Abstract
The point-of-care testing (POCT) of miRNA has significant application in medical diagnosis, yet presents challenges due to their characteristics of high homology, low abundance, and short length, which hinders the achievement of quick detection with high specificity and sensitivity. In this study, a lateral flow assay based on the CRISPR/Cas13a system and MnO2 nanozyme was developed for highly sensitive detection of microRNA-21 (miR-21). The CRISPR/Cas13a cleavage system exhibits the ability to recognize the specific oligonucleotide sequence, where two-base mismatches significantly impact the cleavage activity of the Cas13a. Upon binding of the target to crRNA, the cleavage activity of Cas13a is activated, resulting in the unlocking of the sequence and initiating strand displacement, thereby enabling signal amplification to produce a new sequence P1. When applying the reaction solution to the lateral flow test strip, P1 mediates the capture of MnO2 nanosheets (MnO2 NSs) on the T zone, which catalyzes the oxidation of the pre-immobilized colorless substrate 3,3′,5,5′-tetramethylbenzidine (TMB) on the T zone and generates the blue-green product (ox-TMB). The change in gray value is directly proportional to the concentration of miR-21, allowing for qualitative detection through visual inspection and quantitative measurement using ImageJ software. This method achieves the detection of miR-21 within a rapid 10-min timeframe, and the limit of detection (LOD) is 0.33 pM. With the advantages of high specificity, simplicity, and sensitivity, the lateral flow test strip and the design strategy hold great potential for the early diagnosis of related diseases.
Graphical abstract

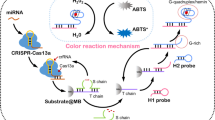
This work integrates the chromogenic reaction of TMB catalyzed by MnO2 NSs, the high specificity and signal amplification capability of the CRISPR/Cas13a cleavage system, and the simplicity and visual clarity of the lateral flow test strip.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNA-21 (miR-21) is a small, non-coding RNA molecule comprising around 22 nucleotides. It is ubiquitous in human cells and plays a critical role in various cellular processes [1]. High expression level of miR-21 is strongly correlated with the occurrence, progression, and prognosis of numerous cancers [2]. By detecting the expression levels of miR-21 in blood, tissue, or other samples, it is possible to assess disease severity, and offers an adjunct diagnostic tool [3].
So far, conventional methods for miRNA detection have some limitations. For instance, RT-qPCR, while widely used, is cumbersome to operate, and requires specialized instruments and personnel. The long detection process makes it unsuitable for rapid testing [4]. Microarrays enable high-throughput detection, but this technology can be costly and may exhibit elevated background noise caused by nonspecific hybridization [5]. In contrast, lateral flow test strip-based miRNA detection offers several advantages. Firstly, the lateral flow assay is simple and rapid, which requires no complex equipment or reagents, making it suitable for field use [6, 7]. Secondly, the lateral flow test strip provides visual results, eliminating the need for specialized equipment for data analysis and interpretation [8, 9]. Additionally, miRNA detection using lateral flow test strip is cost-effective, making it suitable for large-scale screening and primary diagnosis [10]. However, there are some limitations to lateral flow test strip-based miRNA detection. The accuracy and sensitivity of lateral flow test strip may be low, potentially leading to difficulties in detecting low-level expressed miRNAs or in the presence of interfering factors [11]. Therefore, the development of high-sensitive, high-specific lateral flow test strip remains a significant challenge.
CRISPR/Cas13a has gained widespread use in life science research in recent years [12]. As a diagnostic tool, CRISPR/Cas13a offers numerous advantages. Firstly, it exhibits high specificity. The CRISPR/Cas13a system can achieve precise recognition and binding of target sequences through the design of crRNA. This capability reduces the likelihood of misjudgment and false positive results, thereby enhancing diagnostic accuracy [13]. Secondly, it demonstrates high sensitivity. The CRISPR/Cas13a system can detect target sequences even at very low concentrations [14], making it a promising option for early disease diagnosis and the detection of biomarkers at low levels [15]. Additionally, it offers fast and convenience. CRISPR/Cas13a diagnostic methods typically involve simple and rapid experimental procedures and do not necessitate complex reagents and instruments [16]. In comparison to traditional diagnostic approaches, CRISPR/Cas13a yields results in a shorter timeframe, thereby expediting the disease diagnosis and monitoring process. Moreover, it boasts versatility. The system can be tailored to design different crRNA sequences as required to accommodate various target sequences and detection needs [17, 18]. This adaptability enables CRISPR/Cas13a to detect a wide array of targets, including DNA, RNA, and proteins [19, 20]. Lastly, it offers visualization. CRISPR/Cas13a diagnostic methods are frequently combined with dyes, fluorescent probes, and lateral flow test strips [21], enabling direct visual observation of test results with the naked eyes or interpretation using simple instruments. This visualization feature facilitates the effective use of CRISPR/Cas13a in low-resource settings or outside laboratory conditions [22].
