar et al. 2018).
Statistical analysis
The difference in microbial abundance was determined using PAST (Paleontological Statistics) (Hammer et al. 2001) with the Kruskal–Wallis test. The statistical analysis was considered significant for p values below 0.05.
Results
Fungal diversity of Drass metagenome
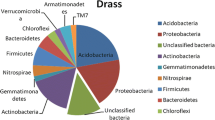
Eight phyla, including unclassified fungi (0.43%), represented the Drass soil metagenome. Ascomycota (67.49%) and Basidiomycota (29.07%) represented over 95% of the fungal abundance (Fig. 1a). Other representative phyla were Mortierellomycota (1.8%), Basidiobolomycota (0.37%), Zoopagomycota (0.18%), and Chytridiomycota (0.062%). Ascomycota was represented by 92 genera (Fig. 1b) and Basidiomycota was represented by 22 genera (Fig. 1c). Genus Mortierella represented Mortierellomycota, while Basidiobolomycota, Chytridiomycota, Glomeromycota, and Zoopagomycota could not be classified at the genus level (Fig. 1d). The most abundant genus Conocybe (20.46%) was represented under phylum Basidiomycota (Fig. 2). Other dominant genera were Rodentomyces (4.42%), Trichocladium (4.24%), Gibberella (3.43%), and Ilyonectria (2.37%) including unclassified genera under higher taxonomic classification such as Ascomycota (3.55%), Glomerellales (3.55%) Helotiales (3.49%), and Nectriaceae (4.36%). Overall, the Drass metagenome was represented by 118 fungal genera (Supplementary data S1). However, a large portion (n = 39) of the genera, amounting to 28.88% of the overall abundance, was unclassified, some of which could be classified only at the phylum level.
Global cold desert fungal alpha diversities
The Chao1 alpha diversity (R) among the global cold deserts showed a strong difference. For instance, the lowest (R = 32) and the highest (R = 162) Chao1 diversity was observed in Antarctic_1 (McMurdo Dry Valleys) and Antarctic_2 (Windmill Islands) respectively. Chao1 diversity of the Arctic datasets followed a similar trend with over two-fold difference between the two Arctic metagenome, i.e., J = 35 and 77 for Arctic_1 (Midtre Lovénbreen Glacier) and Arctic_2 (Ny-Ålesund Region) respectively (Fig. 2a). Similar variations in Shannon index (H) and Pielou’s evenness (J') were observed with Antarctic datasets from McMurdo Dry Valleys and Windmill Islands (Fig. 2b, c). Interestingly, the Drass metagenome was among the highest Chao1, Shannon index, and Pielou’s evenness. However, it should be noted that a major proportion of the OTUs in the datasets could not be classified even at the phylum level. The relative abundance of unclassified OTUs for Antarctic_1, Antarctic_2, Arctic_1, Arctic_2, and Drass were 33.25%, 2.06%, 58.33%, 12.54%, and 0.44% respectively. Furthermore, rarefaction curves were not saturated, suggesting a requirement for extensive sequencing depth (Fig. 3). Hence, the dominance of unclassified OTUs, especially in Antarctic_1 and Arctic_1, and the lack of strong plateau in the rarefaction curves could be a major factor for their low alpha diversities.
Similarities and differences in Drass, Antarctic and Arctic cold desert
The global cold desert was represented by 10 phyla, out of which 5 phyla, i.e., Ascomycota, Basidiomycota, Chytridiomycota, Unclassified, and Mortierellomycota were common to all cold deserts (Fig. 4). In all datasets, Ascomycota was the dominant phylum followed by Basidiomycota or Chytridiomycota. Monoblepharomycota and Rozellomycota were represented in the Antarctic and the Arctic but absent in Drass. Interestingly, phylum Basidiobolomycota (0.37%) was detected only in Drass. The differences among the cold deserts were prominent at the genus level. The predominant genera in Drass, Conocybe (20.46%) and Rodentomyces (4.42%) under phylum Basidiomycota and Ascomycota, were not detected in Antarctic or Arctic datasets (Fig. 5). Similarly, Amandinea in Antarctic_2 (7.17%) and Exophiala (18.77%) in Antarctic_1 were exclusive to Antarctic datasets. Likewise, among the classified genera, genus Thelidium was exclusive to Arctic_1 (2.12%) and Arctic_2 (0.06%) (Supplementary data S2). The similarities among the cold deserts were represented by 21 common genera, out of which 10 genera were unclassified (Fig. 6).
