Abstract
A haloalkaliphilic solvent-tolerant lipase was produced from Alkalibacillus salilacus within 48 h of growth in liquid medium. An overall 4.9-fold enhanced production was achieved over unoptimized media after medium optimization by statistical approaches. Plackett–Burman screening suggested lipase production maximally influenced by olive oil, KH2PO4, NaCl, and glucose; and response surface methodology predicted the appropriate levels of each parameter. Produced lipase was highly active and stable over broad ranges of temperature (15–65 °C), pH (4.0–11.0), and NaCl concentration (0–30 %) showing excellent thermostable, pH-stable, and halophilic properties. The enzyme was optimally active at pH 8.0 and 40 °C. Majority of cations, except some like Co2+ and Al3+ were positive signals for lipase activity. In addition, the presence of chemical agents and organic solvents with different log P ow was well tolerated by the enzyme. Finally, efficacy of lipase-mediated esterification of various alcohols with oleic acid in organic solvents was studied.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
During the last decades, the range of reactions in which enzymes are employed has been extended due to the discovery of extremophilic versions of enzymes. The huge interest in extremozymes stems from their properties superior to those synthesized by mesophilic microorganisms, i.e., activity under unique conditions in various biotechnological and industrial processes, which are conducted under specific parameters, pH, ionic strength, temperature, etc. Among extremozymes, extracellular degrading enzymes, hydrolases (especially lipases) from extremophiles have considerable biotechnological and, therefore, commercial significance (Enache and Kamekura 2010; Pérez et al. 2011).
With the increasing use of lipases in various industries, extracellular microbial lipases have emerged as the key enzymatic constituent in biotechnology, owing to their multifaceted properties, broad substrate specificity, and catalyze the numerous different reactions (Hasan-Beikdashti et al. 2012; Jaeger et al. 1999; Schmid and Verger 1998; Sharma et al. 2011; Treichel et al. 2010). Apart from all the advantages, industrial applications of lipases are often hampered by their low stability in the processes, including low thermostability and loss of activity at alkaline pH values and in the presence of the organic solvents, where most of reactions are performed. To overcome these limitations, it would be highly desirable to identify, characterize, and produce of these enzymes, with special emphasis on those that have better catalytic efficiency and specific properties compatible with extreme reaction conditions (Moshfegh et al. 2013; Enache and Kamekura 2010; Mevarech et al. 2000).
Lipases from halophiles have the unique properties making them the robust biocatalysts choice for enzymatic processes performed at high salt concentrations, extreme pH and temperatures, and aqueous–organic or non-aqueous media, where most enzymes display a severe reduction in their activity (de Lourdes Moreno et al. 2013; Enache and Kamekura 2010; Mevarech et al. 2000). When organic solvent reaction systems offer many advantages over aqueous reaction systems, such as use of hydrophobic substrate, easy separation and recovery of product, reusability of enzyme, reduction of side reactions and microbial contamination, and thermodynamic equilibrium favoring synthesis, use of halophilic lipases in these reactions becomes more critical (de Lourdes Moreno et al. 2013; Hun et al. 2003; Sinha and Khare 2014). These reasons justify the direction of intensive research to obtain lipases, suitable for industrially applications, by screening of new halophilic strains.
In the present work, a halophilic strain showing lipolytic activity was isolated and identified via molecular, morphological, and physiological tests as Alkalibacillus salilacus SR-079 Halo. Also, it was undertaken to optimize the key process nutritional and physico-chemical variables, for cost effective production of lipase from the isolate using both non-statistical and statistical approaches. Additionally, characterization of produced extracellular lipase, especially its activity and stability in the presence of salts, organic solvents, di- and trivalent cations, and chemical agents were reported. Moreover, the lipase was applied for esterification of oleic acid and 3 alcohols in various organic solvents.
Materials and methods
Chemicals
p-Nitrophenyl esters [p-nitrophenyl palmitate (p-NPP), p-nitrophenyl decanoate (p-NPD), p-nitrophenyl octanoate (p-NPO), p-nitrophenyl butyrate (p-NPB), and p-nitrophenyl acetate (p-NPA)], as lipase substrates, p-nitrophenol (p-NP), and Rhodamine B were purchased from Sigma (St. Louis, USA). Media components, surfactants, and organic solvents were obtained from Merck (Darmstadt, Germany). Different oils used for lipase production were locally available. All other chemicals were of analytical grade quality.
Strain isolation, identification, and culture maintenance
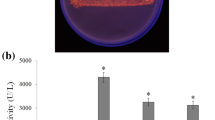
The bacterial strain SR-079 Halo used in this study was isolated from the saline water of Shoor-Mast Lake (36°08′N and 53°04′E), located in the north of Iran, which was maintained on agar slants containing modified Luria–Bertani (LB) medium: 10.0 g L−1 peptone, 5.0 g L−1 yeast extract, 175.3 g L−1 NaCl, 15.0 g L−1 agar, pH 7.4, incubated for 48 h at 37 °C, and then stored at 4 °C and sub-cultured at monthly intervals. A plate assay to detect lipase activity in modified LB agar medium containing olive oil 3 % v/v and the fluorescent dye Rhodamine B 0.001 % w/v was applied. Substrate hydrolysis causes the formation of orange fluorescent halos around bacterial colonies visible upon UV irradiation (350 nm) (Kouker and Jaeger 1987). The isolate was identified by morphological, physiological, biochemical, and molecular assays as A. salilacus. The 16S rDNA gene was amplified using the general bacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1525R (5′-TTCCTCCACTAGGTCGG-3′). Total genomic DNA of the isolated microorganism was extracted using a DNA Extraction Kit (Bioneer, Korea) and PCR was carried out in a total volume of 100 µL containing PCR buffer with 1.5 mM MgCl2, 200 µM dNTPs, Taq DNA polymerase (5.0 U), 1.0 µM each primer, and DNA template (50 ng). The amplification program was as follows: initial denaturation at 94 °C for 5 min, 30 cycles of denaturation at 94 °C for 1 min, annealing at 56 °C for 1 min, and extension at 72 °C for 1.5 min, with final extension at 72 °C for 10 min. The PCR products were examined on 1 % agarose gel. For sequencing, the amplified PCR products were purified and sequenced. The obtained sequence was aligned with Clustal W and was compared with the relevant sequences in GenBank (http://www.ncbi.nlm.nih.org/ available May 03, 2015) and using BLASTN through the NCBI server. To confirm the identity of the isolate, morphological, physiological, and biochemical characteristics of the strain SR-079 Halo were studied in both the LB agar and broth media. The shape and color of the colony, Gram reaction, catalase and oxidase activities, nitrate reduction, H2S production, acid production from glucose, fructose, lactose, sucrose, glycerol, and maltose, hydrolysis of casein, starch, urea, and gelatin, Voges-Proskauer, motility, and methyl red tests were performed (Cappuccino and Sherman 2002). The 16S rDNA gene sequence of the isolate was submitted to GenBank under the accession number KF738177. The tree constructed using the neighbor-joining method was based on a comparison of approximately 1413 nucleotides from 16s rDNA gene of the strain to its closest relatives.