The proper signal output mode is of great significance for lateral flow test strips [23, 24]. Traditionally, the color change resulting from the accumulation of spherical gold nanoparticles (GNP) on the T zone has been commonly utilized for quantitative detection [25]. However, despite the intensive colorimetric signal provided by GNP, the sensitivity of such lateral flow test strips is still too low, which restricts their application in detection of low-concentration targets. To address the demand for enhanced sensitivity, researchers have put forward various signal label alternatives, including fluorescent dyes, quantum dots, upconversion luminescence materials, AuNPs-coated silica nanorods, carbon nanotubes [26], and enzyme-linked labeling. The chromogenic reaction induced by 3,3′,5,5′-tetramethylbenzidine (TMB) under the action of peroxidase offers high sensitivity and easy operation, making it a popular choice for quantitative analysis of biomolecules in enzyme-linked immunosorbent assay (ELISA) and other immunological methods. Compared with native enzymes, nanozymes have smaller size and larger specific surface area, and are able to maintain catalytic activity over wider range of temperature and acid–base conditions [27, 28]. Therefore, more researches tend to use nano-enzymes, such as Fe3O4 nanoparticles [29], Pt-Au nanoparticles [30], carbon nanodots, graphene oxide [31], and gold nanoparticles, to replace native peroxidase for chromogenic reactions. However, this process requires the participation of hydrogen peroxide (H2O2) [32], which is easy to decompose under high temperature, light, and other conditions. The change of its concentration will directly affect the progress of the chromogenic reaction, thereby affecting the sensitivity of detection and limiting the applications in the field detection. Recent researches indicate that MnO2 NSs can serve as a replacement for peroxidase in oxidizing TMB for color reactions, offering distinct advantages. Firstly, MnO2 NSs are cost-effective compared to peroxidase. Peroxidase requires relatively complex and costly preparation, while MnO2 NSs are less expensive and easier to synthesize. Secondly, MnO2 NSs exhibit anti-interference properties, boasting superior stability in various storage conditions, while peroxidase is susceptible to environmental changes, such as temperature and pH variations. Additionally, the controllability of MnO2 NSs is noteworthy, as their structure and catalytic performance can be optimized through various synthesis and modification methods, thus enhancing detection sensitivity and accuracy. Most importantly, MnO2 NSs can catalyze the oxidation of TMB in the absence of hydrogen peroxide, simplifying the reaction process and reducing the test time. These advantages offer new possibilities for utilizing TMB chromogenic reaction in detection applications.
To enhance the specificity and sensitivity of the lateral flow test strip, this work integrates the chromogenic reaction of TMB catalyzed by MnO2 NSs, the highly specific and signal amplification capabilities of the CRISPR/Cas13a cleavage system, and the simplicity and visual clarity of the lateral flow test strip. The designed lateral flow test strip demonstrates the capacity for base mismatch recognition and signal amplification for the target miR-21, enabling qualitative identification with the naked eyes and rapid quantitative diagnosis through software processing. This innovation holds significant potential for disease diagnosis and gene therapy.
Experimental
Materials and reagents
1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide·HCl (EDC), streptavidin (SA), and N-hydroxysulfosuccinimide sodium salt (sulfo-NHS) were obtained from Sigma-Aldrich (St Louis, MO, USA). Triton X-100, sucrose, Tween-20, sodium acetate (NaAc), and acetic acid (HAc) were obtained from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China). Sodium citrate, DEPC-treated water, bovine serum albumin (BSA), and 3,3′,5,5′-tetramethylbenzidine (TMB) were all obtained from Sangon Biotech Co., Ltd (Shanghai, China). Sample pad, conjugate pad, absorption pad, nitrocellulose membrane (NC membrane), and polyvinyl chloride (PVC) backing were all purchased from Jieyi Biotechnology Co., Ltd (Shanghai, China). LwaCas13a nuclease and 10 × Cas13a Reaction Buffer were purchased from Novoprotein (Shanghai, China). DEPC-treated water was used to prepare all aqueous solutions.
All oligonucleotides were synthesized by Sangon Biotech Co., Ltd and purified through high-performance liquid chromatography (HPLC). The sequences are listed in Table S1.
Cas13a/crRNA-based fluorescence assay
The mixture of 2 µL Cas13a (400 nM), 2 µL 10 × Cas13a Reaction Buffer, and 1 µL crRNA (200 nM) was incubated at 25°C for 5 min. Then, 1.7 µL miR-21 (120 nM), 6 µL FQ5U (2000 nM), and 7.3 µL DEPC-treated water were added to the mixture, resulting in a total volume of 20 μL. After the addition of the target and FQ5U, the mixture was quickly transferred into a qPCR instrument and incubated at 37 °C for 60 min. The fluorescence intensity was measured every minute throughout the incubation period.
Synthesis andmodification of MnO2 NSs
A bottom-up approach was employed to synthesize ultrathin MnO2 NSs. In a synthesis vessel, 32 mL SDS (0.1 M) and 1.6 mL H2SO4 (0.1 M) were added into 283.2 mL water and the mixture was subjected to heating at 95 °C for 15 min. The reaction was initiated by rapidly adding 3.2 mL KMnO4 solution (0.05 M) to the mixture, followed by heating the mixture for 60 min to yield a homogeneous solution with a dark brown color. Subsequently, the harvest of MnO2 NSs involved centrifugation at 15,000 rpm for 10 min and repeated wash with ultrapure water until the supernatant became colorless and clear. Finally, the nanosheets were dried under vacuum at 50 °C.
MnO2 NSs-biotin-probe (MnO2 NSs-BP) is synthesized through amide reaction between the EDC/NHS-activated carboxyl group on the surface of MnO2 NSs and the amino group of biotin-probe. EDC (50 mg mL−1) and NHS (50 mg mL−1) were firstly prepared using the coupling buffer (50 mM MES, 0.01% Triton X-100, pH 6.0). Then, 1.2 mL coupling buffer, 400 µL newly formulated EDC solution, and 400 µL newly formulated NHS solution were added to MnO2 NSs, and the mixture was vortexed and mixed before incubation at 25°C for 1 h. Another 80 µL of 100 µM biotin-probe sequence was added, and the mixture was mixed and the reaction was incubated at 25 °C for 2 h. The mixture was subjected to centrifugation at 13,000 rpm for 10 min using ultrafiltration. After centrifugation, the pellet was resuspended in an aqueous solution, resulting in a final concentration of 50 µg mL−1 of MnO2 NSs-BP complex.