Discussion
Composition of fungal assemblages in Drass soil
Most of the reported fungi in literature are mesophiles that thrive at an optimum temperature of 25 °C–37°C (Magan 2007). However, cold deserts are inhabited by cold-tolerant fungi (psychrotolerant). Understanding the ecology of such fungi is essential for a better understanding of cold desert fungal ecology. In this study, we have analyzed the fungal community of Drass cold desert using next-generation sequencing (NGS) and further compared to publicly available datasets of Antarctic and Arctic cold deserts (AACD). Ascomycota (67.49%) and Basidiomycota (29.07%) dominated more than 95% of the Drass metagenome (Fig. 1). The diversity of phylum Ascomycota, represented by 92 genera, was four-fold higher than that of Basidiomycota, which was represented by 22 genera. Ascomycota is the largest fungal phylum predominant in several aquatic and terrestrial ecosystems (Kendrick 2003; Schoch et al. 2009). This could explain the high abundance and diversity of Ascomycota in Drass. Ascomycota and Basidiomycota were also identified as dominated phyla in previous studies on Antarctic soils (Connell et al. 2008; Arenz and Blanchette 2011). In temperate environment, Basidiomycota also dominates wood decay (Blanchette et al. 2004; Ludley and Robinson 2008). At the genus level, Conocybe (20.46%) predominated the Drass dataset. However, 28.88% overall abundance was unclassified, some of which could be classified only at the phylum level. Nonetheless, such a high proportion of unclassified fungal OTUs have also been previously reported (Hallen-Adams et al. 2015; Nash et al. 2017). Some of the fungal genera such as Candida (0.81%), Dioszegia (0.93%), Penicillium (0.37%), Thelebolus (0.31%), Trichosporon (0.43%), Cadophora (0.06%), and, Cladosporium (1.37%) are reported to be prolific producers of extracellular hydrolytic enzymes (Bradner et al. 1999; Krishnan et al. 2011; Carrasco et al. 2012; Duarte et al. 2013, 2018). Such fungi are of ecological significance because extracellular hydrolytic enzymes degrade soil organic matter that is readily absorbed by the producers as well as surrounding cells (Redmile-Gordon et al. 2015). These microbes also have crucial roles in the food chain and nutrient recycling in a cold environment (Wang et al. 2017). Cadophora has been reported in Antarctic soils, indicating its resilience to the cold environment (Malosso et al. 2006; Bridge and Newsham 2009). Besides, species within the genus Tetracladium (0.81%), mainly involved in plant debris degradation, have been documented from alpine glaciers, snow-covered soil (Kuhnert et al. 2012) and Qinghai Tibet Plateau (Wang et al. 2015). Moreover, genus Epicoccum (1.49%) under phylum Dothideomycetes has been reported to produce a wide array of secondary metabolites with antimicrobial, anticancer, and antioxidant activity (Braga et al. 2018). The vast diversity of the Drass mycobiome shows fungal diversity richness of ecological significance and enzymes of potential application in industrial and pharmaceutical processes.
The dominance of global cold desert mycobiome by Ascomycota and Basidiomycota
The relative abundance and distribution of dominant fungal phyla and genera prevailing across the cold deserts in Drass, Antarctic (McMurdo Dry Valleys, Browning Peninsula), and Arctic (Midtre Lovénbreen Glacier, Ny-Ålesund Svalbard) were compared in order to elucidate their similarities and differences (Dreesens et al. 2014; Zhang et al. 2016; Dong et al. 2016; Pudasaini et al. 2017). Alpha diversity and Shannon index of Antarctic_2 was highest, followed by Drass, indicating their richness and heterogeneity of the fungal community (Fig. 2). Alpha diversity is the measure of the observed OTUs in the dataset, while the Shannon index measures the observed OTUs as well as evenness (Alonso et al. 2019). Similarly, the rarefaction curve of Antarctic_2 and Drass exhibited the highest OTU count despite the lack of a strong plateau formation (Fig. 3). At the phylum level, Dikarya (Ascomycota and Basidiomycota), Mortierellomycota, and Chytridiomycota were detected in all cold desert datasets with vast differences in abundance, indicating selective spatial enrichment (Fig. 4). Ascomycota is known to dominate the fungal community around the globe (Egidi et al. 2019). The dominance of Ascomycota could be attributed to its wide array of stress tolerance and resource acquisition genes that could assist its dominance in the soil (Egidi et al. 2019). It should also be noted that fungal communities vary greatly between ecosystems. For instance, Ascomycota is a dominant phylum in arable soil (Moll et al. 2016). However, forest soil is generally dominated by Basidiomycota (Allison SD and Treseder 2008; Wubet et al. 2012). The dominance of the Ascomycota in the cold desert could be due to the lack of woody materials, which is preferred by Basidiomycota (Moll et al. 2016). Chytridiomycota (chytrids) was also abundant in Antarctic_1 (21.27%) and Artic_2 (8.42%) datasets, but comparatively rarer in Drass (0.06%). Chytrids have been detected in high altitude soil where melting snowpack supports the growth of cyanobacteria and algal populations that, in turn, serves as food-substrate for their growth (Schmidt et al. 2012). Mortierella sp. of phylum Mortierellomycota is reported to release nutrients and decompose pine needles particularly in winters or cold environments (Tokumasu 1998). Weinstein et al. (2000) reported Mortierella species as psychrophiles with intracellular trehalose concentrations and stearidonic acid that confers endurance to cold environments. Mortierella sp. are known to produce Long-chain omega-3 fatty acids, EPA (eicosapentaenoic acid) and DHA (docosahexaenoic acid) in their mycelial biomass under low-temperature stress condition (Vadivelan and Venkateswaran 2014). Thus, the phyla Dikarya, Mortierellomycota, and Chytridiomycota represented as a core phyla among all the cold deserts.