Cultivation medium, inoculum, and the enzyme production
For inoculum preparation, loopful of the stock slant culture was grown in the modified LB broth medium, which was the same as the above slant medium without agar and supplemented with 0.5 % v/v olive oil and 2 % v/v Tween 80, pH 7.4. The cultivation was performed at 37 °C with shaking at 150 rpm till a constant absorbance of 1.0 was attained at 600 nm. Fifty mL of production modified M9 minimal medium (referred to as the basal medium) composed of Na2HPO4, 12.8 g L−1; KH2PO4, 3.0 g L−1; MgSO4.7H2O, 0.1 g L−1; NH4Cl, 1.0 g L−1; glucose, 1.0 g L−1; NaCl, 175.3 g L−1, olive oil, 0.5 % v/v; Tween 80, 2.0 % v/v; and pH 7.4 was taken in 250-ml Erlenmeyer flask and seeded with 1 mL of inoculum (2 % v/v). The incubation was carried out at 150 rpm in an orbital shaker maintained at 37 °C. Cell growth was monitored by recording A600. For estimating lipase production, at regular time intervals withdrawn samples were centrifuged at 8000×g for 10 min and the enzyme activity was assayed in the cell-free supernatant.
Analytical methods
Lipase assay
Lipase activity was determined using p-nitrophenyl palmitate (p-NPP) as substrate according to the following method. The substrate p-NPP was dissolved in absolute ethanol (100 µL) and mixed with 900 µL of solution containing 18 % NaCl and 0.5 % Triton-X100 as emulsifying agent in 100 mM phosphate buffer (pH 8.0) to give a final concentration of 0.5 mM. The reaction was initiated by adding 1 mL of crude enzyme (cell-free culture supernatant), and incubation was carried out at 40 °C for 20 min with orbital shaking. Following the addition of 100 µL of Na2CO3 solution (1 M) to stop the reaction and centrifugation of the mixture at 20,000×g for 5 min, the amount of p-nitrophenol (p-NP) released based on a standard curve of p-NP was recorded at 410 nm against a blank. One unit (U) was defined as the amount of enzyme liberating 1 µmol of p-NP per min under the standard assay conditions. Triplicate experiments were carried out and the mean value was calculated.
Bradford assay
The protein content of supernatant was quantified by the Bradford assay. Bovine serum albumin (BSA) was used to prepare the standard curve (Bradford 1976).
Medium optimization for maximum lipase production by A. salilacus
Medium optimization for maximum lipase production was done in the basal production medium. The components were varied either by “one-variable-at-a-time” approach to primarily selection of significant parameters affecting the enzyme yield or according to the statistical design of experiments employing Plackett–Burman design (PBD) for initial screening of the factors, potentially influencing the response and response surface methodology (RSM) for obtaining the optimum concentration of individual factors.
One-variable-at-a-time approach
In the first step of culture conditions optimization, various nutritional and cultural parameters were optimized by keeping all factors at a constant level in the basal medium, except the one under study. Each subsequent factor was examined after taking into account the previously optimized factor(s). Among carbon sources fructose, galactose, lactose, sucrose, maltose, and soluble starch at 0.1 % w/v level were supplemented individually by replacing glucose in the basal medium. For evaluating the effects of nitrogen sources, the additive nitrogen sources (peptone, yeast extract, asparagine, and ammonium nitrate) were applied in addition to 0.5 % w/v NH4Cl already present in the basal medium at 0.5 % w/v concentration. The effects of various inorganic salts on lipase production also was studied using individually removal of KH2PO4, Na2HPO4, CaCl2, and MgSO4 from basal medium and individually addition of FeSO4 and ZnSO4 to the basal medium at the concentration of 0.1 % w/v. Similarly, the effects of various surfactants and oils (by replacing Tween 80 and olive oil present in basal medium with Triton-X100 and desired oils, respectively, at the same level) on lipase production, and salts (NaCl, KCl, and LiCl at the concentrations of 0–30 %) on strain growth and lipase production were evaluated. Initial pH of the medium (7.0–9.0), temperature (25–45 °C), agitation speed (50–150 rpm), fermentation period (24–96 h), aeration (40–80 %, according to the flask volume occupied by air) and inoculum size (1–10 %) were the physical parameters evaluated before being subjected to statistical optimization.
Statistical approach
Following the initial selection of most effective of cultural and nutritional parameters, lipase production was optimized in the modified medium M9 by statistical approaches including PBD and RSM.
Plackett–Burman design
The effect of 10 factors including peptone, NaCl, glucose, MgSO4, FeSO4, CaCl2, NH4Cl, KH2PO4, Na2HPO4, and olive oil were studied on lipase production by Plackett–Burman design (Aghaie-Khouzani et al. 2012; Haaland 1989; Rao and Divakar 2001). These components were varied in the modified M9 basal medium over two levels and the minimum (−1) and maximum (+1) ranges were designated for the parameters (Table S1). Concentration levels were decided on the basis of literature reports on lipase production. The statistical software package Design-Expert 7.0 (Stat Ease Inc., Minneapolis, USA) was used to construct a set of 12 experimental designs according to the matrix shown in Table S2. Analysis of data obtained from this screening was based on the linear model: Y = β 0 + Σ β i X i; where Y is the response, X i is the coded independent variables estimates, and β 0 and β i are model intercept and constant coefficient, respectively. Five replicates in the center point were also performed to find the curvature that may exist in the model and pure experimental error that shows lack of fit (trials 13–17). To identify the significance level (p value) of each variable, Student’s t test was used and factors with p < 0.05 were considered to significantly influence lipase production. All experiments were carried out in triplicate and the averages were considered to be the responses. For each experiment, the lipase production was calculated in terms of U L−1.
Path of steepest ascent method
In this part of optimization, keeping the experimenting along the direction of steepest ascent until no further improvement occurs in the response was aimed and based on the coefficients from the Plackett–Burman linear equation, the optimal level scope of each selected factor was examined. This point would be near the optimal point and could be considered as center point in the following response surface experiment (Aghaie-Khouzani et al. 2012). As a result, seven experiments were designed along this path, and the response was measured at each point (Table S3).
Central composite design RSM
Based on the results of the Plackett–Burman and steepest ascent designs, the effect of four independent factors including glucose, NaCl, KH2PO4, and olive oil was studied on lipase production using RSM. Each factor in the design was studied at five different levels, coded as −α, −1, 0, +1, and +α. The remaining factors were taken at a central coded value considered as zero throughout. The minimum and maximum ranges of variables investigated and their values in actual and coded forms determined (values are listed in Table S4). A central composite design (CCD) developed by Design-Expert 7.0 software was adopted to optimize the concentration of above four significant factors yielding a set of 16 experiments and 5 replicates at the center point to derive a statistical model for obtaining maximum lipase production (Table S5). All experiments were done in triplicates and the average was taken as the dependent variable or response. Regression analysis was performed on the data obtained. The results of the CCD were then used to fit a quadratic equation by multiple regression procedure. This resulted in an empirical model that related the response measured to the independent variables of the experiment and determined the interrelationships of the variables. Predicted relationship between the independent variables and the response represented by the produced lipase were calculated using the following second-order polynomial equation: Y = βo + Σ β i X i + Σ β ii X 2i + Σ β ij X i X j; where Y is the lipase activity, β o is intercept term, β i, β ii and β ij are linear, squared, and cross-product coefficients, respectively.
Analysis of variance (ANOVA) was performed through Fisher’s test and counter and three-dimensional response surface curves were plotted to study the interaction among various factors. A p value below 0.05 was considered to be statistically significant. The fitness of the equation was evaluated by quality indicators, multiple correlation coefficient (R 2), and adjusted R 2. In order to validate the response surface model and evaluate the precision of the model, a random set of optimized checkpoints experiments was set up according to the conditions predicted by the model.