Preparation of lateral flow test strips
The lateral flow test strip is consisted of five components: conjugate pad, sample pad, absorption pad, NC membrane, and PVC backing. Pretreatment is not necessary for PVC backing and absorption pad.
The conjugate pad and sample pad were submerged in a solution of treatment buffer (0.1 M Tris–HCl, 1% BSA, 2% sucrose, 1% Triton X-100, 1% NaCl, and 0.5% Tween-20, pH 8.2) for 30 min and dried overnight at 37 °C. Afterwards, 50 μL MnO2 NSs-BP complex was uniformly applied onto the conjugate pad, ensuring even coating. The conjugate pad was subsequently dried for a duration of 12 h at 45 °C.
T zone and C zone of the lateral flow test strip were prepared respectively. To prepare the T zone, the capture probe with a final concentration of 1 nM (dissolved in PBS buffer containing 1 M KCl, pH 6.0) was mixed with 12 mM TMB (dissolved in anhydrous ethanol). Then, a micropipette to draw the T zone at a distance of 0.85 cm from the top of the NC membrane was used. To prepare the C zone, streptavidin (SA) with a final concentration of 1.5 mg mL−1 was mixed with 12 mM TMB. Then, a micropipette to draw the C zone at a distance of 6 mm from the T zone was used. The treated NC membrane was dried under vacuum at 25 °C in the dark for 2 h. Then, the NC membrane (2.5 cm in length), sample pad (1.2 cm in length), conjugate pad (1 cm in length), and absorption pad (1.7 cm in length) were all pasted onto the PVC backing glue and overlapped by 1–2 mm. Once assembled, the lateral flow test strips were cut into flowmeter strips that are 0.4 cm in width. Finally, the prepared lateral flow test strips were placed in a bag and stored in a dark environment at 4 °C.
Detection of miR-21 using lateral flow test strips
The CRISPR/Cas13a reaction solution included 4 μL Cas13a (400 nM), 4 μL 10 × Cas13a Reaction Buffer, 2 μL crRNA (200 nM), 10.66 μL LOCK-U (1500 nM), 10.66 μL fuel chain (2000 nM), various concentrations of the target miR-21, and DEPC-treated water, resulting in a total volume of 40 μL. After incubation of the CRISPR/Cas13a reaction solution at 37 °C for 5 min, 40 μL of PBS was added. Then, the whole reaction solution was applied to the sample pad of the lateral flow test strip. The lateral flow test strip was left for 5 min for the detection process. Finally, the images of the lateral flow test strips were recorded using a smartphone. The gray value of the T zone was quantified using ImageJ software for further analysis.
Detection of miRNA-21 in actual samples
To assess the practicability of the developed lateral flow test strip, the miR-21-spiked serum samples and the clinical serum samples were employed. Initially, miR-21 standard solution was spiked to artificial serum samples, enabling the measurement of its concentration. The recovery rate and the relative standard deviation (RSD) were then evaluated to gauge the accuracy and precision of the detection. Furthermore, clinical serum samples were collected from patients diagnosed with gastric, lung, and liver cancers at the Wuxi No.2 People’s Hospital in Jiangsu Province, China. These samples were subsequently subjected to the detection process, allowing for further assessment of the performance of the lateral flow strip in a clinical setting.
Results and discussion
Principle of the CRISPR/Cas13a-based lateral flow test strip for miR-21
As depicted in Scheme 1, the principle of miR-21 detection involves two main stages: (A) miR-21-triggered CRISPR/Cas13a cleavage for target recognition and signal amplification, and (B) rapid detection of miR-21 on the lateral flow test strip.
Scheme 1A demonstrates the target recognition and subsequent signal amplification based on CRISPR/Cas13a. The crRNA consists of a 30-nt repeat sequence and a 22-nt spacer sequence. LOCK-U is a hairpin structure containing two uracil (U) (Fig. S1). Cas13a initially binds with crRNA and forms a complex. When the target miR-21 is present, it fully binds to the spacer sequence of crRNA, which leads to the formation of a ternary complex, and consequent activation of the cleavage activity of Cas13a. The latter cleaves the phosphodiester bond between the U-U of LOCK-U, converting its hairpin structure to a double-stranded product. The fuel chain is then introduced, which triggers the strand displacement and releases the sequence P1 from the double strand. Therefore, the presence of miR-21 is converted to a large number of P1 in this stage, which participates in the subsequent lateral flow test strip process.
Scheme 1B outlines the operational principle of the lateral flow test strip for miR-21 detection. The biotin-probe is covalently linked to the MnO2 NSs via an amide reaction and immobilized on the conjugate pad. The 5′ end of the capture probe was modified with adenine polymer (polyA) to enhance its immobilization efficiency on the NC membrane. A mixture of KCl (1 M) and PBS (pH 6.0) was used to further enhance the stability. The strong hydrophobic interaction between DNA and the NC membrane ensures the stable fixation of the capture probe on the NC membrane [21]. The TMB chromogenic system is pre-fixed to the T zone and the C zone. As the product of the above stage migrates from the sample pad to the absorption pad, the MnO2 NSs-BP detection probe immobilized on the conjugate pad further migrates to the NC membrane together with the solution. During this procedure, P1 in the CRISPR/Cas13a reaction solution initially hybridizes with biotin-probe, and subsequently hybridized and captured by the capture probe on the T zone, forming a sandwich structure. MnO2 NSs are thus deposited on the T zone, which catalyze the oxidation of the pre-fixed TMB and generate a blue-green oxidation product (ox-TMB). The excess MnO2 NSs-BP continue to migrate and are finally captured by SA on the C zone, catalyzing the oxidation of TMB pre-fixed on the C zone and resulting in a color change. Qualitative and quantitative detection can be achieved using the naked eyes and ImageJ software. In the absence of the target, due to the inactivation of Cas13a, no P1 sequence can be produced, and thus, MnO2 NSs cannot be enriched on the T zone. Color change is exclusively observable on the C zone as a result of the immobilization of MnO2 NSs through the interaction between SA and biotin.
