Abstract
We have shown previously that the durum wheat TdSOS1 excludes Na+ and Li+ ions outside cells. Moreover, this protein is activated by Arabidopsis kinase SOS2 through phosphorylation. The elimination of both SOS2 phosphorylation sites and the auto-inhibitory domain produces a hyperactive TdSOS1∆972 form, which have a maximal activity independent from the regulatory SOS2/SOS3 complex. We demonstrated that the expression of TdSOS1 enhances salt tolerance of the transgenic Arabidopsis plants. In this study, we analyzed the response to H2O2-induced oxidative stress of the transgenic Arabidopsis expressing one of the two TdSOS1 forms. Firstly, we showed that the exogenous H2O2 treatment leads to an accumulation of SOS1 transcripts in leaves and roots of the durum wheat and also in the transgenic plants. These transgenic plants showed significant oxidative stress tolerance compared to control plants, especially the plants expressing the hyperactive form. This tolerance was manifested by high proline accumulation and low malonyldialdehyde (MDA), O2˙− and H2O2 contents. Furthermore, the activities of three essential ROS scavenging enzymes (SOD, CAT, and POD) were higher in the transgenic plants under oxidative stress, as compared to control plants. Taken together, these data suggested that TdSOS1 plays a crucial role in response to oxidative stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Various abiotic stresses like drought and salinity generate oxidative damage which occurs through the high accumulation in the reactive oxygen species (ROS), including the superoxide radical (O2˙−), singlet oxygen (1O2), hydrogen peroxide (H2O2), and hydroxyl radicals (OH·). Under stress condition, plant cells accumulate ROS at high level causing harmful effects such as degradation of proteins, inactivation of enzymes, genes alteration, and interfere in various pathways of metabolic importance (Choudhury et al. 2013). To moderate the concentration of ROS in plant cells and consequently to tolerate oxidative stress, plants have to assure a balance between ROS production and the activity of the detoxification systems including ascorbate-glutathione, thioredoxins (TRXs), glutathione S-transferase (GST), catalase (CAT), and superoxide dismutase (SOD). The first enzyme implicated in ROS scavenging system is SOD, which dismutates the superoxide radical (O2˙−) to the hydrogen peroxide (H2O2). There are three SOD isoenzymes in plant cells, which are Cu/Zn-SOD and Fe-SOD that are associated with chloroplasts and cytoplasm, and Mn-SOD isoenzyme associated with mitochondria (Bowler et al. 1994; Mittler 2002). Hydrogen peroxide is a toxic reactive oxygen species and has deleterious effects in plant tissue. It is converted by CAT and different classes of peroxidase (POD) to water and O2. Ascorbate peroxidases (APXs) are thought to be the most important H2O2 scavengers operating both in the cytosol and chloroplasts (Mhamdi et al. 2010). Catalase is a critical enzyme implicated in various physiological functions at different physiological stages or under environmental stresses (Mhamdi et al. 2010). Three CAT proteins CAT1, CAT2, and CAT3 were identified in Arabidopsis thaliana. Similar to Arabidopsis CAT1, TdCAT1 from durum wheat is involved in response to multiple abiotic stresses (Du et al. 2008; Feki et al. 2015a, b).
In plant cells, redox homeostasis is maintained through the activation of the antioxidant enzymes and also the production of non-enzymatic components like ascorbate, tocopherols, carotenoids, glutathione, and the osmoprotectant proline (Mhamdi et al. 2010; Suzuki et al. 2012; Rejeb et al. 2014).
Salinity has a negative impact on agriculture crop production (Qadir et al. 2014). High concentration of sodium in the soil leads to ionic stress, which is manifested by the toxicity of Na+ in the cytoplasm. Salinity tolerance is often achieved by the transport of toxic ions out of the cytoplasm into the soil, the apoplast or the vacuole through the plasma membrane active transport system (Mansour 2014). The plasma membrane Na+/H+ antiporter SOS1 (Salt Overly Sensitive 1) removes Na+ from cells and consequently plays a crucial role in maintaining cellular ion homeostasis under salt stress (Shi et al. 2000, 2003). It was reported that in various plant species, SOS1 regulates the transport of sodium from the root to the shoot via xylem (Shi et al. 2002; Oh et al. 2009; Olías et al. 2009). SOS1 was reported firstly in A. thaliana and then in various plant species such as rice, wheat, and tomato (Shi et al. 2000; Atienza et al. 2007; Olías et al. 2009; Feki et al. 2011). Transgenic plants expressing SOS1 from various plants tolerate salt stress compared to non-transformed plants (Shi et al. 2003; Atienza et al. 2007; Yue et al. 2012; Nie et al. 2015). Importantly, the truncated form of TdSOS1 that lack SOS2 phosphorylation site and the auto-inhibitory domain confers significant ionic stress tolerance to the transgenic Arabidopsis plants (Feki et al. 2014). It was reported that SOS1 gene expression is enhanced by salt stress (Shi et al. 2000; Atienza et al. 2007; Xu et al. 2008). In Arabidopsis, ROS and NADPH oxidase modulate SOS1 mRNA stability under salt stress (Chung et al. 2008). The regulation of the bread wheat TaSOS1 gene was evaluated by the functional analysis of two promoters regions in Arabidopsis under different abiotic stress conditions. In this heterologous system, the activities of these promoters were similar only under normal growth conditions (Feki et al. 2015a, b).