Differences in mycobiome of Drass, Antarctic and Arctic cold desert
Out of the 10 identified Phyla, only Basidiobolomycota was unique to the Drass dataset, while the remaining 9 phyla were common to all cold deserts. As far as we know, there is no information documented regarding the existence and role played by Basidiobolomycota phylum in cold habitats. Phylum Glomeromycota was represented in Drass and Antarctic only. However, in contrast to our findings, Glomeromycota was identified in previous fungal ecological studies from the North American Arctic Transect (Freeman et al. 2009). This disparity could be due to the low sequencing depth indicated by the rarefaction curve. Most of the members of phylum Glomeromycota form arbuscular mycorrhizas (AMs) with the roots of land plants or thalli of bryophytes. Such symbiotic association offers greater opportunity to obtain organic nutrients supplied by their host while living in oligotrophic soils of cold deserts in comparison to other soil-borne fungi (Nichols and Wright 2004; Rillig 2004). Phylum Monoblepharomycota, detected in Arctic and Antarctic datasets, was introduced in 2001 (Doweld 2001; Wijayawardene et al. 2018), but not much information is documented regarding the existence or the role played by this phylum in cold polar habitat. Phylum Rozellomycota, detected in the Antarctic and Arctic datasets, was also reported from Ny-Ålesund harbor seawater, moraine lake water (Picard 2017; Tedersoo et al. 2017). The difference in the mycobiome was more substantial at the genus level (Fig. 5). Although Basidiomycota was represented in all three sample locations, its relative abundance varied significantly.
The similarities among the cold desert datasets were minimal at the genus level (Fig. 6). Genus Naganishia under phylum Basidiomycota was predominant in the Antarctic_1 dataset (21.08%), which was also detected significantly lower in Drass (0.31%). The genus Naganishia has been reported in ample abundance in highly elevated soils of Antarctic (Schmidt et al. 2017). They are resistant to freeze–thaw cycles and UV radiation that could aid in its survival in such harsh climatic conditions (Pulschen et al. 2015). However, it has also been hypothesized that the abundance of genus Naganishia might not signify its functions but rather it can be present as dormant cells (Schmidt et al. 2017). In addition, Exophiala (Ant_1 = 18.77%, Ant_2 = 0.37%) was represented exclusively in the Antarctic dataset. Exophiala spp. are poly extremotolerant black yeast found in several types of environments ranging from Apennines glacier (Branda et al. 2010; Gostinčar et al. 2011), Arctic and Antarctic environments (Vaz et al. 2011) to hot saunas (Blasi et al. 2015). They are also known human pathogens that cause infection even in healthy individuals (Song et al. 2017). The mushroom genus, Conocybe (20.46%), was represented as the most abundant genus in the soil metagenome of Drass followed by Rodentomyces (4.4%). Genus Rodentomyces has been reported as saprophytic coprophilous fungi. There is no information documented regarding the existence and role played by this genus in cold habitats (Doveri et al. 2010). Interestingly, the genus, Conocybe, was represented exclusive and predominant in Drass. It was not detected from Antarctic or Arctic metagenome datasets. Furthermore, to the best of our knowledge, there are no reports supporting the presence of Conocybe in the Antarctic as well as Arctic cold desert soil. Interestingly, members of this genus contain psilocybin that causes intense hallucinations. Literature reports psilocybin as a drug with anticancer potential (Kothari et al. 2018) and a myriad of biological activities against obsessive–compulsive disorder, depression, anxiety, and schizophrenia (Andersson et al. 2009).
Although next-generation sequencing is a powerful tool, it is worth mentioning that the detection of unclassified fungal OTUs in high proportion have been noted in the literature (Hallen-Adams et al. 2015; Nash et al. 2017). The presence of such unclassified OTUs could indicate the presence of uncharacterized species. However, it could also be attributed to the rarity of fungal databases. The development of a well-curated fungal database has received less attention in comparison to bacterial databases (Nash et al. 2017).
Conclusion
The fungal communities in Drass soil were analyzed using high throughput sequencing of the ITS gene. Ascomycota and Basidiomycota predominated the mycobiome of Drass at the phylum level while Conocybe dominated at the genus level. Comparative analysis of the Drass mycobiome against Antarctic and Arctic datasets also revealed the dominance of phyla Ascomycota and Basidiomycota in all three cold deserts, although the relative abundance varied. This study shows that cold deserts at the global scale are prone to high spatial selective enrichment with considerable differences in the relative abundance of ecotypes. Besides, genus Conocybe was represented exclusively and dominantly in Drass soil among the three different geographical regions, i.e., Antarctic, Arctic, and Drass. The genus is of pharmaceutical importance, which could be exploited for novel drug discoveries.