Validation of the cleavage activity of CRISPR/Cas13a and the feasibility of strand displacement
To assess the feasibility of the target-triggered CRISPR/Cas13a-mediated cleavage, a fluorescence assay was conducted using miR-21 as the target. As shown in Fig. 1A, FQ5U is used instead of LOCK-U as the substrate and signal output. In the presence of miR-21, it binds to the spacer sequence of crRNA, triggering the cleavage activity of Cas13a. As a result, miR-21 is cleaved through cis-cleavage, while FQ5U, which is modified with FAM at one end and BHQ1 at the other end, is cleaved in trans-cleavage. The cleavage of FQ5U disrupts the fluorescence resonance energy transfer (FRET) between the fluorophore and quenching group, thereby enabling the recovery of fluorescence. As depicted in Fig. 1B, in the absence of either miRNA or crRNA, Cas13a does not exhibit cleavage activity, and thus, no fluorescent signal from the cleaved product can be observed. Only in the presence of both the target and crRNA, Cas13a/crRNA forms a ternary complex with miR-21, and the trans-cleavage activity of the Cas13a is activated, resulting in the cleavage of the FQ5U and the recovery of fluorescence.
Feasibility analysis of CRISPR/Cas13a-based system for miR-21 detection. A Schematic diagram of CRISPR/Cas13a cleavage principle. B Real-time fluorescence measurement of CRISPR/Cas13a reaction. FQ5U was used as a reporter probe. C Electrophoretic analysis of the feasibility of CRISPR/Cas13a for miR-21 detection and strand displacement. M: 20 bp marker; lane 1: 1 µM fuel chain; lane 2: 1 µM LOCK-U; lane 3: 1 µM LOCK-U + 40 nM Cas13a + 10 nM crRNA + 1 µM fuel chain + 10 nM miR-21; lane 4: 1 µM LOCK-U + 40 nM Cas13a + 10 nM crRNA + 1 µM fuel chain. D Validation of the specificity of CRISPR/Cas13a for different miRNAs
Figure 1C demonstrates that the cleaved hairpin structure of the LOCK-U sequence can undergo strand displacement with the fuel chain. When miR-21 forms a ternary complex with Cas13a/crRNA, the trans-cleavage activity of Cas13a is activated, leading to the cleavage of the U-U within the LOCK-U sequence. The product is a double-stranded structure, which allows for the strand displacement with the fuel chain to generate the product P2 (lane 3) (Fig. S2). This is in accordance with the lightened band of LOCK-U (lanes 2 and 3) as well as the appearance of the band of P1 (lane 3). In the absence of the target, Cas13a is inactive, and no P2 is produced (lane 4), and the band of LOCK-U keeps unchanged (lane 4). Fig. S3 illustrates that in the presence of only LOCK-U and the fuel chain, no P2 or other extraneous bands are generated (lanes 4 and 5), indicating the stability of the hairpin structure of LOCK-U.
Optimization of CRISPR/Cas13a cleavage and validation of specificity
To further evaluate the cleavage efficiency of Cas13a, the influence of the ratio of Cas13a to crRNA on the performance was studied. The FAM and BHQ1-modified signal probe FQ5U was employed as the substrate, and the change in fluorescence intensity over 10 min (ΔF) was utilized as the index. As shown in Fig. S4A, when the concentration of Cas13a is higher than that of crRNA, the ΔF elevates, indicating higher cleavage efficiency. On the contrary, when the concentration of Cas13a is lower than that of crRNA, the cleavage reaction is inhibited. Cas13a usually cannot fully bind with crRNA in a 1:1 ratio. When crRNA is present in excess, Cas13a will cut crRNA, instead of the substrate sequence, resulting in a relatively lower fluorescence intensity. To verify this speculation, the concentration of Cas13a was fixed at 40 nM and the concentration of crRNA was varied in the cleavage system. The results are shown in Fig. S4B. The variation of fluorescence intensity increases with the decrease of crRNA concentration, and the optimal ratio of Cas13a to crRNA was determined to be 4:1.
Furthermore, the specificity of the CRISPR/Cas13a recognition and signal amplification system was evaluated by introduction of miR-21 and other miRNAs (miR-21b, miR-21c, miR-141, miR-199) respectively and determination of the fluorescence intensity within 30 min. Significant change in fluorescence can only be observed when miR-21 is present, proving that Cas13a has excellent base recognition ability to accurately identify the target from other sequences (Fig. 1D).
Characterization of synthesisand modification of MnO2 NSs
Ultrathin MnO2 NSs were prepared using a bottom-up one-step synthesis method, and the synthesized MnO2 NSs were characterized [33]. According to the results from X-ray photoelectron spectroscopy (XPS), the presence of Mn and O was confirmed (Fig. 2A) [34]. The high-resolution XPS spectrum of Mn 2p shows characteristic peaks of Mn 2p3/2 at 642.3 eV and Mn 2p1/2 at 654.3 eV, with a spin energy separation of 12 eV (Fig. 2B), which proves the existence of manganese dioxide. Additionally, the characteristic peaks of Mn2O3 and KMnO4 were not found in the XPS spectra. Based on the observation of Fig. 2C, it can be concluded that the XPS spectrum of O 1 s exhibits a relatively weak peak at 531.7 eV and a sharp peak at 529.9 eV, corresponding to two types of oxygen in the MnO2 NSs: one in the interlayer water and the other in the [MnO6] octahedral lattice, respectively [33]. In the TEM image of MnO2 NSs (Fig. 2D), a clear transparent layered structure can be observed, showing a typical two-dimensional morphology with a large surface area, which provides sufficient reaction area with TMB. The compositional analysis of the prepared MnO2 NSs by energy dispersive X-ray spectroscopy (EDS) shows that the nanosheets are composed of Mn, O, and a small amount of K (Fig. 2E).
