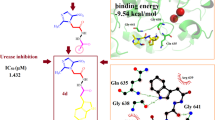
Abstract
Urease is an important virulence factor involved in the colonization and infection of gastric mucosa by Helicobacter pylori. In this work, the urease inhibitory activity of a series of γ-alkylidenebutenolides analogues of natural rubrolides is presented. The compounds were prepared from a commercial 3,4-dibromofuran-(5H)-2-one, as previously reported, including three new derivatives. The rubrolide analogues (at 500 µM) showed percentages of urease inhibition ranging from 20.7 to 99.3%. The most active compounds (IC50 from 111.5 to 306.0 µM) were shown to be more potent than hydroxyurea (IC50 844.4 µM), a standard urease inhibitor. Rubrolide analogues with phenolic hydroxyl groups revealed higher potency compared with other substances evaluated. Their physicochemical parameters (partition coefficient, molecular weight, hydrogen bonding acceptors, number of hydrogen bonding donor groups, number of rotatable bonds, and the number of aromatic bonds) related to general pharmacokinetic requirements showed drug-like properties for the evaluated rubrolide analogues. Docking studies suggest that the presence of hydroxy groups at the ortho position favors both the formation of reversible interactions with urease and the formation of a covalent adduct with the active site causing blocking of the enzyme, like what happens with catechol, its natural inhibitor. The high biological activity herein reported indicates that rubrolides constitute promising leads for the development of a new class of urease inhibitor drugs.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Urease is a nickel-containing metalloenzyme that occurs in a wide range of plants, fungi, and bacteria. It catalyzes the hydrolysis of urea into ammonia and carbon dioxide, a catalytic process of great medical importance [1,2,3]. Urease constitutes an important virulence factor involved in the pathogenicity of Helicobacter pylori, the causal agent of chronic gastric infections with a prevalence of two-thirds in the world population and a mortality rate of 2–4% [4]. Ammonia produced by urease activity is responsible for H. pylori enduring in the acidic stomach environment, allowing bacteria to colonize the gastric mucosa. In the gastric epithelium, ammonia causes direct cytotoxic damage, and monochloramine results from immunological oxidative bursts, generating mutagenic DNA effects. Because of this process, the infection induces an inflammatory response resulting in chronic gastritis, gastric and peptic ulcers, and gastric and duodenal cancer [5].
Currently, the association of a proton-pump-inhibitor with antibiotics, mainly amoxicillin and clarithromycin, is the first-line therapy for the treatment of H. pylori infections. However, this standard triple therapy has provided low success rates in eradicating H. pylori, mainly due to antimicrobial resistance. Prescription of multiple antibiotics to overcome bacterial resistance has been adopted, but with a high incidence of adverse effects that frequently led to the patient withdrawing from therapy and, consequently, contributing to the emergence of resistant bacterial strains [6]. Thus, the development of novel urease inhibitors drugs has become an important target to treat H. pylori infections [7]. Many chemical classes of natural products have been investigated for their urease inhibitory activity, and some of the active natural molecules have been used as models for the synthesis of urease inhibitor analogues [8, 9].
Marine organisms have attracted growing interest as a promising source of novel bioactive metabolites for drug discovery and development [10,11,12]. A variety of such bioactive compounds belong to chemical classes that have in common a butenolide ring in their structure, and their biological properties have attracted interest in the synthesis of analogues in the search for novel drugs [13,14,15,16,17]. Among these natural butenolides of marine origin, rubrolides constitute a class of natural γ-alkylidenebutenolides isolated from various species of marine ascidia. Some of these compounds (Fig. 1) were found to present various biological activities of pharmaceutical interest, including antimicrobial and fosfatase inhibition (e.g. rubrolide A (3), Fig. 1), antioxidant and anti-influenza A (H1N1), anti-inflammatory, and antibacterial properties [18,19,20,21,22].
Inspired by the chemistry and biological activities of marine natural metabolites, our research group focused on synthesis and investigation of a series of biological properties of compounds analogues to different classes of marine butenolides [23,24,25,26,27,28,29]. We have found that rubrolide analogues are potent inhibitors of bacterial biofilm formation [23, 24] and, especially those lacking the β-aromatic ring attached to the lactone ring, are cytotoxic toward cancer cell lines [25].
Now, in line with our current interest in natural butenolide chemistry and the diverse and versatile biological profile of natural rubrolides as well, we started investigating the potential of such compounds as urease inhibitors. The present study details the initial findings in this area.