SOS1 is the direct target of SOS signaling pathway, which contains two other components SOS2 and SOS3. SOS3 is a calcium binding protein with four EF-hands (Liu and Zhu 1998) and SOS2 is Ser/Thr protein kinase (Liu et al. 2000). SOS3 binds to SOS2 via the auto-inhibitory FISL motif, and SOS2 became active (Halfter et al. 2000; Guo et al. 2001; Quintero et al. 2002). The complex SOS2/SOS3 relieves SOS1 from auto-inhibition through phosphorylation (Quintero et al. 2011). SOS2 regulates also the activity of proteins located on the tonoplast such as the Ca2+/H+ antiporter CAX1 and the vacuolar proton ATPase (V-ATPase), indicating that SOS2 plays a central role as a regulator of transport activities (Cheng et al. 2004; Batelli et al. 2007). In addition to the regulation of ion homeostasis in plant cells, the components of SOS signaling pathway play a role in some physiological mechanism involved in salt stress response (Wu et al. 1996; Ji et al. 2013). The connection between SOS signaling and ROS signaling involves the kinase SOS2, which interacts with nucleoside diphosphate kinase 2 and with catalases 2 and 3 (Verslues et al. 2007). Moreover, it has been shown that Arabidopsis SOS1 functions in oxidative stress through its interaction with RCD (radical-induced cell death 1) (Katiyar-Agarwal et al. 2006). To our knowledge, little is known about the implication of durum wheat TdSOS1 in oxidative stress response. In this study, the results showed that the expression of the wild-type TdSOS1 or the hyperactive TdSOS1∆972 form improves oxidative stress tolerance of the transgenic Arabidopsis plants. This tolerance was manifested by high proline accumulation, maintenance of the redox homeostasis, and activation of three essential ROS scavenging enzymes (SOD, CAT, and POD) in transgenic Arabidopsis plants.
Materials and methods
Plant materials and stress treatments
Durum wheat (Triticum turgidum L. subsp. durum) cultivar Om Rabia3 seeds provided by the Tunisian Agronomic Research Institute were germinated in sterile conditions and kept in darkness for 3 days. Seedlings were then transferred to containers with modified half-strength Hoagland’s solution (Epstein 1972) and kept growing under greenhouse conditions (25 ± 5 °C, 16-h photoperiod and 60 ± 10% relative humidity). After 10 days of growth, seedlings were subjected to oxidative stress by adding 15 mM H2O2. Shoots and roots were sampled after 2, 4, and 6 days of treatment and immediately stored at −80 °C for RNA extraction and TdSOS1 gene expression analysis.
Evaluation of transgenic Arabidopsis plants for oxidative stress
The Arabidopsis plants expressing the wild-type TdSOS1 (MW lines) or the hyperactive form TdSOS1∆972 (MH lines) were described previously by Feki et al. (2014). These transgenic plants were grown on MS agar medium (Murashige and Skoog, 1962), with Arabidopsis sos1-1 mutant transformed with the empty binary vector (called pBI line) and the wild-type Arabidopsis plants (gl1), for 1 week under light/dark cycle condition of 16 h light/8 h dark at 22 °C, and then transferred to MS agar medium containing or not 1 mM or 3 mM H2O2. After 10 days of stress application, the effect of H2O2-induced oxidative stress was evaluated firstly by measuring root elongation, secondary root number, and fresh weight of whole seedlings. Each experiment was repeated three times using four different plants of each line.
Semi-quantitative RT-PCR analysis
Total RNA was isolated from frozen shoots and roots of 14-day-old durum wheat seedlings treated or not with 15 mM H2O2 for 1, 2, and 4 days. Semi-quantitative RT-PCRs were carried out as described by Feki et al. (2011) and using the specific TdSOS1 gene primers SOSF (5′-GCAAGGGCCATCATATTTGAAAT-3′) and SOSR (5′-TTTTGAAGTCGCCACAACCT-3′). The wheat’s actin gene (GenBank Accession No. AY663392) was used as an internal gene expression. The two actin gene primers are AF (5′- CTGACGGTGAGGACATCCAGCCCCTTG-3′) and AR (5′-GCACGGCCTGAATTGCGACGTACATGG-3′).
To analyze the expression of the transgene in MW and MH lines treated or not to H2O2-induced oxidative stress, total RNA was isolated and the cDNA was synthesized using M-MLV reverse transcriptase (Invitrogen) and the oligo-dT (18 mer) primer at 37 °C for 1 h. An aliquot (1 μl) of the synthesized cDNA was used as a template for PCR amplifications with the specific TdSOS1 gene primers S1 (5′-GGCATCTTATTGGAAGGATT-3′) and S2 (5′-ATGGTCCGAGGGTTGAAGACAGCAA-3′), and TubF (5′-GTCCAGTGTCTGTGATATTGCA-3′) and TubR (5′-GCTTACGAATCCGAGGGTGCC-3′) for A. thaliana β-tubulin gene that was used for the normalization of the RT-PCR analysis. The PCR amplifications were performed in a final volume of 50 ml and following these PCR conditions: 2 min at 96 °C followed by 30 cycles for 30 s at 96 °C, 45 s at 55 °C, and 2 min at 72 °C, followed by a final extension of 5 min at 72 °C. The amplified products were resolved in 2% agarose gel.