FT-IR was employed to characterize the modification of the biotin-labeled probe on MnO2 NSs. The FT-IR spectra of MnO2 NSs before and after modification were recorded (Fig. 2F). The spectra demonstrate several characteristic bands that indicate the presence of specific molecules on MnO2 NSs. The band at 3421 cm−1 corresponds to the stretching vibration of H2O molecules, suggesting the existence of H2O and H3O+ species [33]. Additionally, the symmetric stretching vibration peak of COO− can be detected at 1400.42 cm−1, indicating the presence of the carboxyl group. At 1629.45 cm−1, a peak similar to the C = O bond peak of the carboxyl group can be observed, although with weaker intensity (black line). The peak at 1646 cm−1 corresponds to the C = O stretching vibration, while the peaks at 1560 cm−1 and 1227 cm−1 represent the N–H bending vibration and C-N stretching vibration, respectively (red line). These findings provide evidence that the linkage between MnO2 NSs and the biotin-probe was successfully achieved through an amide bond [35, 36].
Catalytic capacity of MnO2 NSs
The ability of catalytic oxidation of TMB by MnO2 NSs was assessed, both in the solution and on the NC membrane.
Figure 3A demonstrates the reaction of between MnO2 NSs and TMB in PBS, where the changes in color and UV absorption spectra can be observed. In the UV–vis spectra, no distinct absorption peaks can be observed within the wavelength range of 330 to 760 nm for TMB, PBS, and MnO2 NSs (curves a, b, and c in Fig. 3A). However, when MnO2 NSs are mixed with TMB in PBS, a color change from colorless to blue occurs (Fig. 3A, insets), accompanied by a strong absorption peak at 652 nm in UV–vis spectra (curve d in Fig. 3A). This indicates that MnO2 NSs effectively catalyze the oxidation of the colorless substrate TMB, resulting in the formation of a blue-green product, ox-TMB.
The conventional visual lateral flow test strips generally use gold nanoparticles (AuNPs) as signal labels. However, the sensitivity of the lateral flow test strips is thus restricted by the intrinsic plasmon activity of AuNPs. Therefore, MnO2 NSs-catalyzed chromogenic reaction is employed in this work. Considering the post-addition of the substrate TMB on the T zone will lead inconvenience of the operation, pre-fixation of TMB on the T zone is employed. And the performance of the chromogenic reaction on the NC membrane was firstly investigated. The results are shown in Fig. 3B; compared with the control group (add water), both color change and gray value change can be observed when MnO2 NSs are added to the NC membrane pre-fixed with TMB (Fig. S6), indicating the successful catalysis of TMB by MnO2 NSs. The color of the bands on the NC membrane changes from colorless to light yellow and green, and finally to blue, with the corresponding increased concentration of pre-fixed TMB. Therefore, the chromogenic reaction catalyzed by MnO2 NSs and the pre-fixation of the substrate on the NC membrane is feasible.
Optimization of the lateral flow test strip
The lateral flow test strip utilizes MnO2 NSs to catalyze the oxidation of TMB and generate the signal output. However, when TMB solution is exposed to air, TMB can be slightly oxidized by oxygen, even without any catalysts, leading to a slight change of the solution color or the appearance of a background color on the NC membrane. Therefore, precise control of fixation and reaction conditions of TMB is crucial to minimize the generation of the background color.
The chromogenic system operates in PBS solution, and the mixed droplets of PBS and TMB were added to the T zone and then dried under vacuum during the preparation of the lateral flow test strip. In order to reduce the background color after TMB fixation and drying, (G − G0)/G0 was employed as the reference index for optimization. G represents the gray value in the presence of MnO2 NSs + TMB + PBS, while G0 represents the gray value in the presence of Water + TMB + PBS. The pH of the buffer was firstly optimized. As shown in Fig. 4A, (G − G0)/G0 is the highest at pH 6.0, which indicates the most significant change in gray value between the experimental and control groups. The background color is also the lightest at this pH (Fig. S5). However, the background color of the control group is darker at pH 6.7, which may lead to false positive results. By contrast, compared to pH 6.0, there is little difference in the background color at pH 5.3. Considering the requirement for both DNA hybridization and chromogenic reaction occurring on the T zone, PBS with pH 6 was selected as the optimal reaction solution.
After the application of TMB solution on the NC membrane, the drying of the lateral flow test strip was conducted. The drying temperature is also a critical factor. As shown in Fig. 4B, when the drying temperature is 25 °C, (G − G0)/G0 reaches the maximum value and differs significantly from the values at 18 °C and 35 °C. The color change after the reaction is also most pronounced at this temperature (Fig. S6).
The concentration of TMB is a key factor in determining the performance of the experiments. As depicted in Fig. S6 with the increase in TMB concentration, the color intensity in the experimental group (T zone) becomes more pronounced. However, when the TMB concentration is excessively high, the control group (T zone) also displays noticeable color change, increasing the risk of false positives. Figure 4C indicates that at the TMB concentration of 12 mM, the value of (G − G0)/G0 is maximized, indicating the most significant change in gray values between the experimental and control groups, along with a notable contrast in color in the T zone (Fig. S6).