Functional and biochemical characterization
Effects of temperature, pH, and salt concentration
In order to evaluate the simultaneous effects of temperature and pH on the lipolytic enzyme stability, experiments were performed at temperature range of 25–65 °C with a stepwise increase of 10 °C in the reaction mixtures adjusted at pH range of 3.0–11.0. The buffers (100 mM) used were as follows: citrate buffer (pH 3.0–5.5), phosphate buffer (pH 6.0–8), Tris–HCl buffer (pH 8.5–9.5), and glycine–NaOH buffer (pH 10.0–11.0). The enzyme was pre-incubated in relative buffer containing 18 % NaCl under different temperatures for 1 h, and then residual activity was measured using p-NPP method. To determine the halostability, the lipase was pre-incubated in phosphate buffer (100 mM, pH 8.0) containing different NaCl, KCl, and LiCl concentrations (0–30 %) at 40 °C for 1 h. Residual activity was measured under the assay conditions as described above. In all of these experiments initial lipolytic activity was considered as 100 %.
Effects of organic solvents
Cell-free culture supernatant was mixed with organic solvents in different ratios to achieve range of 20–80 % v/v of selected organic solvents with different log P ow values including hexadecane (8.8), dodecane (6.6), nonane (5.1), decanol (4.9), heptane (4), hexane (3.5), cyclohexane (3.2), octanol (2.9), chloroform (2), cyclohexanol (1.5), hexanone (0.96), butanol (0.8), ethanol (−0.24), acetone (−0.24), and acetonitrile (−0.3). The mixture was incubated at 40 °C for 1 h with constant shaking at 150 rpm. After incubation, samples were withdrawn from the aqueous phase and residual specific lipase activity was assessed under assay conditions. The enzyme incubated without solvent was considered as control and the stability was expressed as the residual specific lipolytic activity.
Effects of metal ions and chemical reagents
To investigate the stability of lipase in presence of metal ions including Fe3+, Fe2+, Ni2+, Co2+, Mg2+, Mn2+, Zn2+, Cu2+, Hg2+, and Al3+ at concentrations of 1, 5, and 10 mM and chemical reagents including ethylenediamine tetraacetic acid (EDTA) (2, 5, and 10 mM), phenylmethylsulfonyl fluoride (PMSF) (1 and 5 mM), β-mercaptoethanol (0.1, 0.25, and 0.5 %), and urea (1 and 5 mM), lipase activity were examined by pre-incubating the enzyme with the ions or reagents in 100 mM phosphate buffer (pH 8.0) contained 18 % NaCl at 40 °C for 1 h. Residual activity was determined under the assay conditions. Lipase activity in the absence of any additives was taken as 100 %.
Effects of surfactants
To study the effect of surfactants, lipase was pre-incubated with the selected surfactants at a concentrations range of 1–10 mM for cetyltrimethylammonium bromide (CTAB) and 0.1–0.5 % for Tween 80 and Triton-X100, and sodium dodecyl sulfate (SDS) for 1 h at 40 °C prior to lipase assay. The control, lipase without the addition of surfactants was set as 100 %.
Substrate specificity
To determine the substrate specificity of lipase, p-nitrophenyl (p-NP) esters with different chain lengths (C2: acetate, C4: butyrate, C8: octanoate, C10: decanoate, and C16: palmitate) were used as the substrates at the final concentration of 0.25 mM, and then the released amount of p-nitrophenol was measured at 410 nm using spectrophotometric method as described above and data were expressed as U L−1 activity. In addition, specificity of natural oils (olive oil, sunflower oil, and sesame oil) for lipase was tested with titrimetric method. Assay mixture was prepared in final volume of 6 mL consisting of 2 mL oil, 2 mL phosphate buffer (100 mM, pH 8.0) contained 18 % NaCl and 2 mL crude enzyme and incubated at 40 °C and after a 20 min-incubation, the reaction was terminated by the adding 8 mL of acetone: ethanol (1:1; v/v). The free fatty acids liberated from oil were titrated by adding sodium hydroxide solution (0.1 N) in the presence of 25 µL of phenolphthalein (0.2 % w/v in ethanol). One unit of lipase activity was defined as the amount of the enzyme which released 1 μmol of fatty acid per min under test conditions.
Application of the lipase for long-chain esters production
The esterification reaction was carried out to analyze the performance of lipase from A. salilacus for the production of long-chain esters. The reaction mixture consisted of 10 mL solvent and concentrations of 1 M from alcohols (butanol, decanol, and octanol) and oleic acid as acyl donor in 50-mL Erlenmeyer flasks. The esterification reaction was initiated by addition of lipase (0.5 U mL−1) to the reaction mixture and flasks were incubated in orbital shaker at 40 °C and 150 rpm for 6 h. Control experiments were conducted in parallel without lipase under similar conditions. Samples were withdrawn to evaluate the esterification rate. The residual oleic acid content was assayed by titration with 0.1 N sodium hydroxide, using phenolphthalein (25 µL from 2 % w/v solution in absolute ethanol) as an indicator and 5 mL of ethanol as a quenching agent. The amount of ester was calculated as being equivalent to the consumed acid. To study the interactive effect of solvents as medium for reaction on esterification, esterification carried out in various organic solvents. Solvents used were acetone, acetonitrile, chloroform, and n-hexane at the final concentration of 80 % v/v.
Statistical analysis
One-way analysis of variance followed by Holm–Sidak multiple comparison test was done to show the significant difference between groups. A probability level of p < 0.05 was considered statistically significant.
Results and discussion
Strain identification and production of the extracellular lipase
Lipase production was observed by 12 out of 170 halophilic isolates collected from different saline areas of Iran, as indicated by formation of orange fluorescent halos visible at 350 nm on screening agar plates contained olive oil and Rhodamine B, and the strain SR-079 with the most lipolytic activity was selected for further studies. Based on the morphological and physiological assessments, the bacterial strain A. salilacus SR-079 Halo was confirmed as a Gram-positive, motile, rod, and aerobic spore-form. The colonies were circular, smooth, and creamy on LB agar. Growth was observed at salt concentrations of 0–30 % (w/v) with an optimum at 12 % NaCl. Also, growth occurred at the pH and temperature ranges 5.0–10.0 (optimum pH 8.0) and 15–55 °C (optimum 40 °C), respectively. Catalase and nitrate reduction tests were positive, while urease, Voges-Proskauer, oxidase, gelatinase, caseinase, amylase, H2S production, and methyl red tests were negative. Experiments for the utilization of different carbon sources such as glucose, fructose, glycerol, lactose, sucrose, and maltose were performed and acid was produced only from fructose and lactose. Observed properties were all typical characteristics of the genus Alkalibacillus. In addition, the partial 16S rRNA sequence of the isolate was obtained. Phylogenetic analysis based on 16S rRNA gene sequence comparisons revealed the isolate SR-079 Halo belonged to the genus Alkalibacillus, and was most close to A. salilacus R559 with accession No. HM179185 (99 % 16S rRNA gene sequence similarity). Based on morphological, physiological, and phylogenetic analysis, the isolated strain of SR-079 Halo was determined to be A. salilacus (Accession No. KF738177). Finally, a tree depicting the phylogenetic affinity of strain SR-079 Halo with its closest species was drawn (Fig. S1).
The substantial extracellular lipase was produced from the middle exponential phase of the isolate growth (12 h), and reached maximum level in the early-stationary phase (48 h) as shown in Fig. 1a. Moreover, produced lipase had optimum activity at the pH, temperature, and salt ranges from 7.0 to 9.0, 25 to 45 °C, and 0 to 30 %, respectively (Fig. 1b–f). The bacterium produced 175 U L−1 of lipase in the basal production medium. Medium optimization for maximum lipase production was carried out using one-variable-at-a-time and statistical approaches.
