Abstract
Golden-angle radial sparse parallel (GRASP) magnetic resonance imaging (MRI) is a recent MR image reconstruction technique which integrates parallel imaging, compressed sensing and golden-angle radial scheme to reconstruct the dynamic contrast-enhanced MRI (DCE-MRI) data. Conventionally, GRASP exploits non-uniform fast Fourier transform to grid and de-grid the golden-angle radial data and employs nonlinear conjugate gradient method to recover the unaliased images. GRASP performs gridding and de-gridding operations of golden-angle radial data in every iteration which increases the computational complexity of the conventional GRASP and takes a long image reconstruction time. In this paper, self-calibrated GRAPPA operator gridding (SC-GROG) followed by iterative soft thresholding (IST) is proposed for faster GRASP reconstruction of the golden-angle radial DCE-MRI data. In the proposed method, firstly SC-GROG maps the undersampled golden-angle radial data to a Cartesian grid and then reconstructs the solution image using the IST technique. The proposed method does not require gridding and de-gridding in each iteration; therefore, it is computationally less expensive as compared to the conventional GRASP reconstruction approach. The proposed method is tested for undersampled DCE golden-angle radial liver perfusion data (at acceleration factors 11.8, 19.1 and 30.9). The reconstruction results are assessed visually as well as using mean square error, line profiles and reconstruction time. The reconstruction results are compared with the conventional GRASP reconstruction. The results show that the proposed method provides better quality reconstruction results in terms of reconstruction time and spatio-temporal resolution than the conventional GRASP approach.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The main objective of dynamic contrast-enhanced (DCE) MRI is to perceive the enhancement pattern of the contrast agent in tissues to identify tumors and malignant tissues [1]. DCE-MRI requires quick data acquisition to offer a suitable balance of spatio-temporal resolution and volumetric coverage for clinical studies, e.g., in cardiac/liver perfusion MRI. In parallel magnetic resonance imaging (pMRI), undersampled Cartesian and non-Cartesian trajectories are used to accelerate the DCE-MRI data acquisition process while maintaining the spatio-temporal resolution; but the resultant image may have aliasing artifacts [2,3,4]. A variety of pMRI Cartesian and non-Cartesian reconstruction techniques have been developed in the recent past to remove these artifacts while maintaining the image quality, e.g., k-t SENSE [6], k-t GRAPPA [7] and SPEAR [8], etc.
Compressed sensing (CS) is another promising technique for rapid DCE-MR imaging [9]. CS methods use spatio-temporal sparsity constraint and reconstruct the unaliased images using nonlinear reconstruction techniques, e.g., nonlinear conjugate gradient [6], iterative soft thresholding [10] and projection over the convex set (POCS) [11].
DCE-MRI data acquisition in a non-Cartesian fashion is more advantageous than the Cartesian acquisition scheme. Non-Cartesian trajectories are less sensitive to motion artifacts and offer efficient exposure of the k-space in lesser time with high spatio-temporal resolution which makes it perfect for DCE-MRI [5]. A major limitation of the non-Cartesian trajectories is that they require a further processing step called “gridding” [14]. Gridding translates the acquired non-Cartesian data points to a Cartesian grid before employing fast Fourier transform (FFT) for image reconstruction [12]. Many methods have been presented in the recent past to re-sample the acquired non-Cartesian data onto a Cartesian map which include SC-GROG [14] and non-uniform fast Fourier transform (NUFFT) [13].
NUFFT was proposed by Jeffrey A. Fessler et al. [13] and is based on min–max interpolation to translate non-Cartesian data onto a Cartesian grid. NUFFT gridding requires appropriate kernel size, shape, and density compensation function (DCF) to re-sample the non-Cartesian data onto a Cartesian grid.
Self-calibrated GRAPPA operator gridding (SC-GROG), a recent gridding technique, maps the non-Cartesian points in k-space onto adjacent Cartesian points using self-calibrated weight sets. SC-GROG does not map all the points to Cartesian locations, and therefore leaves a few empty spaces in the gridded k-space data [14]. Furthermore, SC-GROG performs only local averaging upon the Cartesian grid rather than calculating and applying DCF like NUFFT [15].