Results and discussion
Synthesis of rubrolide analogues
For the synthesis of rubrolide analogues 10–19 a methodology described in the literature [26,27,28] and summarized in Scheme 1 was employed. The synthesis of compounds 11–15 and 17–19 has already been reported by our group [26,27,28]. Briefly, the required intermediates 7–9 were prepared via a regioselective Suzuki–Miyaura cross-coupling between commercially available 3,4-dibromofuranone (6) and the arylboronic acids in the presence of Ag2O, AsPh3 and the catalytic amount of PdCl2(MeCN)2.

The intermediates 7–9 were then submitted to a vinylogous aldol condensation with several aldehydes under the conditions developed by Boukouvalas et al. [30]. In this method, the butenolides 7–9 react with tert-butyltrimethylsilyltriflate (TBDMSTf) and diisopropylethylamine (DIPEA) to produce silyl enol ethers as intermediates that, in turn, react with the aldehydes. Further reaction of the intermediate silyl-protected alcohol with 1,8-diazabicyclo[5.4.0]undec-8-ene (DBU) resulted in the required γ-alkylidenebutenolides 10–15 in variable yields (16–87%). The methoxy derivatives were further treated with BBr3 to afford the corresponding phenols 16–19 in good yields (90–98%) (Scheme 1).
Considering that this aldol condensation procedure usually requires expensive reagents, we have further investigated the use of a simpler, lower cost methodology to prepare γ-alkylidenebutenolides as reported by Xu et al. [31]. Such an approach involves an in situ aldol condensation followed by a β-elimination, using Na2CO3 as the base and methanol as the solvent. Although this protocol has been successfully employed in the synthesis of the antibiotic enhygrolide [32], in our hands the treatment of 7 with Na2CO3 in the presence of 3,4-dimethoxybenzaldehyde aiming to produce 10, only afforded compound 21 (61% yield at room temperature and 70% under reflux) (Scheme 2).

We hypothesized that the presence of a bromine atom at the α-position of the lactone ring may be the reason for the failure of the vinylogous aldol condensation. Since bromine can act as a good leaving group, an addition/elimination reaction in which MeOH attacks the γ-carbon at the lactone ring can take place as illustrated in Scheme 3.

These unexpected results are not without precedent, as exemplified by the failed attempt to promote the aldol condensation/β-elimination in the synthesis of pulvinone analogues using K2CO3 as a base [33]. The structure of 21 was supported by spectroscopic data.
In vitro urease inhibitory activities of rubrolide analogues
The effects of the rubrolide analogues and the simple lactone 21 on urease activity were assessed and compared with those of the standard urease inhibitor hydroxyurea (HU). Initially, all compounds were screened for their inhibitory activities at a concentration of 500 µmol/dm3. The results showed that the ureolytic activity of the rubrolide analogues varied from 20.7 to 96.3% (Table 1). Compounds 14–19 and 21 were the most active, showing higher activity than the standard inhibitors HU or TU.
From the data obtained, we can gain some insight into the structural-activity relationship. As observed, compound 10 bearing three methoxy groups has a very low activity (28.1% inhibition). Also, the analogue 12, which only has methoxy groups as substituents at the aromatic rings, presented relatively low activity (20.7% inhibition). The presence of the electron-withdrawing fluorine group in compound 11 did not improve the activity (23.0% inhibition). Such results show that methoxy or fluorine groups in the aromatic rings do not positively affect the urease activity of such compounds.