Preliminary optimization using “one-variable-at-a-time”
Initial pH (7.0‒9.0) of the medium, temperature (25‒45 °C), fermentation time (24‒96 h), agitation speed (50‒150 rpm), aeration (40‒80 %), inoculum volume (1‒10 %), and salt concentration (0‒30 %), were the cultural parameters evaluated before being subjected to statistical optimization. The kinetic of lipase production, with various agitation speeds and aeration, showed that extracellular lipase production reached maximum level when the cell growth attained a constant level at the early-stationary phase of strain growth period. The highest lipase production obtained with agitation speed of 150 rpm and 80 % aeration. To evaluation of incubation time, the amount of lipase activity was observed daily during a period of 4 days. After approximately 48 h, lipase production reached the maximum level and after 72 h, lipase activity began to decrease rapidly (Fig. 1a) which might be due to the depletion of nutrients, accumulation of toxic end products, and the change in pH of the medium, or proteolysis of lipase by proteases which were produced during the same growth phase.
As shown in Fig. 1c–f, the maximum growth and lipase production by the strain SR-079 Halo occurred at 40 °C and pH 8. Most halophilic lipases were reported to show maximal production at alkali pH and temperatures above 40 °C (Esakkiraj et al. 2014; Khunt and Pandhi 2012; Li et al. 2014), although some studies suggest the opposite (Pérez et al. 2011). The isolated lipase can be considered as a thermoactive and haloalkaliphilic nature lipases, according to the results reported in literature (Gupta et al. 2007; Marques et al. 2014; Yoo et al. 2011). In addition, growth and lipase production was measured at different NaCl, KCl, and LiCl concentrations, and the best results obtained for NaCl (Fig. 1b). Whereas, there was no significant difference between the effect of NaCl and KCl on the production of lipase, NaCl was subjected for subsequent experiments, as it is the most common salt in saline areas and is more available than KCl. Also, the optimum NaCl concentrations for optimal growth and lipase production were different. The optimum NaCl concentration for growth of the isolate SR-079 Halo was about 18 %, whereas the optimum concentration of NaCl for extracellular lipase production was about 12 %. Similar results have been reported in case of other lipases from halophiles (Esakkiraj et al. 2014; Li et al. 2014; Pérez et al. 2011). Different levels of the inoculum were studied to find the optimum inoculum level in the fermentation process. In this study, the maximum growth and lipase production was obtained with 10 % v/v inoculum level. The reason may be that too much biomass leads to the rich product formation. In order to reduce the impact of inoculum medium on composition of lipase production medium and prevention of a rapid drop of lipase production and growth, inoculum volume of 2 % was selected for the next experiments. Various investigators have used different levels of inoculum for lipase production employing diverse microorganisms (Ebrahimpour et al. 2008; Mehta et al. 2012).
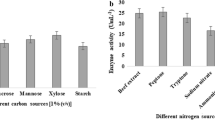
The major factor affecting expression of lipase is the carbon source; hence, a number of carbohydrates (glucose, fructose, galactose, lactose, sucrose, maltose, and soluble starch at 0.1 % w/v concentration) and oils (olive oil, sunflower, and sesame oil 0.5 %) were tested as carbon sources for lipase production. Before supplementation, oils were emulsified with 2 % w/v Tween 80. The results showed that glucose and olive oil were the best carbohydrate and lipid sources, respectively. The production of lipases is mostly inducer-dependent and various inducers have different stimulation effects on lipase production based on the physiological and biochemical pathways of the bacterium. Olive oil as an inducer enhanced lipase production significantly. This result was also followed by sunflower and sesame oils. High levels of lipase production were reported from various microorganisms in the presence of olive oil as carbon source (Ahmed et al. 2010; Balan et al. 2013; Burkert et al. 2004; Gupta et al. 2007; Mehta et al. 2012; Zaliha et al. 2006). Most published experimental data have shown that lipid carbon sources especially natural oils stimulate lipase production, whereas carbon sources that are easily broken down and used by bacteria, have an inhibitory effect and contradict the results obtained by many studies (Burkert et al. 2004; Gupta et al. 2007; Volpato et al. 2008; Zaliha et al. 2006). In this study, lipase production enhancement was observed by addition of glucose at low concentration (0.5 %) and 1 % concentration of disaccharides, which could be due to enhanced growth of the bacterium and probably excretion of lipase was directly related to biomass production. This has also been reported for the other organisms where the maltose is the enhancer of lipase production (Volpato et al. 2008). It is notable that increased glucose concentrations resulted in decreased production of the enzyme. Thus, olive oil and glucose were applied as the best inducer and carbohydrate, respectively, for lipase production. The latter carbon source and concentration were employed in the subsequent experiments.
The type of nitrogen in the medium also regulates the growth and fermentation process for lipase production, thus different types of nitrogen sources were tested. Replacement of NH4Cl with organic nitrogen sources including peptone, yeast extract, and asparagine led to a further marked increase in growth and lipase production. In addition, ammonium nitrate and ammonium chloride as inorganic nitrogen sources were tested and results showed these nitrogen sources supported similar levels of growth and lipase production compared to the other tested nitrogen sources. The best lipase activity was obtained using combination of organic and inorganic nitrogen sources, i.e., peptone in combination with additive NH4NO3 or NH4Cl that was presented as the component of basal medium, tremendously boosted the lipase production. Thus, peptone was selected as the additive nitrogen in the subsequent experiments and NH4Cl as the component inorganic nitrogen source of basal medium was kept intact. Generally, biomass and lipase production is enhanced by sources containing nitrogen especially organic nitrogen sources, which supply powerful source of nitrogen for the growth of bacteria and increases their yields of proteins. These findings confirm relevant data reported in similar studies (Balan et al. 2013; Burkert et al. 2004; Cheng et al. 2011; Ebrahimpour et al. 2008; Mahanta et al. 2008; Mehta et al. 2012; Zaliha et al. 2006).
Effects of KH2PO4, Na2HPO4, CaCl2, MgSO4, FeSO4, and ZnSO4 on growth and lipase production were being examined. CaCl2, MgSO4, FeSO4, and ZnSO4 had enhancing effect and the phosphate salts of KH2PO4 and Na2HPO4 had depressive effect on growth and lipase production. Different microorganisms have various requirements for inorganic salts and metal ions. Calcium ions play essential roles for many microbial species, because they are important in maintaining cell wall rigidity and stabilizing oligomeric proteins. Lipase production by various microorganisms was stimulated in the presence of Ca2+ alone or in combination with other ions such as Mg2+, Fe2+, Mn+2, and Zn+2 (Dandavate et al. 2009; Ebrahimpour et al. 2008; Esakkiraj et al. 2014; Gupta et al. 2007; Mehta et al. 2012; Wu et al. 2013; Zaliha et al. 2006). The most divalent cations are known to play an important role in lipase production, mostly stimulatory though a few reports about their inhibitory role also exist (Boran and Ugur 2010; Dandavate et al. 2009; Esakkiraj et al. 2014; Gupta et al. 2007). It seems that most cations enhance the lipase activity due to the formation of salt from liberated fatty acids by lipase.
Moreover, effects of two surfactants, Tween 80 and Triton-X100 on lipase production were assessed. Since surfactants probably are able to enhance the lipase production due to the emulsification of culture media containing oil to increase the lipid surface (interfacial area between oil and water) for lipase action and detachment of lipase from the oil surface (Reis et al. 2009; Verger 1997), when two types of surfactants were added to the culture, lipase activity and cell growth increased and from amongst them Tween 80 at a concentration of 2 % w/v was found to be the most suitable surfactant and was chosen to be used in the further optimization of lipase production.