Golden-angle radial scheme [5] is used for fast DCE-MRI data acquisition. In GRASP approach, each radial spoke is attained at a continuously increasing angle called golden angle, i.e., 111.246°. Golden-angle sampling allows a high degree of freedom in the imaging experiments because temporal resolution is selected retrospectively by combining a certain number of spokes in a single time frame. The trajectory formed for each time frame using this technique is unique as adjacent spokes are not repeated in any time frame. This results in incoherent streaking artifacts which makes the golden-angle radial trajectory particularly suitable for compressed sensing (CS) [16] reconstruction.
Golden-angle radial sparse parallel MRI (GRASP) [17] is a blend of parallel imaging (pMRI), compressed sensing (CS) and the golden-angle radial sampling scheme for DCE-MRI reconstruction. Acquired golden-angle radial data cannot be directly used for image reconstruction, as the radial data do not lie on a regular Cartesian map, and therefore it requires gridding. Conventionally, GRASP [8] uses NUFFT [13] to grid the golden-angle radial data, followed by nonlinear conjugate gradient (NLCG) algorithm [9] to reconstruct the solution (unaliased) image.
A major disadvantage of NLCG reconstruction scheme is its computational complexity, because it requires many iterations to find the solution image. Conventional GRASP performs gridding and de-gridding of the golden-angle radial data using NUFFT in each iteration to update the data consistency value [8]. This process is repeated until the consistency value meets the specified threshold cost. Gridding and de-gridding of the golden-angle radial data increase the reconstruction time and computational complexity of NLCG, thus making this type of reconstruction unsuitable for clinical applications, especially where the reconstructed data are required immediately after acquisition [5].
This paper proposes to use self-calibrated GRAPPA operator gridding (SC-GROG) [14] with iterative soft-thresholding (IST) technique [10] to reconstruct the golden-angle radial DCE-MRI data. The proposed method does not require gridding and de-gridding operations in each iteration and therefore it is computationally less complex as compared to conventional GRASP reconstruction approach.
2 Theory
2.1 Self-Calibrated GRAPPA Operator Gridding
Nicole et al. proposed a novel gridding technique named SC-GROG [19] which maps the acquired non-Cartesian k-space points onto a Cartesian grid. SC-GROG employs self-calibrated coil-by-coil weight sets to place the non-Cartesian data onto the Cartesian grid. SC-GROG maps the non-Cartesian data \(s\left( {k_{x} ,k_{y} } \right)\) to the adjacent Cartesian position \(s\left( {k_{x} + n\Delta k_{x} ,k_{y} + n\Delta k_{y} } \right)\) with a small shift in the x-direction (\(n\Delta k_{x}\)) and y-direction (\(n\Delta k_{y} )\), i.e.,
In Eq. (1), \(G_{n}\) represents coil-by-coil weight sets which can be expressed as a function of \(G_{x}\) and \(G_{y}\) with dimensions \(N_{c} \times N_{c}\) as:
Here, \(a\) and \(b\), respectively, represent the random shifts along \(k_{x }\) and \(k_{y }\) directions to place the acquired non-Cartesian points to the adjacent Cartesian positions. SC-GROG is incapable of finding all the probable adjacent moves in the x- and y-directions; thus, some empty positions are left in the gridded k-space data [15]. SC-GROG effectively grids the non-Cartesian k-space data onto the Cartesian places without any need of DCFs or other gridding parameters, e.g., window widths, convolution shapes or oversampling factors, while sustaining a low computational complexity [15].
2.2 Iterative Soft Thresholding (IST)
The vital concepts underlying the CS theory are sparsity and nonlinear reconstruction [8]. In the recent past, iterative soft-thresholding (IST) technique has been presented in CS MRI to recover the unaliased images from the undersampled MRI k-space data.