Removal of the methyl groups from 10 resulted in the corresponding tri-hydroxylated compound 16, the most active (99.3% inhibition) among all the analogues tested. The same effect was observed in the case of conversion of 11 (23.0% inhibition) into the OH-free analogue 17 (88.5% inhibition). Additionally, the position of –OH groups seem not to affect the potency of the rubrolides toward urease as attested by the results obtained with compounds 17 and 19 (88.5% and 85.4% inhibition, respectively). The importance of the hydroxyl group for the activity can be clearly observed when comparing the results for compounds 18 and 19 which show 85.1% and 85.4% inhibition of urease. Both compounds present a benzylidene ring bearing two OH groups and while 18 has a phenyl ring with one OH, 19 has this phenyl moiety bearing one fluorine and one methoxy group. These results show also that the presence of fluorine or methoxy does not have a significant effect on the activity. On the other hand, the introduction of a bromine atom in the aromatic ring attached at the β-position of the lactone ring caused an increase in activity, as observed for compound 16 (99.3% inhibition), compared with the activities of the other tri-hydroxylated compounds 17–19 (inhibition in the range from 85.4 to 88.5% when tested at 500 µmol/dm3). Compound 21 also showed a good urease inhibitory activity (76.6% of inhibition) when compared with the standard HU (38.2% of inhibition). It is worth mentioning the potential of 21, a structurally simple rubrolide lacking the benzylidene group, whose inhibitory effect of the ureolytic activity was slightly lower than that of more complex rubrolide analogues (16–19). Therefore, simple lactone 21 is another new prototype for further investigation in terms of potential antiureolytic activity. Among the non-hydroxylated compounds, only 14, bearing the electron-withdrawing group CF3 at the para position in the benzylidene ring, caused a significant urease inhibitory effect (70.0%). The analogue 13, also bearing an electron-withdrawing group with NO2 group at the same position showed a negligible effect (20.7%).
Considering the variable effects observed, all compounds were then subjected to additional assays to determine the minimum concentration necessary to inhibit urease by 50% (IC50; Table 1). The IC50 values for rubrolide analogues 14–21 ranged from 110.0 to 310.0 µmol/dm3 (Table 1). Compounds 10–13 exhibited IC50 values higher than 500 µmol/dm3, which demonstrates that they are poor urease inhibitors as observed in the initial screening (Table 1). Differently, and as expected from the screening, compounds 14–21 were more active than HU or TU, in which notably the hydroxylated analogues 16 and 18 were about 7- and 14-fold more potent than the reference inhibitors HU and TU, respectively. The higher activity of the hydroxylated analogues may be due to the presence of the phenolic hydroxyls at ortho position to each other. These hydroxyl groups might coordinate to the nickel atom that constitutes the active site of urease. Indeed, the positive relationship between the presence of phenolic hydroxyl groups and urease inhibitory activity has also been observed for other classes of molecules [34, 35]. To the best of our knowledge, this is the first study to report the urease inhibitory activity of rubrolide analogues.
Predicted drug-related physicochemical parameters
Before reaching its target site, a therapeutic agent needs to overcome a series of biological barriers. Thus, in vivo activity is not only determined by drug potency but also depends on drug solubility and permeability across cell membranes [36, 37]. Lipinski’s rule of 5 [36] is a set of empirically derived rules that delineate physicochemical parameters and molecular features related to the adsorption, distribution, metabolism, and excretion (ADME) of oral drugs. Such a concept of drug-likeness is considered very useful at the early stages of drug development, allowing us to predict that molecules violating two or more criteria are more likely to show poor pharmacokinetic properties, thus being considered poor candidates [36, 37]. To assess the drug-likeness of the investigated compounds according to ‘Lipinski’s rule of 5’, the physicochemical parameters such as octan-1-ol/water partition coefficient (LogP), number of hydrogen bond donors (HBD), number of hydrogen bond acceptors (HBA), molecular weight (MW), number of rotatable bonds (nRotb), and topological polar surface area (TPSA) were calculated for the rubrolide analogues synthesized using the Molinspiration software package (http://www.molinspiration.com). The obtained data are presented in Fig. 2.
Graphs providing the physicochemical parameters for the rubrolide analogues. Partition coefficient prediction (CLogP), molecular weight (MW), hydrogen bonding acceptors (HBA), number of hydrogen bonding donor groups (HBD), number of rotatable bonds (ROB), and the number of aromatic bonds (ARB). ClogP**, MW**, HBA**, HBD**, ROB** and ARB** are the predict physicochemical properties of drugs by [36]
From the data presented in Fig. 2, except for 21 and 14, which have one rule violation (miLogP), all other compounds are within the limits set by the rules and, thus, are predicted to have good pharmacokinetic profiles.
Docking studies of the interaction between rubrolides and urease
Table 2 presents the key findings from the docking of the compounds against urease, both using the full protein as the target and focusing on the catechol site.