Data availability
Raw metagenomic ITS sequences from the present study were submitted in NCBI SRA as a "Cold desert Metagenome" project with accession PRJNA260660 and experiment number SRX700597.
References
Ahmad B, Javed I, Shah AA, Hameed A, Hasan F (2010) Psychrotrophic bacteria isolated from – 20 °C freezer. African J Biotechnol 9:718–724
Allison SD, Treseder KK (2008) Warming and drying suppress microbial activity and carbon cycling in boreal forest soils. Glob Chang Biol 14:2898–2909. https://doi.org/10.1111/j.1365-2486.2008.01716.x
Alonso A, de Celis M, Ruiz J, Vicente J, Navascués E, Acedo A, Ortiz-Álvarez R, Belda I, Santos A, Gómez-Flechoso MÁ, Marquina D (2019) Looking at the Origin: Some Insights into the General and Fermentative Microbiota of Vineyard Soils. Fermentation 5:78. https://doi.org/10.3390/fermentation5030078
Andersson C, Kristinsson J, Gry J (2009) Occurrence and use of hallucinogenic mushrooms containing psilocybin alkaloids. Nordic Council of Ministers, Copenhagen
Arenz BE, Blanchette RA (2011) Distribution and abundance of soil fungi in Antarctica at sites on the Peninsula, Ross Sea Region and McMurdo Dry Valleys. Soil Biol Biochem 43:308–315. https://doi.org/10.1016/j.soilbio.2010.10.016
Blanchette RA, Held BW, Jurgens JA, McNew DL, Harrington TC, Duncan SM, Farrell RL (2004) Wood-destroying soft rot fungi in the historic expedition huts of Antarctica. Appl Environ Microbiol 70:1328–1335. https://doi.org/10.1128/AEM.70.3.1328-1335.2004
Blasi B, Tafer H, Tesei D, Sterflinger K (2015) From Glacier to Sauna: RNA-Seq of the Human Pathogen Black Fungus Exophiala dermatitidis under Varying Temperature Conditions Exhibits Common and Novel Fungal Response. PLoS ONE 10:e0127103. https://doi.org/10.1371/journal.pone.0127103
Bradner JR, Gillings M, Nevalainen KMH (1999) Qualitative assessment of hydrolytic activities in antarctic microfungi grown at different temperatures on solid media. J Microbiol 15:131–132. https://doi.org/10.1023/A1008855406319
Braga RM, Padilla G, Araújo WL (2018) The biotechnological potential of Epicoccum spp.: diversity of secondary metabolites. Crit Rev Microbiol 44:759–778. https://doi.org/10.1080/1040841X.2018.1514364
Branda E, Turchetti B, Diolaiuti G, Pecci M, Smiraglia C, Buzzini P (2010) Yeast and yeast-like diversity in the southernmost glacier of Europe (Calderone Glacier, Apennines, Italy). FEMS Microbiol Ecol 72:354–369. https://doi.org/10.1111/j.1574-6941.2010.00864.x
Bridge PD, Newsham KK (2009) Soil fungal community composition at Mars Oasis, a southern maritime Antarctic site, assessed by PCR amplification and cloning. Fungal Ecol 2:66–74. https://doi.org/10.1016/j.funeco.2008.10.008
Carrasco M, Rozas J, Barahona S, Alcaíno J, Cifuentes V, Baeza M (2012) Diversity and extracellular enzymatic activities of yeasts isolated from King George Island, the sub-Antarctic region. BMC Microbiol 12:251. https://doi.org/10.1186/1471-2180-12-251
Connell L, Redman R, Craig S, Scorzetti G, Iszard M, Rodriguez R (2008) Diversity of Soil Yeasts Isolated from South Victoria Land, Antarctica. Microb Ecol 56:448–459. https://doi.org/10.1007/s00248-008-9363-1
Dong K, Tripathi B, Moroenyane I, Kim W, Li N, Chu H, Adams J (2016) Soil fungal community development in a high Arctic glacier foreland follows a directional replacement model, with a mid-successional diversity maximum. Sci Rep 6:26360. https://doi.org/10.1038/srep26360
Doveri F, Pecchia S, Sarrocco S, Minnocci A, Vannacci G (2010) Rodentomyces, a new hypocrealean genus from Italy. Fungal Divers 42:57–69. https://doi.org/10.1007/s13225-010-0028-2
Doweld A (2001) Prosyllabus tracheophytorum: tentamen systematis plantarum vascularium (Tracheophyta). Geos Mosc 33–110
Dreesens L, Lee C, Cary S (2014) The Distribution and identity of edaphic fungi in the Mcmurdo dry valleys. Biology (Basel) 3:466–483. https://doi.org/10.3390/biology3030466
Dresch P, Falbesoner J, Ennemoser C, Hittorf M, Kuhnert R, Peintner U (2019) Emerging from the ice-fungal communities are diverse and dynamic in earliest soil developmental stages of a receding glacier. Environ Microbiol. https://doi.org/10.1111/1462-2920.14598