A simple iterative soft-thresholding algorithm used by Xiaobo [10] for CS-MRI reconstruction is defined as:
where \(x\) is a sparse complex MR image and \(x_{i}\) is the \(i^{\text{th}}\) element in \(x\). Since \(x\) is complex, a complex thresholding operator \(\lambda\) is used. Iterative soft-thresholding technique solves the problem when the solution is sufficiently sparse. The optimization problem with the regularization term for the IST algorithm is given as:
Here, \(F_{\text{u}}\) represents the Fourier transform with sampling mask, the solution image is represented by \(x\) and \(y\) is the acquired k-space data, \(\psi\) is the sparsifying transform and \(\lambda\) represents the regularization parameter. IST provides a fast and robust method in CS-MRI to reconstruct the MR images from the acquired highly undersampled data.
2.3 SC-GROG with IST (Proposed Method)
In this work, SC-GROG [14] in conjunction with IST [10] is proposed to reconstruct the GRASP DCE-MRI data. Figure 1 shows a block diagram of the proposed method (SC-GROG with IST).
GRASP reconstruction using the proposed method: Golden-angle radial data are first sorted into time frames and then gridded in a frame-by-frame manner using SC-GROG. The receiver coil sensitivity maps are estimated from the gridded data. Finally, image reconstruction is performed by combining the receiver coil sensitivity profiles and the gridded data using iterative soft thresholding
Initially, the GRASP DCE-MRI data is fixed into time frames by adjusting the Fibonacci number of the consecutive spokes into time frames. The resulting temporal frames are incoherent and enforce sparsity, as no two spokes are repeated in any time frame. The resulting sorted golden-angle radial data are gridded frame by frame using SC-GROG. SC-GROG has a property of mapping the non-Cartesian data to the adjacent Cartesian places and some empty spaces are left in the gridded k-space where the exact Cartesian values cannot be estimated. The resulting SC-GROG gridded data have incoherent artifacts that make GRASP DCE-MRI suitable for CS reconstruction.
Since GRASP exploits the joint sparsity in multi-coil images, the receiver coil sensitivity profiles estimated from SC-GROG gridded data using Walsh method are used to enhance the sparsity of the gridded data.
Finally, IST [10] is used to remove the artifacts from the SC-GROG gridded DCE-MRI golden-angle radial data to provide the solution image.
The proposed method has less computational complexity as compared to conventional GRASP because it uses SC-GROG to map the GRASP DCE-MRI data instead of NUFFT [14], which does not require some extra computations, e.g., density compensation function (DCF), convolution shapes and windows sizes. Also, the use of iterative soft thresholding to solve the convex regularized optimization problem without the need of gridding and de-gridding makes the SC-GROG with IST more efficient than the conventional GRASP approach.
3 Materials and Methods
The validity of the proposed method is examined on free breathing dynamic contrast-enhanced liver perfusion golden-angle radial MRI data downloaded from “http://cai2r.net/resources/software/grasp-matlab-code”. The data set was acquired using 12 channel receiver coils with 256 readouts and 600 spokes. In the golden-angle radial DCE-MRI data acquisition, multiple frame data are usually acquired to quantify the contrast agent uptake by the tissues. Golden-angle radial trajectories have the advantage of continuous data acquisition in 3D space and retrospective sorting into time frames to form 3D + t data. This is particularly possible if the number of spokes in each time frame belongs to a Fibonacci series which is defined as: \(F\left( {n + 2} \right) = F\left( n \right) + F\left( {n + 1} \right) {\text{where }}n \ge 0\). The golden-angle radial data sorted in time frames using Fibonacci series introduces incoherent artifacts that make golden-angle radial data suitable for CS recovery [18].
In the proposed method, firstly the acquired golden-angle radial DCE-MRI data are sorted into time frames according to the Fibonacci series [i.e., 1, 2, 3, 5, 8, 13, 21, 34….]. For example, if 600 spokes with 256 readouts are acquired at a golden-angle increment, then by selecting 21 spokes (a Fibonacci number) in each time frame, this will generate 28-time frames, i.e., 600/21 = 28 frames. The resulting frames are incoherent, as even not a single spoke is repeated in any sorted time frame. The Nyquist sampling requirement for this case is \(256 * \frac{\pi }{2} \approx 402\) projections, conforming to a simulated acceleration rate of 19.1, i.e., 402/21 = 19.1 [15].
