Abstract
Infectious diseases are illnesses caused by harmful pathogens from the exterior. Vaccines could prevent many people from disease infection. Over the past several years, the development and application of molecular diagnostic techniques have launched a revolution in monitoring infectious diseases. Polymerase chain reaction (PCR)-based systems to diagnose the etiologic agents of disease from clinical samples have been applicable in pathogen detection. We demonstrate a microfluidic chip for continuous flow PCR. The PDMS/glass bonding chip provides a miniaturized, cheap, and disposable material for pathogen diagnosis. The homemade thermal control module integrated with two cartridge heaters and one Peltier element supports the denaturation, extension, and annealing regions created inside the chip. Due to the large surface-to-volume ratio in the microchannel, the surface characteristics might augment the protein adsorption onto the channel surfaces and reduce the PCR amplification efficiency. We measure the hydrophilic properties, roughness of the wall and the structure of the surface under various surface treatments and express the PCR amplification efficiency. Results show that the most detailed is the PDMS sheet with the modification of Tween 20 of a concentration of 20% for the reduction of methyl peak of the absorption spectrum, the lowest contact angle, and the minor surface roughness. Next, the continuous flow PCR system amplifies a 385-bp segment of Q fever virus DNA to evaluate the performance of the DNA amplification. The overall product of Tween 20 is good because Tween 20 is an emulsifier, and it has excellent performance in both adhesion properties and coated samples. The need for point-of-care test (PoCT) devices has increased rapidly since the outbreak of COVID-19. The current portable device for PoCT will provide essential tools for real-time diagnosis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
During infectious disease outbreaks, the centers for disease control need to monitor particular areas. Regular monitoring ensures effective therapy and detects treatment failure as early as possible to minimize the emergence of drug-resistant microbes. In well-resourced settings, the cornerstone of monitoring requires standard assays, usually performed on a large laboratory platform device. Most of the devices require a desktop instrument and laboratory processing. However, it is difficult to diagnose many infectious cases in remote areas due to limited access to diagnostic tests, insufficient sensitivity, or long turnaround times. More rapid and accurate detection at lower levels of care is necessary to reach these lost and missed cases. Developing portable, user-friendly, and cost-effective systems for real-time diagnostics is helpful.
Since Saiki et al. (1985), and Mullis and Faloona (1987) introduced the polymerase chain reaction (PCR) assay, nucleic acid amplification has become one of the most reliable techniques in molecular diagnostics of infectious diseases. It offers speed and sensitivity for pathogen detection. Commercial PCR devices typically comprise a computer-controlled thermocycler and several reaction chambers. In these PCR machines, a Peltier element is commonly the heating and cooling source to support the required temperatures for PCR. Though PCR is a widely used technology for DNA amplification, temperature cycling during PCR increases the system cost and the operation time. Three consecutive steps (denaturation step at about 95 °C, annealing step at about 55 °C, and extension step at about 72 °C) per cycle are necessary for DNA replications. Usually, many researchers use 35 thermal cycles to complete the process. However, the PCR chip introduces the possibilities of cheapness and rapidity by using a diverse choice (thermoelectric modules (Chiou et al. 2013), thin film resistors (Felbel et al. 2008), and cartridge heaters (Chen et al. 2013).
The chamber-type PCR chip is a miniaturized version of the conventional thermal cycler. The temperature of the stationary mixture inside the chip is cyclic. On the other hand, the amplification mixture repeatedly moves through two or three isothermal regions to complete thermal cycles in the continuous-flow PCR (CFPCR) chip. The geometry of channels and the arrangement of heating regions determine the residence time and the number of thermal processes. Rapid thermal cycling is beneficial compared with the typical chamber-type PCR.
There are three main types of CFPCR chips: unidirectional (Kwak et al. 2021), closed-loop (Wang et al. 2009), and oscillatory (Kopparthy and Crews 2020) chips. Each one has its advantages. A syringe pump pushes the reactive sample through a channel among several constant temperature regions for the unidirectional chip. The mixture flows into the inlet, performs the PCR process, finishes the requisite thermal cycles, and then flows out the outlet for detection. However, the long sample moving distance could result in high PCR inhibition. So, the surface of the microchannels was modified with chemicals to fix the inhibition issue.
Several proteins, such as Taq DNA polymerase and template DNA, are inside the PCR mixture. When the free energy between a surface and a protein is less than that of the interface and water, protein adsorption usually occurs on the surface. Because commonly used materials, e.g., hydrophobic polydimethylsiloxane (PDMS), in the microchannel, tend to have relatively higher free energies than bulk water, proteins sometimes absorb on the channel surface to be stable. The PCR mixture flows through the long microchannel, i.e., increased overall contact surface, to complete the PCR process for the unidirectional chip. Thus, the surface chemistry of the inside wall of microfabricated devices critically affects the efficiency of PCR, so many researchers paid much attention to protein absorption during CFPCR.