Statistical optimization
Optimal conditions for enzyme production were observed at temperature 40 °C, pH 8, inoculum 2 %, aeration 80 %, and agitation speed 150 rpm. One-factor-at-a-time experiments revealed that peptone, glucose, NaCl, MgSO4.7H2O, FeSO4.7H2O, CaCl2, olive oil, KH2PO4, NH4Cl, and Na2HPO4 were supposed to have more effect on lipase production. These factors were chosen for further optimization.
Plackett–Burman design
The Plackett–Burman experimental design was employed to evaluate factors, which significantly affect the lipase production, by A. salilacus SR-079 Halo. The Plackett–Burman design for 17 trials with three levels of concentrations for ten different variables were carried out according to the experimental matrix as shown in Table S2 and lipase activity was measured as response. Significance of each variable was determined using Student’s t test and p value. The components were screened at the confidence level of 90 % on the basis of their effects. With respect to F value, p value, and confidence level of each component in the results table of Plackett–Burman trials (Table S6), significance of the model was assessed. The model F value 14.08 for lipase activity, implied that the model was significant and the values of “Prob > F” less than 0.05 illustrated the model terms were significant. Out of ten variables studied, the lipase production was highly influenced by glucose (X 2), NaCl (X 3), olive oil (X 7), and KH2PO4 (X 9) on the basis of the analysis of variance (ANOVA). After the neglect of insignificant terms (on the basis of p values higher than 0.1), a modified first-order equation was developed to describe enzyme activity:
where Y is the response (lipase activity), and X 2, X 3, X 7, and X 9 coefficients are the concentrations of glucose, NaCl, olive oil, and KH2PO4, respectively.
The effect of glucose and olive oil on lipase production was positive signal; meanwhile, the high concentration of KH2PO4 and NaCl were the most important signals affecting lipase production at the negative level. An adequate precision of 11.71 (greater than 4) indicates an adequate signal as it measures the signal-to-noise ratio, and this model could be used to navigate the design space. The multiple correlation coefficient (R 2) for the factorial model was 0.83, which implies that 83 % of the variation in the lipase production could be explained by the model. On the other hand, the predicted R 2 value (57 %) was not reasonable conformity with the adjusted R 2 value (77 %). Thus, a second-order model could be a mathematical equation with adequate precision to express the relationship between the significant independent variables and the response. Further statistical analysis revealed that due to the difference between the means of the center point and factorial tests in this design, the optimum levels for lipase production would be out of the experimental ranges chosen for Plackett–Burman design and thereby the steepest ascent method should be applied.
Steepest ascent path
The Plackett–Burman design experiment proved to be a valuable tool for screening significant variables that affected the lipase activity; however, it was unable to predict the optimum levels of the compositions. Based on the modified first-order equation obtained and regression results, it was predicted that glucose, NaCl, olive oil, and KH2PO4 were significant factors, which meant that changing the concentration of them had positive effect on the lipase production. The path of the steepest ascent (Meyer et al. 2009) was determined to find the proper direction of changing factors above to improve lipase activity; therefore, center point of the Plackett–Burman design has been considered as the beginning of the path of steepest ascent, i.e., glucose (0.55 g L−1), NaCl (2.5 mol L−1), olive oil (1.5 %), and KH2PO4 (5.5 g L−1). The results of the experiments and the directions of change the variables are shown in the Table S3. Regarding the results from the steepest ascent path, lipase activity showed a maximum 852.7 U L−1 at run 4, when X 2, X 3, X 7, and X 9 applied at concentrations of 0.95 g L−1, 1.9 mol L−1, 1.9 %, and 4.7 g L−1, respectively. Consequently, this point was near the region of maximum lipase activity response, so these variables were chosen for further optimization.
Response surface methodology
Once the ranges of relevant factors were selected through Plackett–Burman screening and experiment of steepest ascent path, significant gross curvature had been detected in the design space, central composite design (Box and Wilson 1951) and response surface methodology were employed to estimate main effects, interaction effects, and level of the selected variables on the response and to make a quadratic model, consisting of trials plus a star configuration to appraise quadratic results and central points to estimate the pure process variability and reassess gross curvature, with lipase activity as response. Four compositions, i.e., glucose, NaCl, olive oil, and KH2PO4 that significantly affected lipase activity were optimized by RSM using a 4-factor 5-level CCD. The CCD matrix included 5 central points and 16 axial points. Therefore, a group of 21 treatment combinations with different combinations of glucose, NaCl, olive oil, and KH2PO4 were performed (Table S5). The results were analyzed by standard analysis of ANOVA. The significance of each coefficient was determined by Student’s t test and p values (are listed in Table S7). The larger the magnitude of F value and the smaller p value, the more significant is the corresponding coefficient. Model coefficients were estimated by multiple linear regression and the p value was used for checking the significance of each of the coefficients. Values of “Prob > F” less than 0.05 represent model terms are significant. In the present work, square effects of olive oil and KH2PO4 and interaction effect of olive oil–glucose and olive oil–NaCl were found significant for lipase production. The computed F value of 6.06 implied the model is highly significant, because there is only a 1.79 % chance that the model F value could occur due to noise (Table S7). Regression equation obtained after ANOVA indicated that the R-squared value of 0.93 (a value of R-squared > 0.75 indicated the aptness of the model) was in reasonable agreement with the adjusted R-squared of 0.78 and this ensured a satisfactory adjustment of the quadratic model to the experimental data. Adequate precision measures the signal-to-noise ratio. A ratio greater than 4 is desirable, and the ratio of 7.72 indicates an adequate signal. This model can be used to navigate the design space, and a low coefficient of variation (CV = 11.46 %) demonstrated the experiments were precise and reliable. The lack of fit F value of 494401.15 implies the lack of fit is significant and there is only a 0.01 % chance that a lack of fit F value could occur due to noise. A negative Pred R-squared implies that the overall mean is a better predictor of our response than the current model. The pure error was very low, indicating a good reproducibility of the experimental data. A second-order polynomial function was fitted to the experimental lipase activity, resulting in the following regression equation:
where Y is the response (lipase activity), and X 2, X 3, X 7, and X 9 are glucose, NaCl, olive oil, and KH2PO4, respectively.
Based on the obtained results, the model can be utilized to generate response surfaces for the analysis of the variable effects on lipase activity.
Figure 2 represents the three-dimensional response surface plots for the optimization of conditions for lipase production. The model predicted that a combination of adjusting the concentration of glucose, NaCl, olive oil, and KH2PO4 to 1 g L−1, 4.18 mol L−1, 2 %, and 5 g L−1, respectively, would favor maximum lipase activity, giving 856.9 U L−1. Response surface plots as function of two factors at a time, maintaining all other factors at a constant level, more helpful in understanding both the main and interaction effects of these factors. Figure 2a–f shows the response surface plot effects of NaCl and glucose, olive oil and glucose, KH2PO4 and glucose, olive oil and NaCl, KH2PO4 and NaCl, and KH2PO4 and olive oil concentrations, respectively, while other two variables are kept constant at zero level. The response surface model was validated with a random set of experiments both within and outside the design space. The values observed were very close to the predicted values at conditions within design space. Thus, the optimization of lipase production by strain SR-079 Halo was successfully developed by PBD and RSM.