After sorting the golden-angle radial DCE-MRI data, SC-GROG gridding is performed for each time frame individually until all the frames have been gridded on the Cartesian grid. SC-GROG maps each acquired golden-angle radial data point to the neighboring Cartesian position and leaves some empty spaces in the resulting Cartesian grid.
GRASP exploits multi-coil joint sparsity constraints to get the solution image. It has already been shown by Li. Feng et al. [5] that combining CS with parallel imaging enables a higher acceleration rate than CS or parallel MRI (pMRI) alone. In pMRI, the data are acquired using various receiver coils. Optimal SNR images can be reconstructed from the coil data when receiver coil sensitivity profiles are known [8]. In typical MR studies, the receiver coil sensitivity profiles are unknown and can be estimated from the acquired data. In the proposed method, the receiver coil sensitivity profiles are obtained using the Walsh method by temporal averaging of the GROG gridded data. Finally, the IST technique [10] is applied to get the solution image.
The value of the regularization parameter “\(\lambda\)“ in Eq. 3 affects the quality of the reconstructed images [4, 5, 17]; hence, the value of “\(\lambda\)“ for CS reconstruction is often determined empirically by trial and error. The value of the regularization parameter is adjusted until noise-like artifacts that arise due to undersampling are removed and the fine details in images are preserved. In this paper, the value of \(\lambda\) = 0.001 has been chosen empirically.
The proposed algorithm is implemented in MATLAB R2014a via Intel Core i7-4790, 3.70 GHz processor having 16 GB RAM.
4 Results and Discussion
This work presents an SC-GROG-based IST approach to get the solution image form the golden-angle radial dynamic contrast-enhanced MRI data. The reconstruction results using the proposed method are compared with the conventional GRASP [13] approach which uses NUFFT with NLCG to find the solution image.
The reconstruction of the golden-angle radial DCE-MRI liver perfusion data is performed at acceleration factors (AF) of 11.8, 19.1 and 30.9 that correspond to 34, 21 and 13 radial spokes per frame, respectively.
The reconstruction results are assessed visually as well as by mean square error (MSE) and comparing the line profiles of the arterial and venous phases. In this work, MSE and line profiles are used to find the difference between the images reconstructed using the conventional GRASP and the proposed method. Also, a comparison of the reconstruction time is performed for different acceleration factors.
Golden-angle fully sampled data cannot be acquired, as some empty spaces are always left between spokes due to golden-angle increment; therefore; no fully sampled reference images are available. The conventional GRASP reconstructed images (reconstructed with NUFFT with NLCG) are taken as a reference to calculate the mean square error.
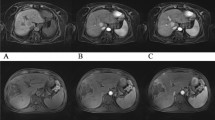
Figure 2 presents the reconstruction results of the proposed method and conventional GRASP at AF = 11.8, 19.1 and 30.9. In Fig. 2a–c, the upper row demonstrates the reconstruction results using the proposed method (SC-GROG with IST) and the bottom row displays the reconstruction results of conventional GRASP [13]. The resultant images show a contrast enhancement in the arterial and venous phases of liver perfusion.
Reconstruction results for the proposed method and conventional GRASP at different acceleration factors (AF): (a) AF = 11.8 with 17 temporal frames having 34 radial spokes/frame; (b) AF = 19.1 with 28 temporal frames having 28 radial spokes/frame; (c) AF = 30.9 with 46 temporal frames having 13 radial spokes/frame of DCE-MRI golden-angle radial data in pre-contrast, arterial and venous phases of liver perfusion
Figure 2a shows the reconstruction results of GROG with IST (the proposed method) and conventional GRASP (NUFFT with NLCG) obtained at acceleration factor 11.8 with 34 golden-angle radial spokes per frame with 17 temporal frames. Figure 2b shows the simulation results of the SC-GROG with IST (the proposed method) and conventional GRASP at acceleration factor 19.1 with 21 golden-angle radial spokes per frame and 28 temporal frames. Figure 2c displays the reconstruction results of the proposed method and conventional GRASP at acceleration factor 30.9 with 13 golden-angle radial spokes per frame and 46 temporal frames. The results illustrate that the proposed method reconstructs the DCE-MRI golden-angle radial liver perfusion data without any degradation in the image quality in the pre-contrast, arterial and venous phases.