Evaluation of oxidative stress parameters
The oxidative stress was evaluated through the determination of malonyldialdehyde (MDA) and H2O2 contents in stressed transgenic Arabidopsis plants. The MDA is considered an indicator of the degree of lipid peroxidation. The leaf tissues were homogenized in 0.1% (w/v) trichloroacetic acid (TCA) solution. After centrifugation at 15,000×g for 30 min, an aliquot of the supernatant was added to 0.5% (w/v) thiobarbituric acid (TBA) in 20% TCA solution. The mixture was heated at 90 °C for 30 min, and then cooled on ice. The MDA equivalent was calculated by measuring absorbance at 532 and 600 nm in reference to a MDA standard curve (Draper et al. 1993).
The quantification of H2O2 levels was determined following the method described by Velikova et al. (2000). Fresh leaf tissues of the transgenic Arabidopsis plants subjected or not to H2O2-induced oxidative stress were homogenized with 0.1% (w/v) TCA, and then centrifuged at 12,000×g for 15 min. An aliquot of the supernatant was added to 10 mM phosphate buffer (pH 7.0) and 1 M potassium iodide (KI), and the absorbance of this mixture was read at 390 nm. These two experiments were repeated three times using four different plants of each line.
Superoxide anion detection
The histochemical NBT (nitro blue tetrazolium) staining was used to detect the accumulation of superoxide anion in seedling of the transgenic Arabidopsis plants as described by Brini et al. (2011). The samples were placed in the NBT solution (0.1 mM NBT, 25 mM HEPES pH 7.6) and subjected to vacuum infiltration for 5 min. After the incubation under dark conditions for 2 h, the samples were treated with 80% ethanol and then observed under binocular loupe and photographed using an Olympus W120 digital still camera. NBT staining was visualized by blue formazan formation. This staining assay was repeated three times using three different plants from each condition.
Quantification of the free proline amount in plant leaves
Small pieces of fresh leaves were placed in 40% methanol solution and then heated at 85 °C for 60 min. After cooling, an aliquot of this solution was added to the solution containing acetic acid, orthophosphoric acid, and ninhydrin (1/1/1) and the mixture was heated at 100 °C for 30 min. The upper phase produced after the addition of 5 ml of toluene was collected in tubes and a pinch of Na2SO4 was added to each tubes. The absorbance of the organic phase was determined at 520 nm. The resulting values were compared with a standard curve constructed using known amounts of proline. This quantification was repeated three times using four different plants of each line.
Preparation of the enzymatic extract
Aliquots of frozen fresh transgenic Arabidopsis seedlings (0.5 g) subjected or not to H2O2-induced oxidative stress were ground to a fine powder with liquid nitrogen and homogenized in a cold solution containing 100 mM Tris-HCl buffer (pH 8.0), 10 mM EDTA (ethylenediaminetetraacetic acid), 50 mM KCl, 0.5 mM PMSF (phenylmethylsulfonyl fluoride), and 2% (w/v) PVP (polyvinylpyrrolidone). The homogenate was centrifuged at 14,000×g for 30 min at 4 °C, and the supernatant was used for determination of the antioxidative enzyme activities. Protein concentration was determined according to Bradford (1976).
Antioxidant enzyme activities
Catalase activity was determined according to Aebi (1984), by monitoring the disappearance of H2O2. An aliquot of crude enzyme extract was added to the reaction mixtures containing 50 mM phosphate buffer (pH 7), 30 mM H2O2. Changes in optical density (OD) of the reaction solution at 240 nm were recorded every 20 s.
The activity of POD was determined according to Maehly and Chance (1954) by the guaiacol oxidation method. An aliquot of crude enzyme extract was added to 50 mM phosphate buffer (pH 7), 20 mM guaiacol, and 40 mM H2O2. Changes in OD of the reacted samples at 470 nm were recorded every 20 s.
SOD activity was determined by measuring the percentage of inhibition of the pyrogallol autoxidation (Marklund and Marklund 1974). An aliquot of crude enzyme extract was added to 10 mM pyrogallol in Tris-cacodylic acid-diethylenetriaminepentaacetic acid buffer (pH 8.2). The rate of autoxidation was taken from the increase in OD of the reaction solution per minute. The activities of SOD, CAT, and POD were expressed as units mg−1 protein. One unit of SOD was defined as the enzyme quantity required causing 50% inhibition of the rate of the pyrogallol autoxidation in comparison with tubes lacking the plant extract. One unit of CAT was defined as μmol ml−1 H2O2 decomposed per minute. One enzyme unit of POD is defined as change in one unit of absorbance min−1. These activity analyses were performed using four different plants of each line and were repeated four times.
Statistical analysis
All data were subjected to one-way ANOVA implemented in the SPSS software 14, and treatment means separations were performed using Student’s t test. Comparisons with P values of <0.05 were considered significantly different.