Bovine serum albumin (BSA) is the most widely used protein for surface passivation applications. BSA coats the channels very well and keeps proteins of interest from surface adsorbing (Qin et al. 2016). Nakayama et al. (2006) utilized 0.2 mg/mL BSA to prevent the adsorption of PCR reagents on the PDMS channel surface. Due to the PCR enzyme binding non-specifically to the PDMS channel walls and becoming inactive in a high surface-to-volume ratio reaction, Cao et al. (2012) used BSA as a blocking agent to improve amplification in microchannels. They optimized the PCR assay for 0.03% (w/v) BSA concentration to push the detection limit to 104 copies/mL. Jiang et al. (2014) pre-passivated the inner surface of the PDMS chip channel before PCR amplification under a continuous BSA flow (0.05 mg/mL) to alleviate the Taq-polymerase adsorption. Han et al. (2014) did an acid/base solution treatment to prevent the binding of biomolecules on the PDMS surface. They injected HCl and NaOH solution to neutralize the inner charge of the PDMS surface. BSA was then incubated for 30 min inside the microchannel for efficient surface treatment. Their results demonstrated that the PCR amplification in untreated channels decreases the product concentration due to the binding of biomolecules to the PDMS surface. Kopparthy and Crews (2020) used a 2.5 mg/mL BSA in the PCR mixture. BSA binds to the amino silane PDMS channel walls and prevents the intercalating dye, LC Green, and DNA binding. Polytetrafluoroethylene (PTFE), consisting of carbon and fluorine, is an excellent hydrophobic material for microfluidic applications. Peham et al. (2011) included BSA in the master mix for dynamic passivation of the microfluidic tubing consisting of transparent PTFE. Although the PTFE tubing has inert properties, 1 mg/mL, BSA stabilized DNA polymerases and increases PCR efficiency.
Some researchers utilized other surface treatment reagents with BSA to reduce the surface adsorption effect. The surfactant Tween 20 (C58H114O26) served as a dispersant, emulsifier, and solubilizer to protect the enzyme (Coutlée and Voyer 1998). Schneegaß et al. (2001) found BSA to be very effective in preventing surface denaturation of polymerase on glass channel surfaces. In addition to the surface treatment of the chip device and the supplement of BSA and Tween 20, mineral oil presented a good combination for the efficient amplification of DNA in the chip device. Felbel et al. (2004) used a chemically oxidized silicon ((CH3)2SiCl2)-modified PCR chip made in silicon in combination with dynamic coating with 1.5 µM BSA to repress the surface-mediated inhibition of Taq DNA polymerase and template DNA. It demonstrated that the PCR’s specificity and product yield were equivalent to a conventional PCR machine. Chen et al. (2007) used a silanized glass capillary with dimethyldichlorosilane (DMDCS) in combination with dynamic surface passivation comprising 0.1% (v/v) Tween 20, 0.01% (w/v) poly(vinylpyrrolidone) (PVP), and 0.2 mg/mL BSA to reduce reagent adsorption and PCR inhibition and enable efficient PCR.
The other treatment methods also positively affect the modification of the PDMS surface. Kwak et al. (2021) demonstrated a consecutive serial analysis for multiple amplified DNA samples in a CFPCR microchip. The isobornyl acrylate (IBA, a highly hydrophobic monomer) mixture (poly(ethylene glycol) dimethacrylate and Irgacure included) with a UV irradiation modifies the surfaces of PDMS microchannels to fix polyacrylamide gels. 2-methacryloyloxyethyl phosphorylcholine (MPC, C11H22NO6P) has a zwitterionic phosphorylcholine headgroup and can prevent nonspecific protein adsorption very well (Seetasang and Xu 2022). Fukuba et al. (2004) coated the surface of the PDMS channel with an MPC-based polymer with a silane coupler. The MPC-based polymer coating terminates the adsorption of DNA polymerase onto the entire surface of the PDMS microchannels. Kuan et al. (2008) studied the effects of grafting poly(ethylene oxide) (PEO, C2nH4n + 2On+1) onto the surface of a PDMS microchannel on the amplification efficiency of a microfabricated device for PCR. The surface treatments with PEO-based surfactants, such as neat silane and Pluronic® F127 ((C3H6O·C2H4O)x), increased in the PCR amplification of a 298-bp DNA product and a 1.1-kb DNA product. Kim et al. (2022) presented the fabrication of a microfluidic PCR device integrated with a PID system and real-time thermal feedback for genetic analysis. The MPC-based polymer was coated to the surface of the PDMS channel to prevent the adsorption of protein molecules onto it.
During PCR, the extension time is longer than the denaturation and annealing time. Previous studies (Schneegaß et al. 2001; Joung et al. 2008; Wu et al. 2015; Kulkarni et al. 2021) used a long microchannel on each extension region to obtain the required extension time for CFPCR. The DNA mixture moves through three isothermal zones, and the length of the extension zone is usually more significant than that of the other two zones. Then, the microfluidic chip size may be increased. The microchannel width increases and then decreases gradually between the denaturation and annealing regions, and the extension region is located in the enlarged part of the channel. Hence, obtaining the required extension time over the microchannel is possible. Li et al. (2006) presented a CFPCR microchip with a serpentine channel of varying width for regional velocity control. The ratio of the widths for denaturation, annealing, and extension zones was predetermined to 1:2:4, respectively. Chen et al. (2015) introduced heat pipes into the cooling module in a CFPCR device. The 30-loop meandering channel is 150 μm in width except for the extension region, which has a maximum width of 540 μm. They can appropriately extend the residence time during the extension stage. Kaprou et al. (2019) fabricated a CFPCR chip to amplify the exon 20 of the BRCA1 gene and the plasmid pBR322. The duration of the extension step increases with the microchannel width increasing by 1.5 at the extension zone.