Duarte AWF, Dayo-Owoyemi I, Nobre FS, Pagnocca FC, Chaud LCS, Pessoa A, Felipe MGA, Sette LD (2013) Taxonomic assessment and enzymes production by yeasts isolated from marine and terrestrial Antarctic samples. Extremophiles 17:1023–1035. https://doi.org/10.1007/s00792-013-0584-y
Duarte AWF, dos Santos JA, Vianna MV, Vieira JMF, Mallagutti VH, Inforsato FJ, Wentzel LCP, Lario LD, Rodrigues A, Pagnocca FC, Pessoa Junior A, Durães Sette L (2018) Cold-adapted enzymes produced by fungi from terrestrial and marine Antarctic environments. Crit Rev Biotechnol 38:600–619. https://doi.org/10.1080/07388551.2017.1379468
Duncan SM, Minasaki R, Farrell RL, Thwaites JM, Held BW, Arenz BE, Jurgens JA, Blanchette RA (2008) Screening fungi isolated from historic Discovery Hut on Ross Island, Antarctica for cellulose degradation. Antarct Sci 20:463–470. https://doi.org/10.1017/S0954102008001314
Durán P, Barra PJ, Jorquera MA, Viscardi S, Fernandez C, Paz C, Mora ML, Bol R (2019) Occurrence of Soil Fungi in Antarctic Pristine Environments. Front Bioeng Biotechnol. https://doi.org/10.3389/fbioe.2019.00028
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Egidi E, Delgado-Baquerizo M, Plett JM, Wang J, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK (2019) A few Ascomycota taxa dominate soil fungal communities worldwide. Nat Commun 10:2369. https://doi.org/10.1038/s41467-019-10373-z
Frank-Fahle BA, Yergeau É, Greer CW, Lantuit H, Wagner D (2014) Microbial functional potential and community composition in permafrost-affected soils of the NW Canadian Arctic. PLoS ONE 9:e84761. https://doi.org/10.1371/journal.pone.0084761
Freeman KR, Martin AP, Karki D, Lynch RC, Mitter MS, Meyer AF, Longcore JE, Simmons DR, Schmidt SK (2009) Evidence that chytrids dominate fungal communities in high-elevation soils. Proc Natl Acad Sci 106:18315–18320. https://doi.org/10.1073/pnas.0907303106
Gangwar P, Alam SI, Bansod S, Singh L (2009) Bacterial diversity of soil samples from the western Himalayas, India. Can J Microbiol 55:564–577. https://doi.org/10.1139/w09-011
Gesheva V, Vasileva-Tonkova E (2012) Production of enzymes and antimicrobial compounds by halophilic Antarctic Nocardioides sp. grown on different carbon sources. World J Microbiol Biotechnol 28:2069–2076. https://doi.org/10.1007/s11274-012-1009-2
Gontcharova V (2010) Black box chimera check (B2C2): a windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene Datasets. Open Microbiol J 4:47–52. https://doi.org/10.2174/1874285801004010047
Gostinčar C, Lenassi M, Gunde-Cimerman N, Plemenitaš A (2011) Fungal adaptation to extremely high salt concentrations. In: Advances in Applied Microbiology. Academic Press, Burlington, pp 71–96
Gupta P, Sangwan N, Lal R, Vakhlu J (2015) Bacterial diversity of Drass, cold desert in Western Himalaya, and its comparison with Antarctic and Arctic. Arch Microbiol 197:851–860. https://doi.org/10.1007/s00203-015-1121-4
Gupta P, Vakhlu J (2015) Culturable bacterial diversity and hydrolytic enzymes from drass, a cold desert in India. African J Microbiol Res 9:1866–1876. https://doi.org/10.5897/ajmr2015.7424
Hallen-Adams HE, Kachman SD, Kim J, Legge RM, Martínez I (2015) Fungi inhabiting the healthy human gastrointestinal tract: a diverse and dynamic community. Fungal Ecol 15:9–17. https://doi.org/10.1016/j.funeco.2015.01.006
Hammer Ø, Harper DAT, Ryan PD (2001) Past: Paleontological statistics software package for education and data analysis. Palaeontol Electron 4:9
Hassan N, Rafiq M, Hayat M, Shah AA, Hasan F (2016) Psychrophilic and psychrotrophic fungi: a comprehensive review. Rev Environ Sci Bio/Technol 15:147–172. https://doi.org/10.1007/s11157-016-9395-9
Heino J, Tolkkinen M, Pirttilä AM, Aisala H, Mykrä H (2014) Microbial diversity and community-environment relationships in boreal streams. J Biogeogr 41:2234–2244. https://doi.org/10.1111/jbi.12369
Kandlikar GS, Gold ZJ, Cowen MC, Meyer RS, Freise AC, Kraft NJB, Moberg-Parker J, Sprague J, Kushner DJ, Curd EE (2018) Ranacapa: An R package and Shiny web app to explore environmental DNA data with exploratory statistics and interactive visualizations. F1000Res 7:1734 . https://doi.org/10.12688/f1000research.16680.1