Results
TdSOS1 transcript levels in response of durum wheat to H2O2-induced oxidative stress
To study the role of TdSOS1 in response of durum wheat to oxidative stress, the expression profile of this gene was analyzed by RT-PCR analysis on shoots and roots of durum wheat cultivar Om Rabia3 exposed or not to 15 mM H2O2 at different times of stress (0, 1, 2, and 4 days). Under standard conditions, basal TdSOS1 expression levels were detected on different organs of the non-treated wheat seedlings. TdSOS1 transcript level increased and reached a maximum in shoot and root subjected to H2O2-induced oxidative stress for 2 days, and then it declined rapidly after 4 days of treatment. In all these organs, this decrease was significant and the TdSOS1 expression levels were lower than those under control conditions (Fig. 1). This decrease could be explained by post-transcriptional regulation. All these data suggested that TdSOS1 may be involved in short-term oxidative stress response of durum wheat plants.
Analysis of the involvement of the two durum wheat TdSOS1 forms in oxidative stress response
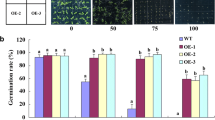
To evaluate the implication of the wild-type TdSOS1 and the truncated TdSOS1∆972 form in oxidative stress response, 1-week old seedling of the two MW lines expressing TdSOS1 and the two MH lines expressing TdSOS1∆972 were transferred to MS medium containing 1 or 3 mM H2O2 and kept in stress for 10 days with the control plants (pBI line and the wild-type Arabidopsis gl1). Under normal conditions, the growth of all these transgenic Arabidopsis lines was almost similar to the control plants. However, the addition of H2O2 to MS agar medium affected dramatically the growth of pBI line compared to the others transgenic lines. Thus, the transgenic lines are more tolerant than pBI line in the presence of oxidative stress. It is worth to note that TdSOS1∆972 transgenic lines showed better tolerance phenotype than TdSOS1 transgenic lines (Fig. 2a). These observations were confirmed by measuring the root elongation, secondary root number, and fresh weight of the control and the transgenic plants under these oxidative stress conditions. Without oxidative stress treatment, the root elongation of these Arabidopsis lines was similar. The addition of 1 mM H2O2 to MS medium decreased spectacularly the root elongation of pBI line, and this reduction was about 65%. By contrast, the roots of MW and MH lines showed growth improvement compared to pBI line. Interestingly, the highest root growth rate was observed on the two MH lines, which was reduced by H2O2 about 40%, and similar to the wild-type Arabidopsis gl1. High concentration of H2O2 (3 mM) in the medium blocked dramatically root growth of all these Arabidopsis lines (Fig. 2b). Under these conditions, we determined the number of secondary root of each Arabidopsis line. H2O2-induced oxidative stress affected the growth of secondary roots of the pBI and the other transgenic lines. This decrease was pronounced in pBI line compared to MW and MH lines. In addition, the number of secondary roots of MH1 and MH6 lines was higher than the one of MW1 and MW7 (Fig. 2c).
Response of transgenic Arabidopsis plants expressing the wild-type TdSOS1 or the truncated TdSOS1∆972 form to oxidative stress. a Effect of H2O2-induced oxidative stress on growth of transgenic Arabidopsis lines expressing TdSOS1 (MW1 and MW7) or TdSOS1∆972 (MH1 and MH6) and the control plants (pBI and wild-type Arabidopsis gl1). This was compared to the growth of the same seedlings placed in normal MS medium (control). After germination on normal MS agar medium for 7 days, seedlings were then transferred to MS medium supplemented with 0, 1, or 3 mM H2O2. Photographs were taken after 10 days of stress application. Determination of root elongation (b), secondary root number (c), and fresh weight (d) of these plants treated or not (control) with 1 or 3 mM H2O2. Values are means ± SE (n = 4). Asterisks indicate statistically significant greater mean values compared to Arabidopsis transformed with the empty binary vector (pBI line) (P < 0.05)
We also measured the whole seedling weight of these four transgenic lines in comparison with the control plants. In the absence of stress, the fresh weight (FW) was similar for all Arabidopsis plants. On the other hand, FW reduced more than 50% only in the case of pBI line exposed to 1 or 3 mM H2O2. Nevertheless, the slight reduction was observed in the transgenic Arabidopsis MH1 and MH6 lines, which was similar to control wild-type Arabidopsis gl1 plants treated with 3 mM H2O2 (Fig. 2d).
All these results showed that the transgenic Arabidopsis plants expressing the wild-type TdSOS1 or the hyperactive TdSOS1∆972 respond differently to the control pBI plants, with significant tolerance phenotype to oxidative stress.
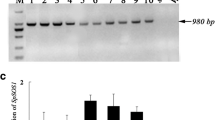
To gain more insights into the implication of the TdSOS1 gene in the tolerance to oxidative stress, the expression profiles of TdSOS1 were analyzed in the two different transgenic Arabidopsis MH and MW lines and the control pBI plants treated with 1 mM H2O2. The semi-quantitative RT-PCR showed that under standard conditions, TdSOS1 expression was higher in MH lines compared to MW lines, but it was absent in pBI line. After H2O2 treatment, the level of TdSOS1 transcript increased in both MW and MH lines (Fig. 3). Thus, the oxidative tolerance of these transgenic Arabidopsis plants was related in part with the expression of the durum wheat TdSOS1 and its truncated TdSOS1∆972 form.