The compounds can be ranked differently based on the criteria used. In terms of urease inhibition percentage, the ranking (from the best to the worst) is as follows: 16–17–19–18–21–14–15–10–12–13–11. Based on the MolDock score using the whole protein as the target, the ranking is 16–12–14–10–13–19–21–15–17–18–11. The ranking changes slightly when considering the re-rank score for the whole protein: 16–12–10–19–13–15–14–17–18–21–11. Choosing the catechol site as the target leads to minor variations in the rankings. The MolDock score ranking becomes 16–12–14–10–13–19–21–15–17–18–11, and the re-rank score ranking is 16–12–10–19–13–15–14–17–18–21–11. A consensus can be observed from these results, with compound 16 being the best fit and 11 being the least favorable, as shown in Fig. 3. Hydrogen bonds play a crucial role in establishing reversible interactions with the site, such as the binding between compound 16 and Asp 596. However, the presence of ortho-hydroxy groups is essential for urease inactivation due to the formation of a covalent adduct with the catechol motif [38].
Compounds 14 and 10 exhibit favorable positions in docking but show moderate urease inhibition. In addition, compounds with hydroxy groups (–OH) in the ortho position display better urease inhibition, likely due to the observed radical mechanism of adduct formation by catechol. Compounds with methoxy groups (–OMe) in the ortho position exhibit better non-covalent interactions with the catechol site, indicating their favorable positioning for reversible interactions without forming adduct-like compounds with OH. The same ranking order is observed when comparing the docking rankings for the whole protein and the catechol site, suggesting that the inhibition is primarily governed by the interaction with this specific site. Compounds with stronger interactions (16, 12, 10, 13, and 19) have fewer steric hindrances at the catechol site. An interesting exception is compound 12, which encounters steric hindrance between the ring containing the two –OMe groups and the residues Arg597 and Glu 598 (Fig. 4).
Compound 12 ranks well in terms of docking but exhibits low urease inhibition. The steric effect likely hampers the interaction between the methoxy groups of the neighboring rings, attenuating the steric hindrance with the enzyme residues. The limited inhibition of urease is attributed to the absence of –OH (as seen in compounds 10 and 11), which is necessary to replicate the radical mechanism of adduct formation observed with catechol. Compound 21 lacks the catechol ring (or its methoxylated derivative) and shows weak interaction with the site. Compounds 13 and 14 possess electron-withdrawing groups on the ring not bound to –OH or –OMe, resulting in a moderate ability to interact with the site. The presence of –OH or –OMe groups appears to be necessary, and the interaction does not seem to be primarily influenced by inductive effects. Compound 15 lacks –OH, –OMe, and electron-withdrawing groups, and its interaction with the site is also modest.
Conclusion
In summary, a series of rubrolide analogues were synthesized and screened for their urease inhibitory activity. Most of them showed very good inhibitory activity and presented IC50 values lower than those for hydroxyurea, a standard inhibitor. The hydroxylated analogues were the most active, which may be due to a greater ability to coordinate with the active site of urease. The drug-likeness of the compounds was assessed based on Lipinski’s rule. Almost all compounds, including the most potent urease inhibitors, showed no rule violations, which allows for predicting that these compounds have the pharmacokinetic profiles required for potential drugs. The docking results suggest that the presence of hydroxy groups at the ortho position favors both the formation of reversible interactions with urease and the formation of a covalent adduct with the active site causing blockage of the enzyme, like what happens with catechol, its natural inhibitor. The results show that rubrolides may constitute a promising lead for the development of novel treatments against urease-producing bacteria.
Experimental
Reagents and solvents were prepared following procedures already reported in the literature [41] or purchased from commercially available suppliers and used without further purification. Melting points were obtained from an MQAPF-301 melting point apparatus (Microquimica, Brazil). Analytical thin-layer chromatography analysis was conducted on aluminum-packed pre-coated silica gel plates. Column chromatography was performed over silica gel 230–400 mesh. The compounds were fully characterized by IR, 1H NMR, 13C NMR, COSY, HETCOR, and NOEDIFF NMR spectroscopy, and the spectra are presented in the Supplementary Material. For new compounds elemental analyses (C, H, N) were conducted using the Elemental Analyser CHN 2400 Perkin-Elmer. Infrared spectra were recorded on a Perkin-Elmer Paragon 1000 FTIR spectrophotometer, using potassium bromide (1% w/w) disks, or as thin liquid film on NaCl plates. Mass spectra were recorded on a Shimadzu GCMS-QP5050A instrument by direct insertion, using EI mode (70 eV). The 1H NMR and 13C NMR spectra were recorded on a Varian Mercury 300 spectrometer at 300 and 75 MHz, respectively, using CDCl3 or (CD3)2CO as solvents and TMS as an internal reference, unless otherwise stated.