Static passivation involves pre-treating the channel surface with a passive coating. Researchers have dealt with efficient surface treatments before biological reactions under a continuous passive substance flow for some time or incubated passivation reagents inside the microchannel. Dynamic passivation involves including passivation reagents in the reactive mixture to make the material compatible with the reaction. Some have dealt with surface passivation using a passive reagent in the PCR mixture. Despite the advantages of PDMS as the materials in microfluidics, its hydrophobicity may cause problems such as difficulty filling aqueous solution and adsorption of biological molecules, especially in miniaturized devices with high surface-to-volume ratios. However, previous studies conducted surface treatment issues in CFPCR devices. The effect of various materials on surface passivation still needs detailed information.
To establish a smooth temperature gradient on the unidirectional CFPCR chip, many researchers set the ordinal position of temperature zones as denaturation, extension, and annealing with three individual heating elements. Hsieh et al. (2022) presented a microfluidic PCR chip and two aluminum blocks with an electric heating film separately defined the temperature zones of 95 and 60 °C. In contrast, the heat transfer from the sides of the aluminum blocks maintains the extension temperature zone in the middle of the device.
This study describes a continuous-flow thermocycling approach and shows that we utilize homemade thermal control sources to accomplish fast temperature ramping. Integrating one Peltier element onto the microfluidic CFPCR chip makes generating and regulating the annealing temperature easy. The infrared absorption spectrum, contact angles and surface roughness demonstrate the comprehensive surface measurements. We use the surface characteristics to examine the protein absorption effect of the hydrophobic PDMS surface. Finally, results show that a 385-bp segment of Coxiella burnetii DNA is amplified successfully in the DNA amplification system.
2 Materials and methods
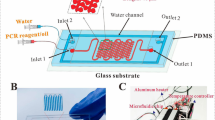
The continuous flow device for polymerase chain reaction consists of one glass/ polydimethylsiloxane (PDMS) bonding microfluidic chip with a poly(methyl methacrylate) (PMMA) structural frame, three homemade thermal control modules for each isothermal region, and a commercial syringe pump for the DNA mixture injection. After finishing the bonding chip, we utilize the surface treatment method to reduce the protein absorption onto the microchannel. We employ two cartridge heaters and one thermoelectric (TEC) cooler to heat up and cool down the temperature of the DNA mixture during thermal cycling, respectively. The DNA mixture moves through the denaturation, annealing, and extension regions to complete one PCR thermal cycle. With the increasing and decreasing width of the microchannel during the extension, the duration time is adequate for PCR thermocycling. After passing through a designated thermal cycle number, we take the DNA product out of the chip for gel electrophoresis.
2.1 Design and fabrication of the reaction chip
We can fabricate the PDMS chip using microelectromechanical system (MEMS) technology. The microfluidic chip is ready after bonding the glass slide onto the PDMS chip. The channel pattern designs the number of cycles performed throughout the chip. The channel layout and the flow rate of the mixture determine the residence time a mixture element spends in each temperature zone.
Our earlier works (Chen et al. 2013, 2015) have depicted the comprehensive fabrication process of the PDMS-glass bonding chip, which includes two steps. The first one is the photolithography process. We clean a silicon wafer by following a specific protocol and dehydrate it on a hotplate. An epoxy-based negative photoresist (SU-8) is spin-coated on a silicon wafer. The photoresist is then soft-baked. The channel pattern exposed to ultraviolet (UV) becomes cross-linked. After development, the master wafer is post-baked to fix the photoresist. Once the mold is complete, the wafer is rinsed in deionized water (DI water) and dried with nitrogen (Fig. 1(a)). The second one is the casting process. The PDMS (Sylgard 184, Dow Corning, Midland, Michigan, USA) mixture thoroughly mixed with the precursor and curing agent in a 10:1 weight ratio is degassed with a mechanical vacuum pump to remove air bubbles. After pouring the PDMS mixture onto the SU-8 patterned master, the PDMS cures in a convection oven, and the replica is peeled off carefully from the master. The hole puncher creates the chip’s inlet and outlet. Methanol is a surfactant that prevents the oxygen-plasma-treated PDMS replica and glass slide from being irreversibly bonded and improperly aligned. The designed chips are ready after surface oxidation and bonding, as shown in Fig. 1(b).
The PDMS chip is 65 mm × 25 mm × 2 mm in outer size. A PDMS channel sheet bonding with a flat glass substrate (75 mm × 25 mm × 1 mm) forms the microfluidic chip. The chip channel is composed of 30 identical cycles. The channel size of the CFPCR chip is 50 μm in-depth (shown in Fig. 2(a)) and 150 μm in width except for the extension region, with a maximum width of 650 μm and 1296.37 mm in length. To ensure enough time for DNA extension, the width of the channel increases to 650 μm and then decreases to 150 μm at the extension region. Figure 2(b) also shows the SEM pictures of the microchannels. We utilize a PMMA housing to assemble the glass slide, the PDMS microchannel sheet, and three heating modules.
2.2 Heating module, cooling module and temperature sensing system
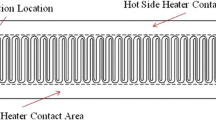
Each homemade thermal control module incorporates one proportional/integral/derivative (PID) controller, cartridge heater or Peltier element, thermocouple, and aluminum block. Two sets of thermal control modules maintain the denaturation and extension temperatures of the chip. Another thermal control module combined with one T-shaped aluminum block is located beneath the annealing region and sustains the annealing temperature, as shown in Fig. 3(a).