The optimized concentrations of mineral nitrogen source, KH2PO4, MgSO4.7H2O, NaCl, and olive oil were higher than their used concentrations in the one-factor-at-a-time experiments. Furthermore, it was found that peptone, FeSO4.7H2O, and CaCl2 were favorable to the lipase production. In addition, Na2HPO4 at lower level was used at optimized medium. These results agreed somewhat with the previous researches (Balan et al. 2013; Burkert et al. 2004; Cheng et al. 2011; Ebrahimpour et al. 2008; Gupta et al. 2007; Mahanta et al. 2008; Mehta et al. 2012). The reason for the difference between various reports is probably due to the characteristic of the each strain. Olive oil was the most significant parameter with a change in its concentration leading to a marked change in lipase production. The importance of olive oil was also evident from its interaction with glucose and NaCl. NaCl, KH2PO4, and glucose were the other most important variables influencing lipase production. The olive oil–glucose and olive oil–NaCl interactions may be due to decreased uptake of glucose and solubility of NaCl in presence of high oil concentration. There have not been many reports on lipase production optimization by statistical methods. Ebrahimpour et al. (2008) have reported 4.7-fold increase in thermophilic lipase yield after optimization. In another study by Gupta et al. (2007), production of an alkaline lipase increased by 12-fold. The other report of RSM in lipase production optimization is related to Cheng et al. (2011), where 4.94-fold enhancement in lipase production was achieved. Thus, the level of lipase production enhancement by SR-079 Halo strain and achieve its activity to about 857 U L−1 can be cost effective for its applications, in particular, there are remarkable features.
Functional and biochemical characterizations
Effects of temperature, pH, and salt concentration
The isochronal effects of temperature and pH on the lipase stability as residual activity are shown in Fig. 3a. The enzyme was optimally stable at pH 8.0 and 40 °C (100 %) and showed, at optimal pH, residual activities of more than 90 % at 25–45 °C, 60 % at 55 °C, and 30 % at 65 °C, respectively, and at optimal temperature the enzyme remaining activities were to be more than 90 % at pH 6–9, 85 % at pH 10, and 55 % at pH 11. pH and temperature optima of the isolated lipase are comparable to the other haloalkaliphilic lipases reported in different studies (Esakkiraj et al. 2014; Khunt and Pandhi 2012; Li et al. 2014). As many thermostable lipases were reported to be neither active nor stable at temperatures above 70 °C (Dheeman et al. 2010, 2011; Ebrahimpour et al. 2008; Marques et al. 2014), the lipase from the strain SR-079 Halo, which showed the excellent thermostability under temperatures ranging from 25 to 65 °C can be rated among the moderately thermostable lipases (Fig. 3a). Thermostability is a desirable property in lipases for application in industrial processes operating under high temperatures. Most halophilic lipases were reported to show thermostability due to the presence of NaCl, which has the capacity to increase the thermostability of enzymes from halophile. As shown in Fig. 3a, optimal pH for the lipase stability was 7.0–9.0. Meanwhile, it was highly stable in the pH range of 5.0–11.0, indicating the alkali-stable nature. Alkaline enzymes have received considerable attention because of their tremendous potential in industrial processes.
Effect of a pH–temperature and b various concentrations of NaCl, KCl, and LiCl on enzyme stability. The residual activity of crude enzyme was determined after 1 h of incubation at various pH–temperatures and salts with shaking at 150 rpm; and the incubation of enzyme for evaluation of salts effect was carried out at 40 °C. The initial activity before incubation was set as 100 %
In addition, lipase stability was measured in the presence of different concentrations of NaCl, KCl, and LiCl, and maximum stability was found to be at 18, 18, and 12 % NaCl, KCl, and LiCl, respectively (Fig. 3b). Meanwhile, the lipase showed strong tolerance to NaCl, KCl, and LiCl as it was highly active and stable over a broad NaCl, KCl, and LiCl concentration ranges from 0 to 30 %. Similar extreme halotolerance has been reported in other lipases from halophiles (Esakkiraj et al. 2014; Khunt and Pandhi 2012). In fact, many halophilic enzymes required the presence of NaCl or KCl for optimal activity and stability (Mevarech et al. 2000). Obtained results showed the lipase from strain SR-079 Halo, unlike most enzymes from halophiles, which lost their stability when exposed to low salt concentrations, retained its stability in the absence of salts.
Effects of additives
High activity and stability of lipases in organic solvents is an essential prerequisite for applications in organic synthesis (Adlercreutz 2013); hence, activity and stability in organic solvents are considered significant factors in a lipase. As described above, the lipase from A. salilacus SR-079 Halo was stable under high salinities, making it quite possible to remain stable in organic solvents. Effect of various organic solvents on the stability of the lipase is shown in Table 1. The residual specific activity was measured in absence of organic solvents and in 6 alkanes, 5 alcohols, 2 ketones, acetonitrile, and chloroform, expressed as U mg−1 protein are given in Table 1. The results obtained indicate the lipase displayed considerable stability in the presence of high concentration (50 % v/v) of hydrophobic organic solvents (log P ow 1.5–8.8) and interestingly, alkanes (except dodecane) and octanol even increased the lipase activity from 106 % in cyclohexane to 190 % in heptane, i.e., water-immiscible organic solvents enhance present lipase activity compared to water-miscible organic solvents which holds a potential application in organic synthesis, fat, and oil modification industries. As there is a more tendency for hydrophilic solvents to cause more significant enzyme inactivation than hydrophobic solvents, the lipase from SR-079 Halo strain that retained more than 50 % of its activity in the presence of hydrophilic organic solvents, such as ethanol and acetone at the high concentration of 80 and 60 % v/v, respectively, can be considered for biotechnological applications. Also, SR-079 Halo strain lipase showed at least 30 % of its optimal lipolytic activity in the presence of 20 % v/v acetonitrile and butanol. Although most lipases due to their general behavior show strong tolerance towards hydrophobic solvents (Boran and Ugur 2010; Dandavate et al. 2009; Essamri et al. 1998; Hun et al. 2003; Ji et al. 2010; Marques et al. 2014; Volpato et al. 2008; Wu et al. 2013), the isolated lipase in this work, as the halophilic and organic solvent-tolerant enzyme, showed the impressive stability towards the broad range of hydrophilic and hydrophobic organic solvents at high concentration up to 80 % v/v.
As may be deduced from the Log P ow (Laane et al. 1987) values of some selected common organic solvents, water-miscible hydrophilic solvents, such as acetone, acetonitrile, ethanol, butanol, and hexanone are usually incompatible with enzymatic activity; whereas, water-immiscible lipophilic solvents, such as alkanes, octanol, decanol, and chloroform retain an enzyme’s high catalytic activity due to the fact that they do not separate the crucial bound water from the enzyme’s surface leading to the unfolding of the enzyme and cause the enzyme to be less stable (Zaks and Klibanov, 1988). Also, the activation of lipase by hydrophobic solvents could be explained that organic solvent molecules could interact with hydrophobic amino acid residues present in the lid that covers the catalytic site of the enzyme, thereby maintaining the enzyme in its open conformation and conducive to catalysis (Rao et al. 1993). The other reason for higher stability in presence of hydrophobic solvents may be the surface–solvent interaction leading to interfacial activation (Reis et al. 2009; Verger, 1997).