Abundance of TdSOS1 transcript in control plant (pBI) and in transgenic Arabidopsis expressing TdSOS1 (MW1 and MW7) or TdSOS1∆972 (MH1 and MH6) exposed or not (0 mM H2O2) to oxidative stress (1 mM H2O2) for 10 days. Amplification in the absence of template (hyphen-minus). A 0.5-kb of β-tubulin gene fragment was amplified by RT-PCR as an internal control
Proline amount in two different TdSOS1 and TdSOS1∆972 transgenic Arabidopsis lines
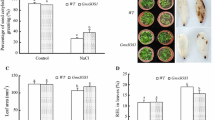
We quantified the amount of proline that is considered as a major index implicated in abiotic stress response, in leaves of the control and transgenic Arabidopsis plants. As shown in Fig. 4, the same proline levels were observed in all these plants. However, H2O2-induced oxidative stress generated an accumulation of proline in both MW and MH lines compared to pBI line. But, the increase of proline amount was higher in MH lines than in MW lines, which was about 2-fold in MH lines and the wild-type Arabidopsis gl1 plants.
Proline contents in leaves of transgenic Arabidopsis plants expressing TdSOS1 (MW1 and MW7) or TdSOS1∆972 (MH1 and MH6) and the control plants (pBI and the wild-type Arabidopsis gl1 plant) under normal growth condition (C) or exposed to oxidative stress (1 mM H2O2) (S) for 10 days. Values are means ± SE (n = 4). Asterisks indicate statistically significant greater mean values compared to Arabidopsis transformed with the empty binary vector (pBI line) (P < 0.05)
Maintenance of the redox homeostasis in transgenic Arabidopsis plants
The production of the superoxide anion was monitored in the control pBI plants and the two different transgenic Arabidopsis lines by NBT staining assay. Under normal growth conditions, a weak NBT staining was similarly detected in leaves of these lines. Exposed to 1 mM H2O2 for 10 days, an intense NBT staining was detected almost in the whole leaves of pBI line. By contrast, O2˙− accumulation detected by NBT stain was minor in transgenic Arabidopsis lines, especially in MH1 line (Fig. 5a).
Revelation of O2 − accumulation by NBT staining (a), determination of MDA concentrations (b), and H2O2 contents (c) in leaves of 7-day-old seedlings of control plants (pBI and wild-type Arabidopsis gl1) and the four transgenic Arabidopsis lines (MW1, MW7, MH1, and MH6) subjected or not (C) to oxidative stress (S) for 10 days. Values are means ± SE (n = 4). Asterisks indicate statistically significant greater mean values compared to Arabidopsis transformed with the empty binary vector (pBI line) (P < 0.05)
The second method used to measure the cellular oxidative stress of these transgenic plants was based on MDA production during the oxidation of polyunsaturated fatty acids. Without oxidative stress, MDA contents were similar in the control and transgenic plants. The presence of 1 mM H2O2 in MS medium increased appreciably the MDA level in pBI line. This increase was about 1.5-fold relative to the non-treated plants. However, the increase was slight in MW lines, whereas the MDA levels were unchanged in MH lines and gl1 plants (Fig. 5b).
We determined also the H2O2 levels in these plants under control and stressed conditions. Contrary to the stressed conditions, the H2O2 levels were almost similar in all these plants. After 10 days of oxidative stress, pBI line showed a marked increase in H2O2 level, which was about 2-fold relative to the non-treated plants. Nevertheless, the amount of H2O2 in MW and MH lines is still lower as compared to pBI line. It is worth to note that the H2O2 levels in MH lines were similar to the wild-type Arabidopsis gl1 plants (Fig. 5c).
All these results suggested that the expression of TdSOS1 or the truncated TdSOS1∆972 form limits the accumulation of ROS in transgenic Arabidopsis plants and consequently improves tolerance to oxidative stress.
The expression of two durum wheat TdSOS1 forms activates the activities of antioxidant enzymes
The activities of three essential ROS scavenging enzymes SOD, CAT, and POD were evaluated in the different transgenic Arabidopsis lines and the control plants treated or not with 1 mM H2O2 for 10 days. The application of oxidative stress increased SOD activities in all these plants. However, the highest SOD increase was observed in MH1, MH6 lines, and wild-type Arabidopsis plants, which was more than 2-fold relative to the non-treated plants (Fig. 6a). Similarly, the two MH lines and gl1 plant showed a higher increase of CAT and POD activities compared to pBI line and the two MW lines exposed to oxidative stress. It is worth to note that the activity of these two antioxidant enzymes in MW lines were higher than pBI line (Fig. 6b, c).
Analysis of the activities of the three antioxidant enzymes SOD (a), CAT (b), and POD (c) in control plants (pBI and wild-type Arabidopsis gl1) and transgenic Arabidopsis plants expressing TdSOS1 (MW1 and MW7) or TdSOS1∆972 (MH1 and MH6) subjected or not (C) to oxidative stress (1 mM H2O2) (S) for 10 days. Values are means ± SE (n = 4). Asterisks indicate statistically significant greater mean values compared to Arabidopsis transformed with the empty binary vector (pBI line) (P < 0.05)
Discussion
Under salinity, the increase of cytoplasmic Na+ and the diminution of K+ impair cell metabolisms by imposing an osmotic stress and by the Na+ toxicity in the cytoplasm. Ion transport via cell membranes is the basic factor determining salinity tolerance (Volkov 2015).