The optimization of the concentration of the capture probe is critical as it affects both the capture efficiency of MnO2 NSs and the color generation of the T zone. When the concentration of miR-21 was fixed at 10 nM, varying concentrations of the capture probe were immobilized on the T zone, followed by detection. The gray value is used as an index. As the concentration of the capture probe increases, the observed color of the lateral flow test strips is gradually deepened (Fig. S7), and the gray value of the T zone exhibits a gradual increase. However, once the concentration of the capture probe surpasses 1 nM, the gray value of the T zone remains relatively constant (Fig. 4D). Therefore, 1 nM capture probe was used to modify the T zone.
Performance of the lateral flow test strip
To test the analytical performance of the lateral flow test strip, miR-21 ranging from 0.1 to 150 pM was applied to the lateral flow test strips, respectively, and tested under the optimal reaction conditions. The gray value of the T zone for each sample was shown after processing the images using ImageJ software. In the absence of miR-21, no band can be observed on the T zone, and the gray value is similar to the blank. At the concentration of 0.8 pM, the T zone displays a slight color change, and the gray value gradually increases with the increasing concentration of miR-21 (Fig. 5A and B). A linear relationship can be derived between gray value and the concentration of miR-21 in the range from 1 to 50 pM, with the regression equation of y = 0.2165x + 10.8261 (R2 = 0.9955), and a detection limit of 0.33 pM (Fig. 5C). Compared to the reported assays for miRNA, the established lateral flow assay has a low detection limit (Table 1).
Results of visual and quantitative detection of miR-21 by the lateral flow test strip. A The image of the lateral flow test strip after detection of different concentrations of miR-21. B The calibration curve of miR-21. C Linear detection range of miR-21. D Validation of the specificity of the lateral flow test strip using different miRNAs. E Stability evaluation of the lateral flow test strip
To assess the specificity of this assay, a series of miRNAs were employed for validation, including miR-141, miR-199, miR-21b, and miR-21c. The results are shown in Fig. 5D; the presence of miR-21 leads to a significant increase in gray value and color change in the T zone (Fig. S8), while other miRNAs do not induce any significant color change and gray value increase in the T zone.
The stability of the flow test strip is important in practical applications. The prepared lateral flow test strips were stored at 4 °C in the dark. The lateral flow test strips were utilized for the detection of various concentrations of the target (1, 10, 30, and 50 pM) after storage of 0, 7, 14, and 21 days. The results of the experiments are presented in Fig. 5E and Fig. S9. A very slight increase in gray value and the background color can be observed within 21 days of storage, which can be ascribed to the self-oxidation of TMB. But it has no significant effect on the detection performance. Therefore, the lateral flow test strips show high stability.
Detection of miR-21 in real samples
In order to assess the practical application of the lateral flow test strip, different concentrations of miR-21 were spiked in 10% diluted human serum and tested using the same protocol. The gray value of the T zone was then read and used to quantify the concentration of miR-21. The results obtained are presented in Table 2. The recovery rate of miR-21 ranges between 101.7 and 105.8%, and the RSD is between 3.37 and 7.62%. These results indicate that the lateral flow test strip has the potential for clinical detection of miR-21.
Clinical serum samples from individuals diagnosed with liver, stomach, and lung cancers, as well as serum samples from healthy people, were employed to assess the effectiveness of the lateral flow test strip. As depicted in Fig. S10, the ratio of miR-21 levels in patients’ serum to those in healthy individuals’ serum was used as the index. A comparative analysis was then conducted between the outcomes of the lateral flow test strips and those generated by quantitative polymerase chain reaction (qPCR). The results show the similarity between the two detection methods, further demonstrating the reliability and practicality of the lateral flow test strip.
Conclusion
In conclusion, this work presents the development of a rapid, accurate, sensitive, and highly specific point-of-care test for miR-21 by combining the CRISPR/Cas13a recognition and signal amplification system with a nanozyme-based high-sensitive lateral flow assay. The components of the chromogenic system including MnO2 NSs and the substrate TMB are both pre-fixed on the lateral flow test strip as the dry reagent, which greatly facilitate the operation of the detection. miR-21 can be detected in less than 10 min, without complicated pretreatment. miR-21 can be determined qualitatively by the naked eyes or quantitatively via ImageJ software, with LOD of 0.33 pM. The dual signal amplification mechanisms including CRISPR/Cas13a-mediated cleavage and strand displacement in the solution and the MnO2 NSs-catalyzed chromogenic reaction greatly increase the sensitivity of the lateral flow test strip. By modifying the LOCK-U, the lateral flow test strip can easily be utilized for detecting other miRNAs. Therefore, the constructed lateral flow test strip offers new possibility for clinical diagnosis and testing in point-of-care settings.
References
Wang X, He Y, Mackowiak B, Gao B. MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut. 2021;70:784–95. https://doi.org/10.1136/gutjnl-2020-322526.
Jenike AE, Halushka MK. Mir-21: A non-specific biomarker of all maladies. Biomark Res. 2021;9:18. https://doi.org/10.1186/s40364-021-00272-1.
Yin F, Cai R, Gui S, Zhang Y, Wang X, Zhou N. A portable and quantitative detection of microRNA-21 based on cascade enzymatic reactions with dual signal outputs. Talanta. 2021;235: 122802. https://doi.org/10.1016/j.talanta.2021.122802.
Wang Y, Hu Y, Xie R, Zeng Q, Hong Y, Chen X, et al. Ultrasensitive label-free miRNA-21 detection based on MXene-enhanced plasmonic lateral displacement measurement. Nanophotonics-berlin. 2023;12:4055–62. https://doi.org/10.1515/nanoph-2023-0432.