As shown in Table S8, the lipase stability was significantly stimulated in the presence of Fe2+, Fe3+, Mg2+, Cu2+, Hg2+, Ni2+, and Mn2+. A possible explanation for this phenomenon is that these cations bind to the active site of the lipase and change the conformation of the protein for more proper function. Other metal ions tested, such as Zn2+, Co2+, and Al3+ showed little depressive effects on the lipase stability. These results indicated that the isolated lipase showed significant tolerance towards metal ions which are toxic to the most enzymes at very low concentrations. Other reports have shown similar and conflicting results (Boran and Ugur 2010; Dandavate et al. 2009; Ebrahimpour et al. 2008; Esakkiraj et al. 2014; Mehta et al. 2012; Wu et al. 2013; Zaliha et al., 2006). The observed discrepancies are probably due to the nature of the microorganisms and their produced enzymes.
After incubation with EDTA, about 93.29 % activity was retained (Table S9), indicating the lipase was not a metalloenzyme, but the slight decrease in enzyme activity may be due to the chelating of the present useful cations for lipase stability. In addition, the presence of SDS led to only marginal reduction in lipase stability. PMSF as the serine inhibitor, at 5 mM concentration decreased the lipase activity that is related to possible reaction of PMSF with the effective serine residues in lipase stability. Urea concentration when raised to 5 mM stimulated the lipase activity probably through its impact on lipid–water interface. Also, it was found that 2-mercaptoethanol increased the lipase stability, more than threefold, interestingly. It seems that 2-ME by inhibiting the oxidation of free sulfhydryl residues and thus maintaining them in a reduced form, enhances the lipase stability. Moreover, all the tested surfactants significantly enhanced lipase stability. There was 472 % increment in lipase activity when pre-incubated with Tween 80, followed by 459 and 367 % in Triton-X100 and CTAB, respectively (Table S9). The positive effect of surfactants could be due to the surfactant’s role in decreasing the surface tension of the liquid and also preventing the aggregation of the enzyme and hence increase the enzyme stability. It is also likely that surfactants increase the accessibility of the substrates to enzyme. The high tolerance of lipase from A. salilacus SR-079 Halo in various surfactants suggests for its application as an additive in detergent industry. These results are consistent with findings from other studies (Boran and Ugur 2010; Dandavate et al. 2009; Esakkiraj et al. 2014; Khunt and Pandhi 2012; Wu et al. 2013).
Substrate specificity
The substrate specificity test showed p-NPP (C16) was most efficiently hydrolyzed by the lipase (Fig. S2). A trend of preferential specificity towards p-NP esters with longer acyl chain lengths is clearly evident. Enzyme activity declined with substrates having shorter chain length, reaching less than 20 % activity with p-NPA (C2). This preference for long-chain fatty acid esters was shown in most lipases (Dheeman et al. 2011; Li et al. 2014).
Application of the lipase for long-chain esters production
Esters of C16–C18 fatty acids with various alcohols are the common component of cleaning and washing products, cosmetics, pharmaceuticals, perfumes, fragrances, air fresheners, polishes, and waxes. Long-chain alcohols and oleic acid have poor solubility in water, and therefore, for increased yields of ester, organic solvents should be selected as reaction media. Since organic solvents produce various physico-chemical effects on enzyme molecules, suspension of enzymes in organic solvents results in conformational changes and thereby the specificity of substrates (Sinha and Khare 2014). For lipase, such changes were usually effectively overcome by the halophilic and organic solvent-tolerant features of the enzyme. Considering that the studied lipase was stable in organic solvents and that its preferential specificity towards long-chain substrates, it was deemed likely to be very suitable for long-chain esters production in organic media, thus its potential for the esterification of oleic acid with 3 alcohols in various organic media was tested.
As shown in Fig. 4, the formation of ester from decanol was maximally obtained in the organic solvent of n-hexane by 80 % conversion after 6 h. In the other non-aqueous solvent media, i.e., acetonitrile, chloroform, and acetone, the lipase was able to esterify 62, 44, and 39 % of oleic acid with decanol, respectively, at the same time. Esterification using two other alcohols also found significant results, so that the reaction efficiency with octanol and butanol in hexane was 65 and 37 %, respectively. Although the reaction conditions were not optimized, SR-079 Halo lipase exhibited significant potential for efficiently esterification of oleic acid in comparison with the other lipases studied (Dheeman et al. 2011; Rao and Divakar 2001). Hence, lipase from A. salilacus SR-079 Halo may prove to be promising in esterification and standardization of reaction parameters may improve its efficiency as a biocatalyst in this process to obtain higher ester yield. This study formed the basic trials conducted to examine the feasibility of halophilic lipases for ester production.
Conclusion
In recent years, the ability of the halotolerant/halophilic bacteria to grow and produce enzymes over a very wide range of salinities, temperature, and pH make them very attractive for research and for the isolation of novel enzymes with unusual properties. An enhanced efficiency in non-aqueous media is always desirable in new lipases and the advantages of halophilic lipases might be exploited for synthesis reactions in organic solvents. In this study, the lipase from A. salilacus SR-079 Halo displayed thermostable, alkali-stable, and halophilic properties, highlighting its great tolerance towards various surfactants, metal ions, and organic solvents, allowing potential application as a biocatalyst in biotechnological processes in various industries, in organic chemistry, and in detergent formulation. Using statistical approach, the lipase production could be enhanced from 174.8 U L−1 in unoptimized media to 856.9 U L−1 giving 4.9-fold increase in lipase production. In addition, the lipase has been shown to be potentially useful for ester production in organic media. These results make the lipase more potentially valuable for biotechnological applications in non-aqueous catalysis.
References
Adlercreutz P (2013) Immobilisation and application of lipases in organic media. Chem Soc Rev 42:6406–6436
Aghaie-Khouzani M, Forootanfar H, Moshfegh M, Khoshayand MR, Faramarzi MA (2012) Decolorization of some synthetic dyes using optimized culture broth of laccase producing ascomycete Paraconiothyrium variabile. Biochem Eng J 60:9–15
Ahmed EH, Raghavendra T, Madamwar D (2010) An alkaline lipase from organic solvent tolerant Acinetobacter sp. EH28: application for ethyl caprylate synthesis. Bioresour Technol 101:3628–3634
Balan A, Ibrahim D, Abdul-Rahim R (2013) Organic-solvent and surfactant tolerant thermostable lipase, isolated from a thermophilic bacterium, Geobacillus thermodenitrificans IBRL-nra. Adv Stud Biol 5:389–401
Boran R, Ugur A (2010) Partial purification and characterization of the organic solvent-tolerant lipase produced by Pseudomonas fluorescens RB02-3 isolated from milk. Prep Biochem Biotechnol 40:229–241
Box GEP, Wilson KB (1951) On the experimental attainment of optimum conditions. J R Stat Soc Series B Stat Methodol 13:1–45
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Burkert JFM, Maugeri F, Rodrigues MI (2004) Optimization of extracellular lipase production by Geotrichum sp. using factorial design. Bioresour Technol 91:77–84