The heating module comprises one heating element, a 5 VDC power, an NPN Darlington power transistor (TIP120), and some electric elements, shown in Fig. 3(b). We have illustrated the detailed circuit diagram in the previous work (Chen et al. 2015). The denaturation or extension region on the chip utilizes a cartridge heater (3.175 mm diameter, 3.8 mm length, 10 W, C1J-9412, Watlow, St. Louis, Missouri, USA) as a contact heating source. The heater is covered by an aluminum rectangular prism, fixed on the frame constructed of extruded aluminum, and powered by a programmable DC voltage supply under the control of a homemade circuit. It increases the mixture temperature inside the microchannel to the desired temperature.
A Peltier element (TEC1-12710, Tun-Hwa Electronic Material Co., Ltd, Taichung, Taiwan) mounted on an aluminum extrusion frame cools the central part of the chip down to the annealing temperature. The supply voltage for the Peltier element is 12 VDC. The Peltier element absorbs the thermal energy from the chip and then creates the annealing region for the DNA mixture. A PMMA block is attached to the aluminum frame to reduce thermal interference, and two air gaps are between adjacent areas.
A LabVIEW program and a data acquisition system (NI 9211, National Instruments, Austin, Texas, USA) are used to measure the mixture temperatures during the thermal cycles to ensure the requirements for PCR. A thermocouple (K30-2-506, Watlow, St. Louis, Missouri, USA) with an outer diameter of 0.01 measures the mixture temperature inside the microchannel during thermal cycling. The thermocouple connects to a data acquisition system that converts analog signals to digital ones. A computer receives the temperature signals through the data acquisition module and records the real-time temperature profiles. The program profiles the temperature within the DNA mixture while the mixture is heated by the cartridge heaters or cooled by the Peltier element. After the temperature measurement, we performed the PCR procedures.
2.3 Surface treatment
Due to the dramatic increase in the surface-area-to-volume ratio in microchannels, controlling the surface properties is crucial in biological microfluidics. For example, the protein adsorption of the channel wall is a significant effect that must be prevented during PCR, as it can significantly degrade the amplification performance. Protein absorption is a critical issue on the hydrophobic surface of the PDMS channel. Furthermore, the hydrophobicity also causes complex filling processes of the aqueous solution inside the microchannel. To reduce the adsorption of enzymes and DNA templates by the PDMS surface and then improve reaction efficiency (Erickson et al. 2005), we use chemical additives and passivate bovine serum albumin (BSA), polyvinylpyrrolidone (PVP, (C6H9NO)n) or Tween 20 (C58H114O26) to coat on the reaction chambers and the flow channels. In our experiments, we add various concentrations of BSA, PVP and Tween 20 to prevent the adsorption of PCR reagents.
In the part of surface structure, we use Fourier transform infrared spectrometer (FTIR) (Thermo Nicolet Avatar 360 FTIR System, Thermo Fisher, USA) to take the Infrared spectra of PDMS surfaces and modified PDMS surfaces. Molecular vibration changes the net dipole moment between atoms so that the frequency of molecular vibration changes accordingly. When this frequency is the same as the frequency of infrared rays, the sample absorbs infrared rays, and the FITR measures the absorbed infrared radiation to determine the functional group of the measured substance. The frequency of molecular vibration is related to elements and bonds. Each bond vibrates at a specific frequency, absorbing a particular amount of energy. For measurements on coated PDMS sheets, we immersed PDMS for 60 min in buffer solutions containing the surfactants.
We use a contact angle meter (FTA-1000B, First Ten Angstroms, USA) to measure the contact angle of the solid surface. The contact angle is formed by the liquid/gas interface to the concrete surface, and its value range is between 0° and 180°. Using this characteristic, we can understand the hydrophilicity/hydrophobicity of PDMS before/after surface treatment (Trinh and Lee 2022). 40 mm × 40 mm × 5 mm PDMS blocks were used, and surface treatments were performed by immersion instead of flowing the reagents.
We utilize an atomic force microscope (AFM) (JPK NanoWizard® V, Billerica, Massachusetts, USA) to examine and observe the surface morphology of the substrate. The detection device uses the measuring element to intercept the height change between the sample and the probe and then send it back to the computer for signal processing. Then, we can obtain the average roughness of the PDMS surface structure. An original, untreated PDMS block was tested as a comparison.
2.4 DNA sample preparation and PCR operations
A hydrostatic pressure created by the syringe pump drives all DNA mixtures through the microchannel. Before the sample injection, we passivate the channel wall to reduce the problem of surface compatibility and avoid PCR inhibition by interacting biomolecules with the walls. Finally, the PCR product is collected in a vial and analyzed using a polyacrylamide gel stained with ethidium bromide to confirm the accuracy of the PCR.
Q fever is a pandemic disease with acute and chronic conditions caused by the bacteria Coxiella burnetii. On the portable device and commercial PCR machine (MJ Mini™ 48-Well Personal Thermal Cycler, Bio-Rad, Hercules, California, USA), we amplify a 385-bp segment of Coxiella burnetii DNA to evaluate the performance of the DNA amplification. The reaction mixture for CFPCR requires 50 µL of a mixed solution, which contains 5 µL of 250 ng/µL of the DNA template, 2.5 µL of 0.5 µM of each forward and reverse primer, 25 µL of 2× reaction buffer, 5 µL of 1.5 mM MgCl2, 1 µL of 0.2 mM of each dATP, dGTP, dCTP, and dTTP, and 1.25 U/µL of Taq DNA polymerase and nuclease-free water.