Synthesis of rubrolide analogues
Detailed synthetic procedures and spectroscopic data for compounds 10, 16, and 21 are following presented. Compounds 11–15 and 17–19 were prepared using the same methodology employed to synthesize compounds 10 and 16. The detailed synthetic procedure, as well as their spectroscopic characterization, have previously been reported in the literature [23,24,25].
(Z)-3-Bromo-4-(5-bromo-2-methoxyphenyl)-5-(3,4-dimethoxybenzylidene)furan-2(5H)-one (10, C20H16Br2O5)
To a two-necked round-bottom flask (25 cm3), under a nitrogen atmosphere, compound 7 (200 mg, 0.58 mmol), dichloromethane (4 cm3), tert-butyldimethylsilyl trifluoromethanesulfonate (TBDMSOTf, 170 mm3, 0.75 mmol), N,N-diisopropylethylamine (DIPEA, 200 mm3, 1.16 mmol) and 3,4-dimethoxybenzaldehyde (106 mg, 0.69 mmol) were added. The resulting solution was stirred at room temperature for 1 h. After this period, 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) (150 mm3, 1.16 mmol) was added, and the resultant mixture was refluxed for 1 h. Then, the reaction was quenched by the addition of dichloromethane (70 cm3) and washing of the organic phase with aqueous HCl solution (3 mol dm−3, 2 × 25 cm3) and brine (2 × 25 cm3). The organic layer was dried over magnesium sulfate, filtered, and concentrated under reduced pressure. The resulting residue was purified by column chromatography on silica gel and eluted with ethyl acetate/hexane (3:5 v/v) to afford the required product 10 (215 mg, 0.44 mmol) as a yellow amorphous residue in 75% yield. IR (KBr): \(\overline{v}\) = 3005, 3005, 2942, 2928, 2831, 1764, 1646, 1595, 1577, 1515, 1482, 1461, 1274, 1256, 1167, 1151, 1026, 886, 970, 811 cm−1; 1H NMR (300 MHz, CDCl3): δ = 3.83 (s, 3H), 3.91 (2 × s, 6H), 5.86 (br s, 1H), 6.86 (d, 1H, J = 8.4 Hz), 6.95 (d, 1H, J = 9.0 Hz), 7.29 (dd, 1H, J = 8.4 Hz, 2.1 Hz), 7.36–7.38 (m, 2H), 7.61 (dd, 1H, J = 9.4 Hz, 2.4 Hz) ppm; 13C NMR (75 MHz, CDCl3): δ = 55.9, 109.9, 111.1, 112.6, 113.0, 113.3, 114.0, 118.0, 112.4, 125.6, 132.5, 134.4, 145.9, 149.0, 150.5, 150.7, 155.7, 165.0 ppm; MS (EI, 70 eV): m/z (%) = 498 ([M + 2]+, 52), 497 ([M + 1]+, 22), 496 (M+, C20H16Br2O5, 100), 494 (53), 178 (24), 163 (47), 135 (31), 107 (20), 92 (19), 79 (19), 77 (28).
(Z)-3-Bromo-4-(5-bromo-2-hydroxyphenyl)-5-(3,4-dihydroxybenzylidene)furan-2(5H)-one (16, C17H10Br2O5)
Compound 3 (100 mg, 0.20 mmol) and dichloromethane (5 cm3) were added to a two-necked round-bottom flask (25 cm3), under a nitrogen atmosphere. The mixture was cooled to − 78 °C before the dropwise addition of BBr3 in dichloromethane (2 M, 0.58 cm3). The reaction mixture was allowed to reach room temperature and stirred for 22 h. It was then quenched by adding water (10 cm3) and ethyl acetate (25 cm3). The organic phase was separated, and the aqueous phase was extracted with ethyl acetate (3 × 25 cm3). The organic extracts were combined and washed with brine (15 cm3), dried over magnesium sulfate, filtered, and concentrated under reduced pressure. The resulting residue was purified by column chromatography on silica gel eluted with ethyl acetate/chloroform (2:3 v/v) to afford the required product 16 (82 mg, 0.2 mmol). IR (KBr): \(\overline{v}\) = 3327, 2922, 2851, 1732, 1604, 1523, 1473, 1370, 1283, 1217, 1181, 1118, 1018, 895, 818, 698 cm−1; 1H NMR (300 MHz, CO(CD3)2): δ = 6.05 (s, 1H), 6.86 (d, 1H, J = 8.4 Hz), 7.06 (d, 1H, J = 8.7 Hz), 7.12 (dd, 1H, J = 8.4 Hz, 2.1 Hz), 7.48 (d, 1H, J = 2.7 Hz), 7.49 (d, 1H, J = 2.1 Hz), 7.55 (dd, 1H, J = 8.7 Hz, 2.7 Hz) ppm; 13C NMR (75 MHz, CO(CD3)2): δ = 108.5, 110.4, 114.9, 116.4, 117.4, 118.5, 119.0, 124.4, 125.9, 132.6, 134.6, 145.0, 146.0, 152.0, 154.5, 165.4 ppm; MS (EI, 70 eV): m/z (%) = 456 ([M + 2]+, 52), 454 (M+, C17H10Br2O5, 54), 452 (27), 376 (50), 375 (24), 374 (87), 266 (21), 163 (28), 150 (34), 123 (87), 122 (33), 121 (29), 89 (28), 82 (100), 81 (32), 80 (79), 79 (36), 76 (26), 63 (27), 44 (54).