Figure 3 shows the line profile comparison between the conventional GRASP reconstruction and the proposed method in arterial and venous phases of the liver perfusion data at AF = 11.8. The line profile displays the pixel intensity values taken from regularly spaced points along a line in an image. The line profiles of the images reconstructed via conventional and the proposed method show a high degree of similarity in terms of pixel intensity distribution which confirms that the proposed method reconstructs the solution images without degrading the image quality.
a A line profile comparison of the conventional GRASP reconstruction (top row) and the proposed method (bottom row) in the arterial and venous phases of liver perfusion data. There is no visual difference between the reconstruction results with the two methods. b The line profiles in the arterial phase (top row) and venous phase (bottom row) show a high degree of temporal correlation that confirms our findings in visual analysis
Table 1 provides a comparison of the reconstruction results for the proposed method and conventional GRASP in terms of MSE and reconstruction time at acceleration factors of 11.8, 19.1 and 30.9. All the reconstructions have been performed in MATLAB R2014a with Intel Core i7-4790, 3.70 GHz processor having 16 GB RAM.
Reconstruction time for 17 frames with the conventional method is 18.7 min, while with the proposed method, i.e., GROG-IST, the reconstruction time is 1.5 min, i.e., \(\approx\) 12 × faster than the conventional GRASP. Reconstruction time for 28 frames with the conventional method is 22.3 min, while with the proposed method reconstruction time is 2.1 min, i.e., \(\approx\) 10× faster. Similarly, the reconstruction time for 46 frames with the conventional method is 29.3 min, while with the proposed method the reconstruction time is 3.3 min, i.e., \(\approx\) 9× faster than the conventional method.
MSE values given in Table 1 show that the proposed method efficiently reconstructs the solution image in lesser time without any quality degradation. The MSE values at AF = 11.8, 19.1 and 30.9 are \(1 \times 10^{ - 5}\), \(2 \times 10^{ - 5}\) and \(2 \times 10^{ - 5}\), respectively. Small MSE values in Table 1 show that the proposed method accurately reconstructs the solution image in lesser time without any quality degradation.
The results given in Figs. 2, 3 and Table 1 prove that the proposed method successfully recovers the solution image with contrast enhancement in the arterial and venous phases of the golden-angle radial DCE-MRI liver perfusion data at different acceleration factors.
A limitation of the current work is that the gridding performed via NUFFT (in conventional GRASP) leads to sharp images, while the gridding performed with GROG (in the proposed method) leads to slightly blurred images, especially at higher acceleration factors. This is due to the fact that the signal-to-noise ratio is preferred over resolution in GROG density compensation filter [19]. This issue can be addressed by decreasing the acceleration factor that subsequently increases the number of spokes per frame. In the current work, the images reconstructed with the proposed method at lower acceleration factors, i.e., 11.8 and 19.1, offer high resolution (Fig. 2a, b), while the images get slightly blurred at acceleration factor 30.9, as can be seen in Fig. 2c.
The validity of the proposed method has been tested on a golden-angle radial liver perfusion data set. The proposed method shows an improvement in image reconstruction time than the conventional GRASP method by \(\approx\) 12×, \(\approx\) 10× and \(\approx\) 9× at acceleration factors 11.8, 19.1 and 30.9 with 17, 28 and 46 temporal frames having 34, 21 and 13 radial spokes per frame, respectively. If the radial data is acquired with a sufficient number of receiver coils and the signal-to-noise ratio is adequate, then GROG can be effectively used as a preprocessing step before iterative reconstruction methods for radial MRI [12]. Thus, the improvement in reconstruction performance via GROG is independent of a particular data set and therefore it can be easily extended to other applications such as prostate, neck perfusion and cardiac MRI [18].