Cappuccino JG, Sherman N (2002) Microbiology: a laboratory manual, 6th edn. Benjamin Cummings, Menlo Park, pp 133–198
Cheng SW, Wang YF, Liu FF (2011) Optimization of medium compositions using statistical experimental design to produce lipase by Bacillus subtilis. Chem Biochem Eng Q 25:377–383
Dandavate V, Jinjala J, Keharia H, Madamwar D (2009) Production, partial purification and characterization of organic solvent tolerant lipase from Burkholderia multivorans V2 and its application for ester synthesis. Bioresour Technol 100:3374–3381
de Lourdes Moreno M, Pérez D, García MT, Mellado E (2013) Halophilic bacteria as a source of novel hydrolytic enzymes. Life 3:38–51
Dheeman DS, Frias JM, Henehan GT (2010) Influence of cultivation conditions on the production of a thermostable extracellular lipase from Amycolatopsis mediterranei DSM 43304. J Ind Microbiol Biotechnol 37:1–17
Dheeman DS, Henehan GTM, Frías JM (2011) Purification and properties of Amycolatopsis mediterranei DSM 43304 lipase and its potential in flavour ester synthesis. Bioresour Technol 102:3373–3379
Ebrahimpour A, Zaliha Abd Rahman RN, Ean Ch’ng DH, Basri M, Salleh AB (2008) A modeling study by response surface methodology and artificial neural network on culture parameters optimization for thermostable lipase production from a newly isolated thermophilic Geobacillus sp. strain ARM. BMC Biotechnol 8:15
Enache M, Kamekura M (2010) Hydrolytic enzymes of halophilic microorganisms and their economic values. Rom J Biochem 47:47–59
Esakkiraj P, Prabakaran G, Maruthiah T, Immanuel G, Palavesam A (2014) Purification and characterization of halophilic alkaline lipase from Halobacillus sp. DOI, Proc Natl Acad Sci India, Sect B. doi:10.1007/s40011-014-0437-1
Essamri M, Deyris V, Comeau L (1998) Optimization of lipase production by Rhizopus oryzae and study on the stability of lipase activity in organic solvents. J Biotechnol 60:97–103
Gupta N, Sahai V, Gupta R (2007) Alkaline lipase from a novel strain Burkholderia multivorans: statistical medium optimization and production in a bioreactor. Process Biochem 42:518–526
Haaland PD (1989) Experimental design in biotechnology. Marcel Dekker Inc, New York, pp 1–18
Hasan-Beikdashti M, Forootanfar H, Safiarian MS, Ameri A, Ghahremani MH, Khoshayand MR, Faramarzi MA (2012) Optimization of culture conditions for production of lipase by a newly isolated bacterium Stenotrophomonas maltophilia. J Taiwan Inst Chem Eng 43:670–677
Hun CJ, Abd Rahman RNZ, Salleh AB, Basri M (2003) A newly isolated organic solvent tolerant Bacillus sphaericus 205y producing organic solvent-stable lipase. Biochem Eng J 15:147–151
Jaeger K-E, Dijkstra BW, Reetz MT (1999) Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol 53:315–351
Ji Q, Xiao S, He B, Liu X (2010) Purification and characterization of an organic solvent-tolerant lipase from Pseudomonas aeruginosa LX1 and its application for biodiesel production. J Mol Catal B Enzym 66:264–269
Khunt M, Pandhi N (2012) Purification and characterization of lipase from extreme halophiles isolated from little rann of Kutch, Gujarat, India. Int J Life Sci Pharma Res 2:L55–L61
Kouker G, Jaeger KE (1987) Specific and sensitive plate assay for bacterial lipases. Appl Environ Microbiol 53:211–213
Laane C, Boeren S, Vos K, Veeger C (1987) Rules for optimization of biocatalysis in organic solvents. Biotechnol Bioeng 30:81–87
Li X, Qian P, Wu SG, Yu HY (2014) Characterization of an organic solvent-tolerant lipase from Idiomarina sp. W33 and its application for biodiesel production using Jatropha oil. Extremophiles 18:171–178
Mahanta N, Gupta A, Khare SK (2008) Production of protease and lipase by solvent tolerant Pseudomonas aeruginosa PseA in solid-state fermentation using Jatropha curcas seed cake as substrate. Bioresour Technol 99:1729–1735
Marques TA, Baldo C, Borsato D, Buzato JB, Celligoi MAPC (2014) Production and partial characterization of a thermostable, alkaline and organic solvent tolerant lipase from Trichoderma atroviride 676. Int J Sci Technol Res 3:77–83
Mehta A, Kumar R, Gupta R (2012) Isolation of lipase producing thermophilic bacteria: optimization of production and reaction conditions for lipase from Geobacillus sp. Acta Microbiol Immunol Hung 59:435–450
Mevarech M, Frolow F, Gloss LM (2000) Halophilic enzymes: proteins with a grain of salt. Biophys Chem 86:155–164
Meyer RH, Montgomery DC, Anderson-Cook CM (2009) Response surface methodology: process and product optimization using designed experiments. Wiley, Canada
Moshfegh M, Shahverdi AR, Zarrini G, Faramarzi MA (2013) Biochemical characterization of an extracellular polyextremophilic a-amylase from the halophilic archaeon Halorubrum xinjiangense. Extremophiles 17:677–687
Pérez D, Martín S, Fernández-Lorente G, Filice M, Guisán JM, Ventosa A, García MT, Mellado E (2011) A novel halophilic lipase, LipBL, showing high efficiency in the production of eicosapentaenoic acid (EPA). PLoS One 6:1–11
Rao P, Divakar S (2001) Lipase catalyzed esterification of α-terpineol with various organic acids: application of the Plackett–Burman design. Process Biochem 36:1125–1128
Rao PV, Jayaraman K, Lakshmanan CM (1993) Production of lipase by Candida rugosa in solid state fermentation. 2: medium optimization and effect of aeration. Process Biochem 28:391–395
Reis P, Holmberg K, Watzke H, Leser ME, Miller R (2009) Lipases at interfaces: a review. Adv Colloid Interface Sci 147–148:237–250
Schmid RD, Verger R (1998) Lipases: interfacial enzymes with attractive applications. Angew Chem Int Ed 37:1608–1633
Sharma D, Sharma B, Shukla AK (2011) Biotechnological approach of microbial lipases: a review. Biotechnol 10:23–40
Sinha R, Khare SK (2014) Effect of organic solvents on the structure and activity of moderately halophilic Bacillus sp. EMB9 protease. Extremophiles. doi:10.1007/s00792-014-0683-4
Treichel H, de Oliveira D, Mazutti MA, Di Luccio M, Oliveira JV (2010) A review on microbial lipases production. Food Bioprocess Technol 3:182–196
Verger R (1997) Interfacial activation of lipases: facts and artifacts. Trends Biotechnol 15:32–38
Volpato G, Rodrigues RC, Heck JX, Ayub MAZ (2008) Production of organic solvent tolerant lipase by Staphylococcus caseolyticus EX17 using raw glycerol as substrate. J Chem Technol Biotechnol 83:821–828
Wu G, Wu G, Zhan T, Shao Z, Liu Z (2013) Characterization of a cold-adapted and salt-tolerant esterase from a psychrotrophic bacterium Psychrobacter pacificensis. Extremophiles. doi:10.1007/s00792-013-0562-4
Yoo HY, Simkhada JR, Cho SS, Park DH, Kim SW, Seong CN, Yoo JC (2011) A novel alkaline lipase from Ralstonia with potential application in biodiesel production. Bioresour Technol 102:6104–6111
Zaks A, Klibanov AM (1988) The Effect of water on enzyme action in organic media. Biol Chem 263:8017–8021
Zaliha RN, Rahman RA, Baharum SN, Salleh AB, Basri M (2006) S5 lipase: an organic solvent tolerant enzyme. J Microbiol 44:583–590
Acknowledgments
This work was supported financially by the Grant No. 93-01-90-25247 from Tehran University of Medical Sciences, Tehran, Iran to M.A.F.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Driessen.
M. R. Khoshayand is the correspondence for the section of statistical experimental design.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Samaei-Nouroozi, A., Rezaei, S., Khoshnevis, N. et al. Medium-based optimization of an organic solvent-tolerant extracellular lipase from the isolated halophilic Alkalibacillus salilacus . Extremophiles 19, 933–947 (2015). https://doi.org/10.1007/s00792-015-0769-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-015-0769-7