Kendrick B (2003) Ainsworth Bisbys dictionary of the fungi a review. Mycologist 17:17–19. https://doi.org/10.1017/S0269915X03001204
Kothari D, Patel S, Kim S-K (2018) Anticancer and other therapeutic relevance of mushroom polysaccharides: A holistic appraisal. Biomed Pharmacother 105:377–394. https://doi.org/10.1016/j.biopha.2018.05.138
Krishnan A, Alias SA, Wong CMVL, Pang KL, Convey P (2011) Extracellular hydrolase enzyme production by soil fungi from King George Island, Antarctica. Polar Biol 34:1535–1542. https://doi.org/10.1007/s00300-011-1012-3
Kuhnert R, Oberkofler I, Peintner U (2012) Fungal growth and biomass development is boosted by plants in snow-covered soil. Microb Ecol 64:79–90. https://doi.org/10.1007/s00248-011-0001-y
Lahti L, Shetty S (2017) Microbiome R package. Biocond 1–71. https://doi.org/10.18129/B9.bioc.microbiome
Ludley KE, Robinson CH (2008) ‘Decomposer’ Basidiomycota in Arctic and Antarctic ecosystems. Soil Biol Biochem 40:11–29. https://doi.org/10.1016/j.soilbio.2007.07.023
Magan N (2007) Fungi in extreme environments. The Mycota 4:85–103
Malosso E, Waite IS, English L, Hopkins DW, O’Donnell AG (2006) Fungal diversity in maritime Antarctic soils determined using a combination of culture isolation, molecular fingerprinting and cloning techniques. Polar Biol 29:552–561. https://doi.org/10.1007/s00300-005-0088-z
Margesin R, Miteva V (2011) Diversity and ecology of psychrophilic microorganisms. Res Microbiol 162:346–361. https://doi.org/10.1016/j.resmic.2010.12.004
McMurdie PJ, Holmes S (2013) Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Moll J, Hoppe B, König S, Wubet T, Buscot F, Krüger D (2016) Spatial distribution of fungal communities in an arable soil. PLoS ONE 11:e0148130. https://doi.org/10.1371/journal.pone.0148130
Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, Ajami NJ, Petrosino JF (2017) The gut mycobiome of the human microbiome project healthy cohort. Microbiome 5:153. https://doi.org/10.1186/s40168-017-0373-4
Nichols K, Wright S (2004) Contributions of fungi to soil organic matter in agroecosystems. In: Magdoff F, Weil RR (ed) Soil organic matter in sustainable agriculture, 1st edn. CRC Press, Boca Raton, pp 179–198
Nilsson RH, Larsson K-H, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L, Saar I, Kõljalg U, Abarenkov K (2019) The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47:D259–D264. https://doi.org/10.1093/nar/gky1022
Picard KT (2017) Coastal marine habitats harbor novel early-diverging fungal diversity. Fungal Ecol 25:1–13. https://doi.org/10.1016/j.funeco.2016.10.006
Pietikäinen J, Pettersson M, Bååth E (2005) Comparison of temperature effects on soil respiration and bacterial and fungal growth rates. FEMS Microbiol Ecol 52:49–58. https://doi.org/10.1016/j.femsec.2004.10.002
Pudasaini S, Wilson J, Ji M, van Dorst J, Snape I, Palmer AS, Burns BP, Ferrari BC (2017) Microbial diversity of browning peninsula, eastern antarctica revealed using molecular and cultivation methods. Front Microbiol. https://doi.org/10.3389/fmicb.2017.00591
Pulschen AA, Rodrigues F, Duarte RTD, Araujo GG, Santiago IF, Paulino-Lima IG, Rosa CA, Kato MJ, Pellizari VH, Galante D (2015) UV-resistant yeasts isolated from a high-altitude volcanic area on the Atacama Desert as eukaryotic models for astrobiology. Microbiologyopen 4:574–588. https://doi.org/10.1002/mbo3.262
R Core Team (2017) A language and environment for statistical computing. R Found Stat Comput Vienna, Austria 2:2017
Redmile-Gordon MA, Evershed RP, Hirsch PR, White RP, Goulding KWT (2015) Soil organic matter and the extracellular microbial matrix show contrasting responses to C and N availability. Soil Biol Biochem 88:257–267. https://doi.org/10.1016/j.soilbio.2015.05.025
Řezáčová V, Slavíková R, Konvalinková T, Zemková L, Řezáč M, Gryndler M, Šmilauer P, Gryndlerová H, Hršelová H, Bukovská P, Jansa J (2019) Geography and habitat predominate over climate influences on arbuscular mycorrhizal fungal communities of mid-European meadows. Mycorrhiza. https://doi.org/10.1007/s00572-019-00921-2