It has been showed that SOS1 protein is the first to be able to transport sodium out of cells under salt stress (Ji et al. 2013). In the absence of stress, SOS1 is maintained in resting state through the intra-molecularly interaction of the auto-inhibitory domain with an adjacent domain of SOS1. In the presence of salt stress, SOS3 activates SOS2 and the complex SOS2/SOS3 releases SOS1 from auto-inhibition via phosphorylation (Halfter et al. 2000; Guo et al. 2001; Quintero et al. 2002, 2011). It was reported recently that Arabidopsis 14-3-3 proteins regulate SOS pathway in plants through interaction with the SOS2 catalytic domain. Indeed, salt stress reduces this interaction and leads to an increase in SOS2 kinase activity generating salt stress tolerance (Zhou et al. 2014). It was reported that the photoperiodical and circadian clock switch Gigantea (GI) sequesters with SOS2, preventing consequently the activation of SOS1 in the absence of stress (Kim et al. 2013).
So far, various studies demonstrated the role of SOS pathway in the regulation of ion homeostasis and salt tolerance in plants. The overexpression of the direct target of SOS pathway SOS1 proteins in transgenic plants conferred tolerance to salt stress (Shi et al. 2003; Atienza et al. 2007; Yue et al. 2012; Nie et al. 2015). In a previous report, we have shown that the heterologous expression of the durum wheat TdSOS1 in yeast strain lacking endogenous Na+ efflux proteins showed complementation of the Na+ and Li+ sensitive phenotype. TdSOS1 contains an N-terminal transmembrane region encompassing 12 putative transmembrane domains followed by a long hydrophilic C-terminal part. The C-terminal part containing the auto-inhibitory domain and SOS2 phosphorylation site is essential for its activation by the Arabidopsis kinase SOS2. The elimination of this part generates a hyperactive TdSOS1∆972 form, which bypass the need of activation by SOS2 protein and becomes a constitutively active exchanger (Feki et al. 2011). It has been shown that the expression of the wild-type TdSOS1 or the hyperactive TdSOS1∆972 form enhances ionic stress tolerance of the transgenic Arabidopsis plants (Feki et al. 2014). Until recently, little is known about the role of durum wheat TdSOS1 in oxidative stress response. To this end, we further analyzed the response to oxidative stress of the transgenic Arabidopsis expressing separately the two TdSOS1 forms. In this study, we showed that the expression of TdSOS1 increased in leaves and roots of durum wheat and also in transgenic Arabidopsis lines exposed to H2O2-induced oxidative stress, suggesting that TdSOS1 may be involved not only in salt stress but also in oxidative stress. These data corroborate other findings which showed that in 35S:SOS1 transgenic Arabidopsis plants, ROS especially H2O2 produces significant accumulation of SOS1 mRNA. It was suggested that this accumulation was due to the increase of the stability of SOS1 transcripts under stress conditions. However, Arabidopsis SOS1 mRNA is unstable in the absence of stress, and the cis-element required for this instability is localized in 500 pb region at the 3′ end of SOS1 mRNA (Chung et al. 2008). Thus, the elimination of 1.2 kb in the 3′ end of TdSOS1 could explain the high amount of TdSOS1 transcripts in TdSOS1∆972 transgenic plants compared to those expressing the wild-type TdSOS1 form under normal growth conditions.
The function of durum wheat plasma membrane Na+/H+ antiporter TdSOS1 in oxidative stress response was demonstrated in the transgenic Arabidopsis lines expressing the wild-type TdSOS1 or its truncated TdSOS1∆972 form. The results showed that the expression of one of these two TdSOS1 forms enhanced tolerance to H2O2-induced oxidative stress compared to control plants (pBI line). Nevertheless, this tolerance was significantly greater for transgenic plants expressing the hyperactive TdSOS1∆972 form than those expressing the wild-type TdSOS1 form. These results are in agreement with other studies which showed that the hyperactive TdSOS1∆972 form enhances considerable tolerance to salt stress in Arabidopsis sos1-1 plants and also in yeast system (Feki et al. 2011, 2014). Indeed, this form is active independently of the presence of Arabidopsis SOS2/SOS3 complex (Feki et al. 2011). In addition, it is possible that the activation of the wild-type TdSOS1 form in plants requires additional events beside phosphorylation by SOS2/SOS3 complex, like the phosphorylation by mitogen-activated protein kinase (MPK6). Indeed, SOS1 is a target of the phospholipase D (PLD) signaling pathway in response to salt stress. In Arabidopsis, the activation of PLDα1 generates accumulation of the lipid second messenger phosphatidic acid, which in turn activates MPK6 (Yu et al. 2010).