4-(5-Bromo-2-methoxyphenyl)-5-methoxyfuran-2(5H)-one (21, C12H11BrO4)
Compounds 7 (50 mg, 0.15 mmol), 3,4-dimethoxybenzaldehyde (44 mg, 0.29 mmol), Na2CO3 (4.6 mg, 0.043 mmol), and methanol (3 cm3) were added in a two-necked round-bottom flask (25 cm3), under a nitrogen atmosphere. The resulting mixture was refluxed for 20 h and the solvent was removed under reduced pressure and the resultant residue dissolved in chloroform (15 cm3). The chloroform solution was washed with water (2 × 10 cm3), dried with Mg2SO4, and concentrated under reduced pressure to afford the crude product 21 as a solid in 70% yield. The solid was further recrystallized in dichloromethane and hexane (1:5 v/v). This reaction was repeated using Na2CO3 (15 mg, 0.145 mmol) at room temperature (20 h) instead of the reflux temperature, affording 21 in 61% yield. M.p.: 101.0–103.5 °C; IR (KBr): \(\overline{v}\) = 3115, 3093, 3005, 2965, 2949, 2908, 2831, 1754, 1612, 1561, 1492, 1389, 1363, 1287, 1267, 1163, 1145, 1021, 993, 974, 829 cm−1; 1H NMR (300 MHz, CDCl3): δ = 3.56 (s, 3H), 3.92 (s, 3H), 6.23 (d, 1H, J = 0.9 Hz), 6.71 (d, 1H, J = 0.9 Hz), 6.89 (d, 1H, J = 9.0 Hz), 7.53 (dd, 1H, J = 9.0 Hz, 2.4 Hz), 7.60 (d, 1H, J = 2.4 Hz) ppm; 13C NMR (75 MHz, CDCl3): δ = 55.7, 55.9, 103.5, 113.0, 113.5, 120.1, 120.2, 132.0, 135.1, 155.7, 158.0, 171.0 ppm; MS (EI, 70 eV): m/z (%) = 300 ([M + 2]+, 15), 298 (M+, C12H11BrO4, 15), 212 (100), 211 (38), 210 (100), 209 (29), 132 (45), 131 (71), 63 (23), 62 (28), 51 (24), 50 (23).
Urease inhibition assay
Screening to identify potential urease inhibitors was performed by incubating each synthesized compound at a final concentration of 500 µM in reactions containing buffer solution (Na2HPO4/NaH2PO4 50 mM, pH 7.4), urea (10 mM), and 125 mU/cm3 jack bean type III urease (Sigma U-1500–100 kU). Each mixture was incubated for 15 min at 25 °C, and the reactions were interrupted following the methodology described by Weatherburn [41] with modifications by Brito et al. [42]. The ammonium concentration was determined by phenol hypochloride assay (636 nm), and the inhibition percentage [INH(%)] was calculated using the following equation: INH(%) = 100 − [(AINH/AB) × 100], where AINH and AB are ammonium concentration in the tubes with and without inhibitor, respectively. The inhibitory potential of the compounds was compared to that of the standard inhibitor hydroxyurea (HU) or thiourea (TU). Compounds that showed inhibitory activity higher than that of the standard HU (the most efficient urease inhibitor used in this study) were further used from 50 to 3,200 µmol dm−3 to determine the concentration necessary to inhibit the enzyme by 50% (IC50). All experiments were performed in triplicate.