The thermal cycling program for the commercial PCR machine involves heating the PCR mixture at 94 °C/3 min to denature the initial DNA, followed by denaturation at 94 °C/30 sec, annealing at 55 °C/30 sec, and extension at 72 °C/30 sec. Upon completion of 30 thermal cycles, the temperature of the DNA product keeps at 72 °C/3 min for the final extension. The negative control experiment replaces the template genomic DNA with nuclease-free water.
After the PCR process, the products are collected in a vial from the chamber and mixed with 1× blue dye. We use an agarose gel electrophoresis (Mini-Sub Cell GT System, Bio-Rad, Hercules, California, USA) to analyze the PCR products. 10 µL of each sample are loaded onto 2% agarose gel (Certified Molecular Biology Agarose, 25 g #1,613,100, Bio-Rad, Hercules, California, USA) and electrophoresed in 10× Tris/Boric Acid/EDTA (TBE) buffer. After electrophoresis for about 40 min at 120 VAC, the gel is stained with 10 mg/mL ethidium bromide solution and imaged under UV illumination.
After the bonding process, we use several reagent mixtures to modify the surface of the PDMS channel to reduce the protein absorption effect. Each mixture flows through the polytetrafluoroethylene (PTFE) tube and stays inside the channel for 24 h for absorbance on the surface. Then, the deionized water flushes the channel to remove the reagent mixture. The DNA mixture moves through three sequential reaction regions corresponding to the denaturation, annealing, and extension regions.
3 Results and discussion
The following sections comprehensively examine the influences of operational parameters on the temperature profiles and the surface characteristics along the PDMS channel. The channel undergoes two heating sources and one cooling element during PCR processes. Finally, the PCR experiments are carried out using a continuous flow device.
We set a PDMS sheet (75 mm × 25 mm × 2 mm) on the glass substrate (75 mm × 25 mm × 1 mm). The structure is 75 mm long, 25 mm wide, and 3 mm thick. We set two constant temperature zones (40 mm × 8 mm) with different temperatures on both sides of the bottom of the substrate and a low-temperature zone in the middle of the backside of the wafer and then present the three temperature zones specified by PCR. The traditional continuous-flow PCR chip sets the middle-temperature zone in the center for the extension. The medium-temperature site is affected by the high-temperature and low-temperature zones at both sides of the chip, and the isothermal area presented is relatively narrow. However, in the PCR process, the sample takes a long time in the extension temperature zone, so we move the medium temperature zone to one side of the chip. We then set the low-temperature zone in the center and simulated the temperature field for this configuration to explore the internal temperature distribution. For the part of the microchannel, we set the size to be 150 μm in width and 50 μm in height, and the width of the flow channel at the widened part is 650 μm.
3.1 The measurement of the chip temperature
Since PCR mainly utilizes the thermal cycles of DNA samples to achieve DNA amplification, temperature accuracy is essential to PCR technology. Therefore, this study uses three methods for temperature measurement: real-time infrared temperature measurement, chip internal temperature measurement and thermochromic dye.
First, we use a self-made temperature control module to measure the chip’s temperature. The temperature measurement element of the module is DS1821. We set up three DS1821s, attached them to the bottom of the two heating aluminum blocks and the hot end of the cooling chip, and conducted temperature measurements. Then, we use an 8051 single chip to extract the temperature measured by the temperature control module and observe its temperature rise and stability, as shown in Fig. 4.
Figure 4 shows that the high-temperature and medium-temperature zones reach the specified temperature within 600 s and are stable. The part in the low-temperature zone reaches the set temperature initially, and the chip keeps in a state of thermal equilibrium. When the heating element starts to heat up, and the temperature of the central part exceeds the specified temperature, the cooling chip starts to transmit heat energy to the heat dissipation fins. Therefore, we find from the figure that the temperature of the cooling chip is above the requisite temperature. It stabilizes after 30 min, mainly because of the high-temperature and medium-temperature zones. Hence, the energy taken away by the cooling chip gradually increased. Still, the chip temperature was required to reach the PCR temperature, so the self-made temperature control module set can provide the necessary temperature for PCR. After confirming the feasibility of the temperature device, we carry out the chip’s temperature measurements.
We measure the temperature of the chip surface and use an infrared thermal imager to capture the radiant energy of the chip surface and convert it into a picture file. Then, we use the fixed-point temperature measurement function of the thermal imager’s built-in software to obtain the image’s fixed-point temperature of different coordinates on the surface of the chip. We divide the chip’s surface area into upper and lower parts, capturing 15 points up and down in the Y direction to get 30 fixed-point temperatures. Finally, we catch 10 times in the X direction to get 10 temperature curves on the chip surface. A total of 300 fixed-point temperatures are shown in Fig. 5(a).
Since the infrared thermal imaging camera can only measure the surface temperature of the chip, we find from the previous results (Chen et al. 2015) that there exists a slight temperature difference between the surface temperature and the internal temperature. Therefore, it is impossible to correctly determine the internal temperature if only the surface temperature of the chip is measured. Consequently, we use a single-core wire to make a temperature-measuring chip model and then use PDMS molding technology to create a temperature-measuring chip with a measurement channel for temperature measurement with a thermocouple thermometer. We mark on the surface of the self-made chip 18 temperature measurement points to ensure the accuracy and uniformity of temperature, as shown in Fig. 5(b). Then, we compare the 18 fixed-point temperatures measured by the self-made temperature measuring chip with the 30 temperature average curves of the infrared thermal imager. We produce a comparison curve between the surface and internal temperatures, as shown in Fig. 5(c). The figure shows a 6 to 10 K temperature difference between the exterior and interior of the high-temperature zone and the medium-temperature sites. In comparison, the temperature difference of the low-temperature area is about 2 to 4 K, which is consistent with the results of the previous research. The method can confirm the interior temperature and use this result to match it with an infrared thermal imaging camera for instant temperature determination.