5 Conclusion
In this paper, a new method (GROG with IST) is proposed as an alternative to conventional GRASP (NUFFT with NLCG). The proposed method efficiently recovers the solution image with high spatio-temporal resolution in lesser computational time as compared to conventional GRASP approach, as it does not require gridding and de-gridding in each iteration. The results show that the proposed method provides \(\approx\) 12×, \(\approx\) 10× and \(\approx\) 9× faster reconstructions at acceleration factors 11.8, 19.1 and 30.9 with 34, 21 and 13 radial spokes per frame, respectively. The proposed method successfully recovers the solution image with proper contrast enhancement in the arterial and venous phases of the golden-angle radial DCE-MRI liver perfusion MR data.
References
A.B. Rosenkrantz, C. Geppert, R. Grimm, T.K. Block, C. Glielmi, L. Feng, R. Otazo, J.M. Ream, M.M. Romolo, S.S. Taneja, D.K. Sodickson, H. Chandarana, J. Magn. Reson. Imaging 41(5), 1365–1373 (2015)
R. Otazo, E. Candès, D.K. Sodickson, Magn. Reson. Med. 73(3), 1125–1136 (2015)
H. Jung, K. Sung, K.S. Nayak, E.Y. Kim, J.C. Ye, Magn. Reson. Med. 61(1), 103–116 (2009)
C. Prieto, S. Uribe, R. Razavi, D. Atkinson, T. Schaeffter, Magn. Reson. Med. 64(2), 514–526 (2010)
L. Feng, L. Axel, H. Chandarana, K.T. Block, D.K. Sodickson, R. Otazo, Magn. Reson. Med. 75(2), 775–788 (2016)
L. Feng, M.B. Srichai, R.P. Lim, A. Harrison, W. King, G. Adluru, E.V.R. Dibella, D.K. Sodickson, R. Otazo, D. Kim, Magn. Reson. Med. 70(1), 64–74 (2013)
F. Huang, J. Akao, S. Vijayakumar, G.R. Duensing, M. Limkeman, Magn. Reson. Med. 54(5), 1172–1184 (2005)
L. Feng, R. Grimm, K.T. Block, H. Chandarana, S. Kim, J. Xu, L. Axel, D.K. Sodickson, R. Otazo, Magn. Reson. Med. 72(3), 707–717 (2014)
R. Otazo, D. Kim, L. Axel, D.K. Sodickson, Magn. Reson. Med. 64(3), 767–776 (2010)
X. Qu, W. Zhang, D. Guo, C. Cai, S. Cai, Z. Chen, Inverse Probl. Sci. Eng. 18(6), 737–758 (2010)
M. Kaleem, M. Qureshi, H. Omer, Appl. Magn. Reson 47, 13–22 (2016)
N. Seiberlich, F. Breuer, R. Heidemann, M. Blaimer, M. Griswold, P. Jakob, Magn. Reson. Med. 59(5), 1127–1137 (2008)
J.A. Fessler, B.P. Sutton, IEEE Trans. Signal Process. 51(2), 560–574 (2003)
N. Seiberlich, F. Breuer, M. Blaimer, P. Jakob, M. Griswold, Magn. Reson. Med. 59(4), 930–935 (2008)
I. Aslam, F. Najeeb, H. Omer, Appl. Magn. Reson. 49(1), 107–124 (2018)
M. Lustig, D. Donoho, J.M. Pauly, Magn. Reson. Med. 58(6), 1182–1195 (2007)
D.O. Walsh, A.F. Gmitro, M.W. Marcellin, Magn. Reson. Med. 43(5), 682–690 (2000)
“Center for Advanced Imaging Innovation and Research (CAI2R)” created by NYU school of Medicine USA. http://cai2r.net/resources/software/grasp-matlabcode. Accessed 19 Feb 2018
Y. Tian, K. Erb, G. Adluru, D. Likhite, A. Pedgaonkar, M. Blatt, E. DiBella, Med. Phys. 44(8), 4025–4034 (2017)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shahzadi, I., Aslam, I., Qazi, S.A. et al. Golden-Angle Radial Sparse Parallel MR Image Reconstruction Using SC-GROG Followed by Iterative Soft Thresholding. Appl Magn Reson 50, 977–988 (2019). https://doi.org/10.1007/s00723-019-01127-7
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00723-019-01127-7