In this study, the tolerance of the transgenic Arabidopsis lines to oxidative stress was manifested by higher growth rate and proline accumulation. Similarly, the exogenous H2O2 treatment leads to high accumulation of proline in maize seedlings (Yang et al. 2009). It was reported that the accumulation of proline may participate to the scavenging of ROS in addition to its role as an osmolyte (Rejeb et al. 2014). Moreover, the evaluation of some oxidative stress parameters showed that the transgenic Arabidopsis plants expressing one of the two TdSOS1 forms respond differently to the control plant (pBI line). Indeed, the degree of oxidative stress was significantly gentler in transgenic Arabidopsis plants than that of control plants. Thus, TdSOS1 protein plays a crucial role in response to oxidative stress of the transgenic Arabidopsis plants. In other findings, it was showed that the mutation of AtSOS1 in Arabidopsis sos1-1 mutant plants generates an excessively accumulation of ROS under salt stress, and plants become sensitive to apoplastic ROS imposed by H2O2, like rcd1 mutants. It was demonstrated that the implication of Arabidopsis SOS1 protein in oxidative stress is mediated by RCD1 protein, which is a regulator of oxidative stress responses. Indeed, under salt and oxidative stresses, SOS1 interacts with RCD1 via the cytoplasmic C-terminal part. The two domains of SOS1 protein that are localized between the amino acids 440-806 and 936-1103 are essential for this interaction (Katiyar-Agarwal et al. 2006). Sequence analysis of the C-terminal part of TdSOS1 revealed high sequence homology with these two domains. Thus, it is possible that TdSOS1 is recognized by RCD1 protein in the transgenic Arabidopsis plants under oxidative stress. It seems that the first domain is implicated in this interaction, because the second domain is absent in the truncated TdSOS1∆972 form.
Arabidopsis SOS1 controls the expression of some genes that are important for oxidative stress response, like FeSOD and ENH1 genes (Katiyar-Agarwal et al. 2006). In this study, we showed that the transgenic lines expressing the wild-type TdSOS1 form or the hyperactive TdSOS1∆972 form had considerably higher activities of three essential ROS scavenging enzymes SOD, CAT, and POD under H2O2-induced oxidative stress, as compared to control pBI lines. It is worth to note that these activities in TdSOS1∆972-transgenic lines were higher than those in TdSOS1-transgenic lines, but almost similar to those in the wild-type Arabidopsis gl1 plants. Thus, these data can be correlated with the low O2˙− and H2O2 levels in TdSOS1∆972-transgenic lines. These data indicate that the expression of the hyperactive TdSOS1∆972 form leads to a more efficient antioxidant activity in transgenic Arabidopsis plants that counteract the oxidative stress evoked with exogenous H2O2 treatment.
In summary, we showed in this study that the expression of the wild-type TdSOS1 form or the hyperactive form in the transgenic Arabidopsis plants improves oxidative stress tolerance. This tolerance was accompanied by high proline levels in leaves of the transgenic plants, and low accumulation of ROS like O2˙− and H2O2. Moreover, the activities of SOD, CAT, and POD are higher in the transgenic plants as compared to control pBI plants. All these results reveal a previously uncharacterized function of durum wheat TdSOS1 in oxidative stress tolerance.
References
Aebi H (1984) Catalase in vitro. Method Enzymol 105:121–126
Atienza MJ, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, Quintero FJ (2007) Conservation of salt overly sensitive pathway in Rice. Plant Physiol 143:1001–1012
Batelli G, Verslues PE, Agius F, Qiu QS, Fujii H, Pan S, Schumaker KS, Grillo S, Zhu JK (2007) SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol Cell Biol 27:7781–7790
Bowler C, Van Camp W, Van Montagu M, Inzé D (1994) Superoxide dismutase in plants. Crit Rev Plant Sci 13:199–218
Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brini F, Yamamoto A, Jlaiel L, Takeda S, Hobo T, Dinh HQ, Hattori T, Masmoudi K, Hanin M (2011) Pleiotropic effects of the wheat dehydrinDHN-5 on stress responses in Arabidopsis. Plant Cell Physiol 52:676–688
Cheng NH, Pittman JK, Zhu JK, Hirschi KD (2004) The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J Biol Chem 279:2922–2926
Choudhury S, Panda P, Sahoo L, Panda SK (2013) Reactive oxygen species signaling in plants under abiotic stress. Plant Signal Behav 8:23681
Chung JS, Zhu JK, Bressan RA, Hasegawa Paul M, Shi H (2008) Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J 53:554–565
Draper HH, Squires EJ, Mahmoodi H, Wu J, Agarwal S, Hadley M (1993) A comparative evaluation of thiobarbituric acid methods for the determination of malondialdehyde in biological materials. Free Radic Biol Med 15:353–363
Du YY, Wang PC, Chen J, Song CP (2008) Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J Integrative Plant Biol 50:1318–1326
Epstein E (1972) Mineral nutrition of plants: principles and perspectives. John Wiley and Sons, New York
Feki K, Quintero FJ, Pardo JM, Masmoudi K (2011) Regulation of durum wheat Na+/H+ exchanger TdSOS1 by phosphorylation. Plant Mol Biol 76:545–556
Feki K, Quintero FJ, Khoudi H, Leidi EO, Masmoudi K, Pardo JM, Brini F (2014) A constitutively active form of a durum wheat Na+/H+ antiporter SOS1 confers high salt tolerance to transgenic Arabidopsis. Plant Cell Rep 33:277–288
Feki K, Brini F, Amar SB, Saibi W, Masmoudi K (2015a) Comparative functional analysis of two wheat Na+/H+ antiporter SOS1 promoters in Arabidopsis thaliana under various stress conditions. J Appl Genetics 56:15–26