From Fig. 5(c), we can understand the temperature distribution inside the chip through the above method. We use a thermochromic dye to demonstrate the temperature distribution of the sample flowing in the microchannel. We dilute and mix the temperature dye with DI water at a volume ratio of 4:1 and inject it into the microchannel with a constant width for observation, as shown in Fig. 6(a) and (b). When the sample enters the high-temperature zone, it offers a transparent color, as shown in Fig. 6(a). When it enters the low-temperature area, the temperature-changing dye gradually turns black. It also appears transparent as the sample flows into the middle-temperature area, so the sample in the micro-channel reaches the temperature required for PCR. In Fig. 6(b), the temperature of the traditional micro-channel with a constant width changes color at the turning point near the low-temperature area in the middle-temperature area. The dye turned back to black. However, this phenomenon does not occur while the channel expands to widen in Fig. 6(a). Therefore, the microchannel designed in this manuscript provides the temperature required by PCR and is better than the traditional microchannel type.
The effect of the channel geometry on the temperature distribution visualized qualitatively using a thermally sensitive dye. The transition point lies between the heating blocks when the fluid with dye moves. (a) The traditional microchannel with a constant width. (b) The microchannel with various widths
3.2 Surface treatment
The adsorption channel phenomenon of DNA polymerase affects the reaction efficiency of PCR. DNA polymerase is a protein and is composed of amino acids. However, amino acids consist of amino acids through condensation reactions. All amino acids contain the functional groups of amine (-NH2) and carboxylic acid (-COOH). PDMS is a high-molecular organosilicon compound, and the functional groups on the surface of PDMS are all methyl groups (-CH3). Therefore, when the carboxyl group of DNA polymerase reacts with the methyl group of PDMS, it inevitably affects the results of PCR experiments. The common point of the three surface treatment reagents we use is that they are all soluble in water and cover the target object as a thin film to achieve a barrier effect. Therefore, we mix deionized water and surface passivation reagents at different concentrations. We add PDMS sheets to the surface passivation reagent, soak it for 1 h (Fukuba et al. 2004; Joung et al. 2008; Han et al. 2014), and remove the surface liquid. Thus, we compare the surface modification effect of the surface treatment reagent on PDMS.
The spectrum of FTIR used in this work is mid-infrared, ranging from 4000 to 700 cm− 1. FTIR is used to measure the absorption spectrum of untreated PDMS, which can be used as a control group for comparison in subsequent discussions, as shown in Fig. 7. The specific methyl absorption fragments are 3000 ~ 2900 cm− 1, 1700 ~ 1400 cm− 1, 1300 ~ 1200 cm− 1, and 900 ~ 700 cm− 1 in the FTIR spectrum (Johnson et al. 2013). In the follow-up discussion, we compare the absorption intensity of surface passivation reagents at the corresponding range. Then, we discuss various concentrations of surface treatment reagents and their modification effects. Reaching a good reagent concentration with the other two, the most suitable reagent for modifying PDMS can be obtained.
We consider the absorption spectrum of BSA-modified PDMS sheets. BSA owns the characteristics of proteins as DNA polymerase does to form a protein film inside the microfluidic channel, which reduces the contact surface between DNA polymerase and PDMS. Figure 8(a) shows the absorption spectrum of different BSA concentrations for PDMS surface modification. From Fig. 8(b), in the range of 1700 to 1400 cm− 1, it can be found that after BSA modification, when the concentration is above 0.5 mg/mL, the methyl peak of 1412 cm− 1 decreases slightly more than unmodified PDMS. Therefore, BSA has little effect on modifying methyl groups. There are two new peaks at 1652 and 1540 cm− 1. These two peaks are the functional groups of BSA at a concentration of 2 mg/mL and 4 mg/mL. However, when we add the methyl spectrum for comparison, the methyl peak at 2960 and 2905 cm− 1 is better at 4 mg/mL concentration.
Next, we discuss the infrared absorption spectrum of the PDMS sheet modified by PVP. Figure 9(a) shows that the chemical structure of PVP has a functional methyl group. Still, because PVP has pretty good adsorption and biocompatibility, it expresses that PVP uses the functional group to bond with the surface of PDMS to form a thin film and then further avoid sample adsorption on PDMS. Figure 9(b) shows the specific absorption spectrum of the methyl group. The peaks significantly improve, but the others tend to decrease. Although the FTIR spectrum cannot judge whether the PVP reagent’s functional group has modified the microchannel wall, By observing the absorption spectra of PVP and PDMS, the modification effect at a concentration of 4 mg/mL is relatively good.
Finally, we discuss the absorption spectrum of the PDMS sheet modified by Tween 20. Tween 20 is a kind of ester, and its chemical bond structure has the characteristics of hydrophilicity at one end and lipophilicity at the other. After the passage, the sticky properties of esters tend to attach the hydrophobic end to the wall and protect the DNA polymerase with the hydrophilic end, as shown in Fig. 10(a). Figure 10(b) indicates that Tween 20 has a new methyl peak at 2906 cm− 1 at a 5 to 20% concentration. This peak is the functional group of Tween 20, but it differs from other methyl peaks. Compared with this, there is a trend of substantial reduction, so a 20% concentration is better than different concentrations.