Seo SB, Hwang J-S, Kim E, Kim K, Roh S, Lee G, et al. Isothermal amplification-mediated lateral flow biosensors for in vitro diagnosis of gastric cancer-related microRNAs. Talanta. 2022;246: 123502. https://doi.org/10.1016/j.talanta.2022.123502.
Huang Y, Zhang Y, Hao W, Lu H, Dong H, Zhang X. Sensitive microRNA detection based on bimetallic label photothermal lateral flow locked nucleic acid biosensor with smartphone readout. Sensor Actuat B-chem. 2023;375: 132945. https://doi.org/10.1016/j.snb.2022.132945.
Kor K, Turner AP, Zarei K, Atabati M, Beni V, Mak WC. Structurally responsive oligonucleotide-based single-probe lateral-flow test for detection of miRNA-21 mimics. Anal Bioanal Chem. 2016;408:1475–85. https://doi.org/10.1007/s00216-015-9250-9.
Gao X, Xu H, Baloda M, Gurung AS, Xu L-P, Wang T, et al. Visual detection of microRNA with lateral flow nucleic acid biosensor. Biosens Bioelectron. 2014;54:578–84. https://doi.org/10.1016/j.bios.2013.10.055.
Jin X, Chen L, Zhang Y, Wang X, Zhou N. A lateral flow strip for on-site detection of tobramycin based on dual-functional platinum-decorated gold nanoparticles. Analyst. 2021;146:3608–16. https://doi.org/10.1039/d1an00403d.
Gao X, Xu L-P, Wu T, Wen Y, Ma X, Zhang X. An enzyme-amplified lateral flow strip biosensor for visual detection of microRNA-224. Talanta. 2016;146:648–54. https://doi.org/10.1016/j.talanta.2015.06.060.
Zhang J, Tang L, Yu Q, Qiu W, Li K, Cheng L, et al. Gold-platinum nanoflowers as colored and catalytic labels for ultrasensitive lateral flow microRNA-21 assay. Sensor Actuat B-chem. 2021;344: 130325. https://doi.org/10.1016/j.snb.2021.130325.
Xu C, Zhou Y, Xiao Q, He B, Geng G, Wang Z, et al. Programmable RNA editing with compact CRISPR–Cas13 systems from uncultivated microbes. Nat Methods. 2021;18:499–506. https://doi.org/10.1038/s41592-021-01124-4.
Zhou H, Bu S, Xu Y, Xue L, Li Z, Hao Z, et al. CRISPR/Cas13a combined with hybridization chain reaction for visual detection of influenza a (H1N1) virus. Anal Bioanal Chem. 2022;414:8437–45. https://doi.org/10.1007/s00216-022-04380-1.
Cui Y, Fan S, Yuan Z, Song M, Hu J, Qian D, et al. Ultrasensitive electrochemical assay for microRNA-21 based on CRISPR/Cas13a-assisted catalytic hairpin assembly. Talanta. 2021;224: 121878. https://doi.org/10.1016/j.talanta.2020.121878.
Liu FX, Cui JQ, Park H, Chan KW, Leung T, Tang BZ, et al. Isothermal background-free nucleic acid quantification by a one-pot Cas13a assay using droplet microfluidics. Anal Chem. 2022;94:5883–92. https://doi.org/10.1021/acs.analchem.2c00067.
Shan Y, Zhou X, Huang R, Xing D. High-fidelity and rapid quantification of miRNA combining crRNA programmability and CRISPR/Cas13a trans-cleavage activity. Anal Chem. 2019;91:5278–85. https://doi.org/10.1021/acs.analchem.9b00073.
Wessels H-H, Méndez-Mancilla A, Guo X, Legut M, Daniloski Z, Sanjana NE. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat Biotechnol. 2020;38:722–7. https://doi.org/10.1038/s41587-020-0456-9.
Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–44. https://doi.org/10.1126/science.aaq0179.
Zhou T, Huang M, Lin J, Huang R, Xing D. High-fidelity CRISPR/Cas13a a trans-cleavage-triggered rolling circle amplified dnazyme for visual profiling of microRNA. Anal Chem. 2021;93:2038–44. https://doi.org/10.1021/acs.analchem.0c03708.
Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–4. https://doi.org/10.1038/nature24049.
Zhou B, Ye Q, Li F, Xiang X, Shang Y, Wang C, et al. CRISPR/Cas12a based fluorescence-enhanced lateral flow biosensor for detection of staphylococcus aureus. Sensor Actuat B-chem. 2022;351: 130906. https://doi.org/10.1016/j.snb.2021.130906.
Arizti-Sanz J, Bradley AD, Zhang YB, Boehm CK, Freije CA, Grunberg ME, et al. Simplified Cas13-based assays for the fast identification of SARS-CoV-2 and its variants. Nat Biomed Eng. 2022;6:932–43. https://doi.org/10.1038/s41551-022-00889-z.
Liu J, Meng L, Fei Z, Dyson PJ, Jing X, Liu X. MnO2 nanosheets as an artificial enzyme to mimic oxidase for rapid and sensitive detection of glutathione. Biosens Bioelectron. 2017;90:69–74. https://doi.org/10.1016/j.bios.2016.11.046.
Hu L, Yuan Y, Zhang L, Zhao J, Majeed S, Xu G. Copper nanoclusters as peroxidase mimetics and their applications to H2O2 and glucose detection. Anal Chim Acta. 2013;762:83–6. https://doi.org/10.1016/j.aca.2012.11.056.