Rillig MC (2004) Arbuscular mycorrhizae, glomalin, and soil aggregation. Can J Soil Sci 84:355–363. https://doi.org/10.4141/S04-003
Sarmiento F, Peralta R, Blamey JM (2015) Cold and hot extremozymes: industrial relevance and current trends. Front Bioeng Biotechnol 3:1–15. https://doi.org/10.3389/fbioe.2015.00148
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Schmidt SK, Naff CS, Lynch RC (2012) Fungal communities at the edge: Ecological lessons from high alpine fungi. Fungal Ecol 5:443–452. https://doi.org/10.1016/j.funeco.2011.10.005
Schmidt SK, Vimercati L, Darcy JL, Arán P, Gendron EMS, Solon AJ, Porazinska D, Dorador C (2017) A Naganishia in high places: functioning populations or dormant cells from the atmosphere? Mycology 8:153–163. https://doi.org/10.1080/21501203.2017.1344154
Schnecker J, Wild B, Hofhansl F, Alves RJE, Bárta J, Čapek P, Fuchslueger L, Gentsch N, Gittel A, Guggenberger G, Hofer A, Kienzl S, Knoltsch A, Lashchinskiy N, Mikutta R, Šantrůčková H, Shibistova O, Takriti M, Urich T, Weltin G, Richter A (2014) Effects of soil organic matter properties and microbial community composition on enzyme activities in cryoturbated arctic soils. PLoS ONE 9:e94076. https://doi.org/10.1371/journal.pone.0094076
Schoch CL, Sung G-H, López-Giráldez F, Townsend JP, Miadlikowska J, Hofstetter V, Robbertse B, Matheny PB, Kauff F, Wang Z, Gueidan C, Andrie RM, Trippe K, Ciufetti LM, Wynns A, Fraker E, Hodkinson BP, Bonito G, Groenewald JZ, Arzanlou M, Sybren de Hoog G, Crous PW, Hewitt D, Pfister DH, Peterson K, Gryzenhout M, Wingfield MJ, Aptroot A, Suh S-O, Blackwell M, Hillis DM, Griffith GW, Castlebury LA, Rossman AY, Lumbsch HT, Lücking R, Büdel B, Rauhut A, Diederich P, Ertz D, Geiser DM, Hosaka K, Inderbitzin P, Kohlmeyer J, Volkmann-Kohlmeyer B, Mostert L, O’Donnell K, Sipman H, Rogers JD, Shoemaker RA, Sugiyama J, Summerbell RC, Untereiner W, Johnston PR, Stenroos S, Zuccaro A, Dyer PS, Crittenden PD, Cole MS, Hansen K, Trappe JM, Yahr R, Lutzoni F, Spatafora JW (2009) The ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst Biol 58:224–239. https://doi.org/10.1093/sysbio/syp020
Shivaji S, Begum Z, Shiva Nageswara Rao SS, Vishnu Vardhan Reddy PV, Manasa P, Sailaja B, Prathiba MS, Thamban M, Krishnan KP, Singh SM, Srinivas TNR (2013) Antarctic ice core samples: culturable bacterial diversity. Res Microbiol 164:70–82. https://doi.org/10.1016/j.resmic.2012.09.001
Shivaji S, Pratibha MS, Sailaja B, Hara Kishore K, Singh AK, Begum Z, Anarasi U, Prabagaran SR, Reddy GSN, Srinivas TNR (2011) Bacterial diversity of soil in the vicinity of Pindari glacier, Himalayan mountain ranges, India, using culturable bacteria and soil 16S rRNA gene clones. Extremophiles 15:1–22. https://doi.org/10.1007/s00792-010-0333-4
Siciliano SD, Palmer AS, Winsley T, Lamb E, Bissett A, Brown MV, van Dorst J, Ji M, Ferrari BC, Grogan P CH and SI (2014) Polar soil bacterial and fungal biodiversity survey Ver. 1. In: Aust. Antarct. Data Cent. https://doi.org/10.4225/15/526F42ADA05B1, Accessed 2020–06–03
Siles JA, Margesin R (2016) Abundance and diversity of bacterial, archaeal, and fungal communities along an altitudinal gradient in alpine forest soils: what are the driving factors? Microb Ecol. https://doi.org/10.1007/s00248-016-0748-2
Song Y, Laureijssen-van de Sande WWJ, Moreno LF, Gerrits van den Ende B, Li R, de Hoog S (2017) Comparative ecology of capsular exophiala species causing disseminated infection in humans. Front Microbiol 8:1–25. https://doi.org/10.3389/fmicb.2017.02514
Srinivas TNR, Singh SM, Pradhan S, Pratibha MS, Kishore KH, Singh AK, Begum Z, Prabagaran SR, Reddy GSN, Shivaji S (2011) Comparison of bacterial diversity in proglacial soil from Kafni Glacier, Himalayan Mountain ranges, India, with the bacterial diversity of other glaciers in the world. Extremophiles 15:673–690. https://doi.org/10.1007/s00792-011-0398-8
Tedersoo L, Bahram M, Puusepp R, Nilsson RH, James TY (2017) Novel soil-inhabiting clades fill gaps in the fungal tree of life. Microbiome 5:42. https://doi.org/10.1186/s40168-017-0259-5