We compared the influence of the concentration of different surface treatment reagents on the absorption spectrum of PDMS methyl groups. The reagents were 4 mg/mL of BSA, 4 mg/mL of PVP and 20% of Tween 20, as shown in Fig. 11(a). Figure 11(b) shows a significant peak in the 3000 to 2800 cm− 1; here is the functional group of Tween 20. We find two functional groups of BSA in the 1700 to 1400 cm− 1 field. However, the methyl peak of PVP is higher than that of the control group in the part of 1300 to 700 cm− 1. Therefore, Tween 20 is the best surface structure, followed by PVP and BSA.
Because hydrophobic PDMS microchannels have relatively high free energies compared to water, proteins tend to absorb them on the channel surface. This paper’s three surface passivation reagents are soluble in water. We can measure the hydrophilicity and hydrophobicity after surface treatment to consider the adhesion effects between the film produced by surface passivation treatment and PDMS. The measurement uses a contact angle goniometer with DI water as the liquid. When the contact angle exceeds 90°, it is hydrophobic; otherwise, it is hydrophilic. Firstly, the PDMS without surface treatment is measured, and its contact angle is 106°, as shown in Fig. 12(a). After standing for a while, the contact angle remained at hydrophobicity (103°), as shown in Fig. 12(b).
After immersing PDMS in the surface treatment reagent for 1 h, we measure the contact angle immediately after removing the surface liquid. Figure 13(a) to (c) show that the contact angle profile of each surface treatment reagent has a sliding curve within 10 min. When we separate PDMS from the reagent mixture, the mixture still saturates the PDMS surface, and the contact angle profile is not steady. The three surface treatment reagents all recovered their hydrophobic properties after 16 h at room temperature, and the surface film had lost its activity entirely, which led to the reaction that the contact angle gradually increased. The comparison of the three reagents shows that the contact angle of PDMS modified by the Tween 20 reagent is the lowest, and the surface is modified to be hydrophilic. Therefore, the Tween 20 modified film is superior to the other two surface treatment reagents.
The methyl group is the key to adsorbing proteins on the surface of the microchannel. While the surface of the microchannel becomes rough, the contact area between the microchannel and the protein increases, and the adsorption of the sample and the methyl group increases. Therefore, discussing the surface roughness of the PDMS sheet is necessary. In this paper, we use the contact mode of the atomic force microscope (AFM) to measure the average roughness (Ra) of PDMS, and the surface morphology of PDMS can be obtained through the AFM image results, as shown in Fig. 14.
After we know the surface type of untreated PDMS, we can compare different surface treatment methods. We immerse PDMS in the surface treatment reagent for 1 h, remove it from the reagent, and use a rubber air blower to remove the surface liquid. However, we must keep the sample wet to avoid changes in the surface film. Figure 14 shows the AFM measurements of PDMS after surface treatment. According to the figure, we illustrate the average roughness of the surface-modified film, and the roughest is PVP with a concentration of 1 mg/mL. This result may be because PVP is a kind of adhesive that will be adsorbed and bonded to each other during surface modification. The resulting surface is relatively rough. The most detailed is Tween 20 with a concentration of 20%. Since the AFM used in this article will change its unit according to the overall roughness, the 3D image will appear rough. Tween 20 is attached to the surface using the characteristics of esters to form a fine and smooth surface. Therefore, the surface roughness is the least for Tween 20. This result is consistent with FTIR.
3.3 PCR amplification in the microfluidic chip
We verified the feasibility of this experimental device in PCR application. We compare the gel electrophoresis results of nucleic acid products after modifying the microfluidic channels with surface treatment reagents. The DNA fragment used in this work is Q fever virus, and the DNA fragment is 385 bp. The ratio of other added reagents, such as primers and PCR reagents, is shown in Table 1. The steps of the PCR experiment are as follows: first, we inject the surface treatment reagent into the microchannel by using the sample driving system, and after standing for one hour, air flow pushes it out of the channel. We use the temperature control system to control the heating rod and the Peltier element to form the temperature zone required for PCR. Then, we use the self-made temperature measuring chip and an infrared thermal imager to measure the chip’s temperature to avoid the experiment’s failure due to incorrect temperature distributions. After completion, we install the chip on the device and inject 10 µL of the PCR mixture into the microchannel at a 1 µL/min flow rate. Finally, we compared it with the amplification of a commercial PCR thermal cycler.
First, we discuss the effect of microfluidic surface treatment on nucleic acid amplification. Surface treatment can effectively help DNA amplification, so this article compares three treatment reagents. We draw 10 µL of PCR mixture and inject the liquid into a non-surface-treated channel and a surface-treated one with a concentration of 5% Tween 20 at a flow rate of 1 µL/min for PCR reaction comparison, as shown in Fig. 15. From the gel electrophoresis image, lane MK is a DNA marker, and lane M is the amplification result of a commercial PCR thermal cycler. The parameters are set to 367 K preheating zone for 180 s, followed by high temperature 367 K, medium temperature 345 K, and low temperature 328 K. We set a dwell time for each isothermal process of 30 s respectively, repeat 30 cycles, and finally perform a final preheating zone at 345 K for 180 s. Then, lane 1 uses a surface treatment reagent, and lane 2 is the untreated channel. The lane with surface treatment has noticeable results, while the untreated channel is less obvious. Therefore, surface treatment can aid in DNA amplification.