Hizir MS, Top M, Balcioglu M, Rana M, Robertson NM, Shen F, et al. Multiplexed activity of perAuxidase: DNA-capped AuNPs act as adjustable peroxidase. Anal Chem. 2015;88:600–5. https://doi.org/10.1021/acs.analchem.5b03926.
Shi W, Wang Q, Long Y, Cheng Z, Chen S, Zheng H, et al. Carbon nanodots as peroxidase mimetics and their applications to glucose detection. Chem Commun. 2011;47:6695. https://doi.org/10.1039/c1cc11943e.
Liu B, Liu J. Surface modification of nanozymes Nano Res. 2017;10:1125–48. https://doi.org/10.1007/s12274-017-1426-5.
Jiao L, Yan H, Wu Y, Gu W, Zhu C, Du D, et al. When nanozymes meet single-atom catalysis. Angew Chem Int Edit. 2019;59:2565–76. https://doi.org/10.1002/anie.201905645.
Wei H, Wang E. Fe3O4 magnetic nanoparticles as peroxidase mimetics and their applications in H2O2 and glucose detection. Anal Chem. 2008;80:2250–4. https://doi.org/10.1021/ac702203f.
Jiang T, Song Y, Wei T, Li H, Du D, Zhu M-J, et al. Sensitive detection of Escherichia coli O157:H7 using Pt–Au bimetal nanoparticles with peroxidase-like amplification. Biosens Bioelectron. 2016;77:687–94. https://doi.org/10.1016/j.bios.2015.10.017.
Song Y, Qu K, Zhao C, Ren J, Qu X. Graphene oxide: Intrinsic peroxidase catalytic activity and its application to glucose detection. Adv Mater. 2010;22:2206–10. https://doi.org/10.1002/adma.200903783.
Yan X, Song Y, Wu X, Zhu C, Su X, Du D, et al. Oxidase-mimicking activity of ultrathin MnO2 nanosheets in colorimetric assay of acetylcholinesterase activity. Nanoscale. 2017;9:2317–23. https://doi.org/10.1039/c6nr08473g.
Liu Z, Xu K, Sun H, Yin S. One-step synthesis of single-layer MnO2 nanosheets with multi-role sodium dodecyl sulfate for high-performance pseudocapacitors. Small. 2015;11:2182–91. https://doi.org/10.1002/smll.201402222.
Cai X, Liang M, Ma F, Zhang Z, Tang X, Jiang J, et al. Nanozyme-strip based on MnO2 nanosheets as a catalytic label for multi-scale detection of aflatoxin B1 with an ultrabroad working range. Food Chem. 2022;377: 131965. https://doi.org/10.1016/j.foodchem.2021.131965.
Sheng E, Lu Y, Tan Y, Xiao Y, Li Z, Dai Z. Oxidase-mimicking activity of ultrathin MnO2 nanosheets in a colorimetric assay of chlorothalonil in food samples. Food Chem. 2020;331: 127090. https://doi.org/10.1016/j.foodchem.2020.127090.
Shi M, Wang S, Zheng S, Hou P, Dong L, He M, et al. Activatable MRI-monitoring gene delivery for the theranostic of renal carcinoma. Colloid Surface B. 2020;185: 110625. https://doi.org/10.1016/j.colsurfb.2019.110625.
Yao M, Lv X, Deng Y, Rasheed M. Specific and simultaneous detection of microRNA 21 and let-7a by rolling circle amplification combined with lateral flow strip. Anal Chim Acta. 2019;1055:115–25. https://doi.org/10.1016/j.aca.2018.12.040.
Cimmino W, Migliorelli D, Singh S, Miglione A, Generelli S, Cinti S. Design of a printed electrochemical strip towards miRNA-21 detection in urine samples: optimization of the experimental procedures for real sample application. Anal Bioanal Chem. 2023;415:4511–20. https://doi.org/10.1007/s00216-023-04659-x.
Deng K, Zhang Y, Tong X. Sensitive electrochemical detection of microRNA-21 based on propylamine-functionalized mesoporous silica with glucometer readout. Anal Bioanal Chem. 2018;410:1863–71. https://doi.org/10.1007/s00216-018-0859-3.
Zheng W, Yao L, Teng J, Yan C, Qin P, Liu G, et al. Lateral flow test for visual detection of multiple microRNAs. Sensor Actuat B-chem. 2018;264:320–6. https://doi.org/10.1016/j.snb.2018.02.159.
Wei H, Peng Y, Bai Z, Rong Z, Wang S. Duplex-specific nuclease signal amplification-based fluorescent lateral flow assay for the point-of-care detection of microRNAs. Analyst. 2021;146:558–64. https://doi.org/10.1039/d0an01673j.
Funding
This work was financially supported by the National Natural Science Foundation of China (42177212, 62301234, 32371430).
Author information
Authors and Affiliations
Contributions
Mingyuan Wang: ideas, investigation, data curation, writing — original draft. Shixin Cai: investigation, data curation. Yunqing Wu: formal analysis, validation. Qi Li: supervision. Xiaoli Wang: management, funding acquisition. Yuting Zhang: supervision, funding acquisition. Nandi Zhou: writing — review and editing, supervision, funding acquisition. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
The experiments were approved by the Ethics Committee of the Wuxi No.2 People’s Hospital in Jiangsu Province, China.
Source of biological material
All of the clinical serum samples were collected from the Wuxi No.2 People’s Hospital in Jiangsu Province, China.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, M., Cai, S., Wu, Y. et al. A lateral flow assay for miRNA-21 based on CRISPR/Cas13a and MnO2 nanosheets-mediated recognition and signal amplification. Anal Bioanal Chem 416, 3401–3413 (2024). https://doi.org/10.1007/s00216-024-05290-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-024-05290-0