Drug-likeness calculation
Computational calculation was carried out to assess whether the investigated compounds can fulfill the features of candidate drugs, based on Lipinski’s Rule of Five [36, 37]. Physicochemical parameters such as n-octanol/water partition coefficient (miLogP), number of hydrogen bond donors (HBD), number of hydrogen bond acceptors (HBA), molecular weight (MW), number of rotatable bonds (nRotb), and total polar surface area (TPSA) were calculated using Molinspiration software package (http://www.molinspiration.com).
Docking studies of the interaction between rubrolides and urease
To better understand the forces that govern the interaction between the rubrolides and urease, a docking study was performed between them, using plant urease from Jack bean (Canavalia ensiformis) as a target [38]. Two crystallographic structures of ureases isolated from Jack bean were used: the PDB 3LA4 structure, without any ligand bound, and the PDB 5G4H structure, containing a catechol ring as a ligand (available in the RCSB PDB but not published). The interaction catechol-5G4H-binding site involves residues Lys 169, Cys 322, and Val 321. The urease here is inactivated by catechol by forming an adduct (covalent) with Cys 322 [38]. By aligning both enzyme sequences it was possible to find the PDB 3LA4 corresponding residue as a disulfide bridge located at 592 position. A docking analysis performed in the whole protein was done, and a subsequent docking on the Arg 439 to Glu 827 region in 3LA4 (corresponding to the catechol site of 5G4H) was also used. The compounds’ structures were previously optimized using molecular dynamics at 300 K (Boltmann Jump method) with the AM1BCC force field, using Gasteiger-type charges. The docking on the whole protein was performed in Hex Cuda software [39] using the shape-only correlation type, whereas docking on the catechol site was pursued using the shape plus electro correlation type. No post-processing was used in this screening. The analysis of the docking results was aided by the use of the Molegro Molecular Viewer software, with the use of the MolDock scoring function (MolDock score [40]) and the linear combination of some additional energy terms (a re-rank score, using the re-rank weight).
Data availability
Not applicable.
References
Follmer C (2010) J Clin Pathol 63:424
Konieczna I, Żarnowiec P, Kwinkowski M, Kolesińska B, Frączyk J, Kamiński Z, Kaca W (2012) Curr Protein Pept Sci 13:789
Rutherford JC (2014) PloS Pathog 10:e1004062
Sharndama HC, Mba IE (2022) Braz J Microbiol 53:33
Wang Y-K, Li C, Zhou Y-M, Zeng L, Li Y-Y, Huang S-L, Zhu C-Y, Wang Y, Wang S-N, Chen X-D (2022) J Inflamm Res 15:6231
Yang H, Guan L, Bing H (2022) Gastroenterology Res Pract 2022:4710964
Fiori-Duarte AT, Rodrigues RP, Kitagawa RR, Kawano DF (2020) Curr Med Chem 27:3967
Yang W, Feng Q, Peng Z, Wang G (2022) Eur J Med Chem 234:114273
Modolo LV, de Souza AX, Horta LP, Araújo DP, de Fátima A (2015) J Adv Res 6:35
Li J, Li Q-Y, Wu Y-C (2021) Bioorg Med Chem 35:116058
Ghareeb MA, Tammam MA, El-Demerdash A, Atanasov AG (2020) Cur Res Biotechnol 2:88
Lyu C, Chen T, Qiang B, Liu N, Wang H, Zhang L, Liu Z (2021) Nucleic Acids Res 49:D509
Ye Y, Liang J, She J, Lin X, Wang J, Liu Y, Yang D, Tan Y, Luo X, Zhou X (2023) Mar Drugs 21:27
Bao J, Li X, He F, Zhang X, Zhu K, Tao H, Yu J, Liu H, Zhang H (2020) Tetrahedron Lett 61:152193