Feki K, Kamoun Y, Mahmoud RB, Farhat-Khemakhem A, Gargouri A, Brini F (2015b) Multiple abiotic stress tolerance of the transformants yeast cells and the transgenic Arabidopsis plants expressing a novel durum wheat catalase. Plant Physiol Biochem 97:420–431
Guo Y, Halfter U, Ishitani M, Zhu JK (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13:1383–1400
Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci U S A 97:3735–3740
Ji H, Pardo JM, Batelli G, Van Oosten MJ, Bressan RA, Li X (2013) The salt overly sensitive (SOS) pathway: established and emerging roles. Mol Plant 6:275–286
Katiyar-Agarwal S, Zhu J, Kim K, Agarwal M, Fu X, Huang A, Zhu JK (2006) The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc Natl Acad Sci U S A 103:18816–18821
Kim WY, Ali Z, Park HJ et al (2013) Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat Commun 4:1352
Liu J, Zhu JK (1998) A calcium sensor homolog required for plant salt tolerance. Science 280:1943–1945
Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci U S A 97:3730–3734
Maehly AC, Chance B (1954) The assay of catalase and peroxidase. Meth Anal Biochem 1:357–424
Mansour MMF (2014) The plasma membrane transport systems and adaptation to salinity. J Plant Physiol 171:1787–1800
Marklund S, Marklund G (1974) Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur J Biochem 47:469–474
Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F, Noctor G (2010) Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J Exp Bot 61:4197–4220
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nie WX, Xu L, Yu BJ (2015) A putative soybean GmsSOS1 confers enhanced salt tolerance to transgenic Arabidopsis sos1-1 mutant. Protoplasma 252:127–134
Oh SJ, Kim YS, Kwon CW, Park HK, Jeong JS, Kim, JK SJ (2009) Overexpression of the Transcription Factor AP37 in Rice Improves Grain Yield under Drought Conditions. Plant Physiol 150(3):1368–1379
Olías R, Zakia E, Jun L, Paz alvarez D, Marin-Manzano, Mari Carmen M, Pardo JM, Andrés B (2009) The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ 32:904–916
Qadir M, Quillerou E, Nangia V, Murtaza G, Singh M, Thomas RJ, Drechsel P, Noble AD (2014) Economics of salt-induced land degradation and restoration. Nat Res Forum 38:282–295
Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99:9061–9066
Quintero FJ, Martinez-Atienza J, Villalta I, Jiang X, Kim WY, Ali Z, Fujii H, Mendoza I, Yun DJ, Zhu JK, Pardo JM (2011) Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc Natl Acad Sci U S A 108:2611–2616
Rejeb KB, Abdelly C, Savouré A (2014) How reactive oxygen species and proline face stress together. Plant Physiol Bioch 80:278–284
Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci 97:6896–6901
Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14:465–477
Shi H, Lee BH, Wu SJ, Zhu JK (2003) Overexpressing of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nature Biotechnol 21:81–85
Suzuki N, Koussevitzky S, Mittler R, Miller G (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35:259–270
Velikova V, Yordanov I, Edreva A (2000) Oxidative stress and some antioxidant system in acid rain treated bean plants: protective role of exogenous polyamines. Plant Sci 151:59–66
Verslues PE, Batelli G, Grillo S, Agius F, Kim YS, Zhu J, Agarwal M, Katiyar-Agarwal S, Zhu JK (2007) Interaction of SOS2 with nucleoside diphosphate kinase 2 and catalases reveals a point of connection between salt stress and H2O2 signaling in Arabidopsis thaliana. Mol Cell Biol 27:7771–7780
Volkov V (2015) Salinity tolerance in plants. Quantitative approach to ion transport starting from halophytes and stepping to genetic and protein engineering for manipulating ion fluxes. Frontiers in Plant Sci 6:1–25
Wu SJ, Ding L, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8:617–627
Xu H, Jiang X, Zhan K, Cheng X, Chen X, PArdo JM, Cui D (2008) Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch Biochem Biophy 473:8–15
Yang SL, Lan SS, Gong M (2009) Hydrogen peroxide-induced proline and metabolic pathway of its accumulation in maize seedlings. J Plant Physiol 166:1694–1699
Yu L, Nie J, Cao C, Jin Y, Yan M, Wang F, Liu J, Xiao Y, Liang Y, Zhang W (2010) Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol 188:762–773
Yue Y, Zhang M, Zhang J, Duan L, Li Z (2012) SOS1 gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio. J Plant Physiol 169:255–261
Zhou H, Lin H, Chen S, Becker K, Yang Y, Zhao J, Kudla J, Schumaker KS, Guo Y (2014) Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26:1166–1182
Acknowledgments
This study was supported by a grant from the Ministry of Higher Education, Scientific Research and Information Technology and Communication of Tunisia.
Author contributions
KF and FB designed the experiments, KF and ST performed the experiments, KF drafted the manuscript, and KM and FB critically revised the manuscript for important intellectual content and provided the final approval of the manuscript for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Human participants and/or animals are not involved in this research.
Additional information
Handling Editor: Bhumi Nath Tripathi
Rights and permissions
About this article
Cite this article
Feki, K., Tounsi, S., Masmoudi, K. et al. The durum wheat plasma membrane Na+/H+ antiporter SOS1 is involved in oxidative stress response. Protoplasma 254, 1725–1734 (2017). https://doi.org/10.1007/s00709-016-1066-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-016-1066-8