Tokumasu S (1998) Fungal successions on pine needles fallen at different seasons: the succession of surface colonizers. Mycoscience 39:417–423. https://doi.org/10.1007/BF02460902
Vadivelan G, Venkateswaran G (2014) Production and enhancement of omega-3 fatty acid from Mortierella alpina CFR-GV15: Its food and therapeutic application. Biomed Res Int 2014:1–9. https://doi.org/10.1155/2014/657414
Vaz ABM, Rosa LH, Vieira MLA, de Garcia V, Brandão LR, Teixeira LCRS, Moliné M, Libkind D, van Broock M, Rosa CA (2011) The diversity, extracellular enzymatic activities and photoprotective compounds of yeasts isolated in Antarctica. Brazilian J Microbiol 42:937–947. https://doi.org/10.1590/S1517-83822011000300012
Wang M, Jiang X, Wu W, Hao Y, Su Y, Cai L, Xiang M, Liu X (2015) Psychrophilic fungi from the world’s roof. Persoonia Mol Phylogeny Evol Fungi 34:100–112. https://doi.org/10.3767/003158515X685878
Wang M, Tian J, Xiang M, Liu X (2017) Living strategy of cold-adapted fungi with the reference to several representative species. Mycology 8:178–188. https://doi.org/10.1080/21501203.2017.1370429
Wang N, Zang J, Ming K, Liu Y, Wu Z, Ding H (2013) Production of cold-adapted cellulase by Verticillium sp. isolated from Antarctic soils. Electron J Biotechnol 16:1–10. https://doi.org/10.2225/vol16-issue4-fulltext-12
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham A, Hyde ER, Knight R (2017) Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5:27. https://doi.org/10.1186/s40168-017-0237-y
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols. Elsevier, New York, pp 315–322
Wijayawardene NN, Pawłowska J, Letcher PM, Kirk PM, Humber RA, Schüßler A, Wrzosek M, Muszewska A, Okrasińska A, Istel Ł, Gęsiorska A, Mungai P, Lateef AA, Rajeshkumar KC, Singh RV, Radek R, Walther G, Wagner L, Walker C, Wijesundara DSA, Papizadeh M, Dolatabadi S, Shenoy BD, Tokarev YS, Lumyong S, Hyde KD (2018) Notes for genera: basal clades of Fungi (including Aphelidiomycota, Basidiobolomycota, Blastocladiomycota, Calcarisporiellomycota, Caulochytriomycota, Chytridiomycota, Entomophthoromycota, Glomeromycota, Kickxellomycota, Monoblepharomycota, Mortierellomyc. Fungal Divers 92:43–129. https://doi.org/10.1007/s13225-018-0409-5
Wubet T, Christ S, Schöning I, Boch S, Gawlich M, Schnabel B, Fischer M, Buscot F (2012) Differences in Soil Fungal Communities between European Beech (Fagus sylvatica L) Dominated Forests Are Related to Soil and Understory Vegetation. PLoS One. https://doi.org/10.1371/journal.pone.0047500
Yadav AN, Sachan SG, Verma P, Tyagi SP, Kaushik R, Saxena AK (2015) Culturable diversity and functional annotation of psychrotrophic bacteria from cold desert of Leh Ladakh (India). World J Microbiol Biotechnol 31:95–108. https://doi.org/10.1007/s11274-014-1768-z
Zhang T, Wang N-F, Liu H-Y, Zhang Y-Q, Yu L-Y (2016) Soil pH is a key determinant of soil fungal community composition in the Ny-Ålesund Region, Svalbard (High Arctic). Front Microbiol. https://doi.org/10.3389/fmicb.2016.00227
Zimmerman NB, Vitousek PM (2012) Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc Natl Acad Sci U S A 109:13022–13027. https://doi.org/10.1073/pnas.1209872109
Acknowledgements
PG is thankful to DBT, Government of India (BT/PR11727/BCE/08/720/2008) and CSIR (9/100/0177) 2K13-EMR-I, Government of India for funding of this project. RK thanks to DST-SERB/EEQ/2018/001085 for partial financial assistance and IM thanks to UGC-JRF/SRF fellowship Govt. of India and the research facilities supported by the Central University of Kerala.
Author information
Authors and Affiliations
Contributions
PG conceived and designed the experiments. JV contributed reagents, YP provided materials. PG, IM, RK analyzed the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Communicated by A. Oren.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gupta, P., Vakhlu, J., Sharma, Y.P. et al. Metagenomic insights into the fungal assemblages of the northwest Himalayan cold desert. Extremophiles 24, 749–758 (2020). https://doi.org/10.1007/s00792-020-01191-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-020-01191-z