Kim J-Y, Oh G-W, Lee JM, Kim H-S, Ki D-W, Ko S-C, Yim M-J, Kim K-W, Lee D-S, Baek K (2022). Nat Prod Comm. https://doi.org/10.1177/1934578X221137411
Orfali R, Aboseada MA, Abdel-Wahab NM, Hassan HM, Perveen S, Ameen F, Alturki E, Abdelmohsen UR (2021) RSC Adv 11:17116
Barbosa LCA, Varejão JOS, Varejão EVV (2017) Strategies for total synthesis of furanocembranolides and related natural products from marine organisms. In: Atta-ur-Rahman FRS (ed) Studies in natural products chemistry, vol 52, p 115. Elsevier, Amsterdam
Miao S, Andersen RJ (1991) J Org Chem 56:6275
Sikorska J, Parker-Nance S, Davies-Coleman MT, Vining OB, Sikora AE, McPhail KL (2012) J Nat Prod 75:1824
Zhu T, Chen Z, Liu P, Wang Y, Xin Z, Zhu W (2014) J Antibiot 67:315
Pearce AN, Chia EW, Berridge MV, Maas EW, Page MJ, Webb VL, Harper JL, Copp BR (2007) J Nat Prod 70:111
Wang W, Kim H, Nam S-J, Rho BJ, Kang H (2012) J Nat Prod 75:2049
Pereira UA, Barbosa LCA, Maltha CRA, Demuner AJ, Masood MA, Pimenta AL (2014) Bioorg Med Chem Lett 24:1052
Pereira UA, Barbosa LCA, Maltha CRA, Demuner AJ, Masood MA, Pimenta AL (2014) Eur J Med Chem 82:127
Pereira UA, Moreira TA, Barbosa LCA, Maltha CRA, Bomfim IS, Maranhão SS, Moraes MO, Pessoa C, Barros-Neponuceno FWA (2016) Med Chem Commun 7:345
Barbosa LC, Maltha CRA, Lage MR, Barcelos RC, Donà A, Carneiro JWM, Forlani G (2012) J Agric Food Chem 60:10555
Varejão JOS, Barbosa LCA, Maltha CRA, Lage MR, Lanznasterc M, Carneiro JWM, Forlani G (2014) Electrochim Acta 120:334
Varejão JOS, Barbosa LCA, Ramos GA, Varejão EVV, King-Díaz B, Lotina-Hennsen B (2015) J Photochem Photobiol B Biol 145:11
Moreira TA, Antolínez IV, Valença WO, Roy S, Ramirez I, Barbosa LCA, Ren D (2022) Bioorg Med Chem Lett 57:128498
Boukouvalas J, Maltais F, Lachance N (1994) Tetrahedron Lett 35:7897
Xu H-W, Wang J-F, Liu G-Z, Hong G-F, Liu H-M (2007) Org Biomol Chem 5:1247
Muddala R, Acosta JAM, Barbosa LCA, Boukouvalas J (2017) J Nat Prod 60:10555
Xu H-W, Xu C, Fan Z-Q, Zhao L-J, Liu H-M (2013) Bioorg Med Chem Lett 23:737
Chen Y, Liao J, Chen M, Huang Q, Lu Q (2015) J Chem Pharm Res 7:10
Rauf A, Uddin G, Raza M, Patel S, Bawazeer S, Hadda TB, Jehan N, Mabkhot YN, Khan A, Mubarak MS (2017) Nat Prod Res 31:1214
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Adv Drug Deliv Rev 46:3
Bickerton GR, Paolini GV, Besnard J, Muresan S, Hopkins AL (2012) Nat Chem 4:90
Mazzei L, Cianci M, Musiani F, Lente G, Palombo M, Ciurli S (2017) J Inorg Biochem 166:182
Macindoe G, Mavridis L, Venkatraman V, Devignes M-D, Ritchie DW (2010) Nucleic Acids Res 38:W445
Perrin DD, Armarego WLF (2003) Purification of laboratory chemicals, 5th edn. Bodmin, Butterworth–Heinemann, Oxford
Weatherburn MW (1967) Anal Chem 39:971
Brito TO, Souza AX, Mota YCC, Morais VSS, Souza LT, de Fatima A, Macedo Junior FC, Modolo LV (2015) RSC Ad 5:44507
Acknowledgements
We are grateful to Conselho Nacional de Desenvolvimento Científíco e Tecnológico (CNPq) for research fellowships (LCAB and LVM), Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG grant # APQ1557-15) for the financial support and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Grant 001) for the research fellowship (JOSV).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Varejão, J.O.S., Barbosa, L.C.A., Varejão, E.V.V. et al. Rubrolide analogues as urease inhibitors. Monatsh Chem 154, 1177–1187 (2023). https://doi.org/10.1007/s00706-023-03106-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00706-023-03106-y