Then, PCR comparisons are performed for different surface treatment reagents, as shown in Fig. 16. The surface treatment methods are all DNA amplification after standing for 1 h. Lane M is a commercial PCR thermal cycler, T1 and T2 are Tween 20 concentrations of 10% and 20%, B1 and B2 are BSA concentrations of 1 and 2 mg/mL, and P1 and P2 are PVP concentrations of 1 and 2 mg/mL. The results show that the effect of T2 is relatively apparent, but it is still not comparable to the results of commercial PCR thermal cyclers. However, the results of lanes B1 and B2 could be more varied. Although many researchers used BSA as a blocking agent to improve amplification in microchannels, their effect is limited, which will lead to this result. BSA has not significantly affected amplification yield when used in PCR amplification free of any PCR inhibitors (Kramer and Coen 2001). Integrating the surface treatment comparison and the actual PCR results, the overall product of Tween 20 is relatively good because Tween 20 is an emulsifier, and it has excellent performance in both adhesion properties and coated samples. However, PVP is mainly due to its structure having many methyl groups, and no good data is demonstrated in either FTIR or hydrophobicity, so we cannot correctly predict the relevant comparison. Dynamic passivation by adding PVP increased the PDMS/glass microchip PCR product. However, as shown in the studies of Kim et al. (2006), Xia et al. (2007), and Chen et al. (2007), the addition of PVP did not completely solve the problem of enzyme adsorption to the microchannel wall. Furthermore, higher concentrations of PVP might result in reaction inhibition. From the studies of Xia et al. (2007) and Chen et al. (2007) that the native PDMS or glass surface in an aqueous solution is negatively charged, while PVP is positively charged; therefore, if the surface adsorbed PVP before loading the PCR mixture, which might have helped reduce enzyme adsorption on the microchannel surface, thereby improving PCR product yield and specificity. Finally, BSA uses the same characteristics for protection, and its effect is limited.
Effect of the concentrations of surface treatment reagents on the PCR yield. The first lane: marker; lane M: commercial PCR machine; lanes T1 and T2: PCR products from the CFPCR chip for 10% and 20% of Tween 20; lanes B1 and B2: PCR products from the CFPCR chip for 1 and 2 mg/mL of BSA; lanes P1 and P2: PCR products from the CFPCR chip for 1 and 2 mg/mL of PVP
4 Conclusions
This paper uses MEMS process technology to fabricate the PDMS/glass microfluidic chip. Then, we use the 8051 single chip and DS1821 temperature measuring element to create a temperature control system and complete the heating and cooling of the chip. We set up a low-temperature zone in the center of the bottom of the chip, and the two sides are a high-temperature zone and a medium-temperature zone. The low-temperature area blocks the effective influence of medium-temperature and high-temperature. An infrared thermal imager measures the surface temperature, and a thermocouple measuring chip and the temperature-changing dye measure the interior temperature. The difference between the internal and surface temperatures is about 6 to 8 K. The temperature of the sample reaches the specified temperature through each temperature zone, which confirmed that the micro-device designed in this paper could meet the needs of PCR.
Then, we carry out three measurements on the surface treatment reagents in the pretreatment of microchannels. We used FTIR to measure the surface characteristics of PDMS and the spectrum of methyl groups to compare the coating properties of different concentrations of surface treatment reagents. Then, we use AFM to observe the surface morphology. When the surface of PDMS is rougher, it inevitably reflects the attachment of the sample. The surface treatment agent is a hydrophilic solvent in water, and PDMS is a hydrophobic material. Therefore, we can present the effect of the surface agent on the contact angle. Based on the above measurements, when the concentration of Tween 20 is 20%, the modification effect is relatively sound, the roughness of the treated PDMS surface is about 7 nm, and the adhesion is much higher than the other two. Finally, we compare the difference in PCR amplification between the channels with or without surface treatment from the gel electrophoresis pattern. All chips with each reagent can successfully amplify DNA fragments of Q fever by adding different surface treatment reagents and comparing them. The amplification result is the best with Tween 20, followed by PVP, and finally, BSA.
References
Cao Q, Mahalanabis M, Chang J, Carey B, Hsieh C, Stanley A, Odell CA, Mitchell P, Feldman J, Pollock NR, Klapperich CM (2012) Microfluidic Chip for Molecular Amplification of Influenza A RNA in human respiratory specimens. PLoS ONE 7(3):e33176
Chen L, West J, Auroux PA, Manz A, Day PJ (2007) Ultrasensitive PCR and real-time detection from human genomic samples using a bidirectional Flow Microreactor. Anal Chem 79(23):9185–9190
Chen JJ, Shen CM, Ko YW (2013) Analytical study of a microfludic DNA amplification chip using Water cooling Effect. Biomed Microdevices 15(2):261–278
Chen JJ, Liao MH, Li KT, Shen CM (2015) One-heater Flow-through polymerase chain reaction device by heat pipes cooling. Biomicrofluidics 9(1):014107
Chiou CH, Shin DJ, Zhang Y, Wang TH (2013) Topography-assisted electromagnetic platform for blood-to-PCR in a Droplet. Biosens Bioelectron 50:91–99
Coutlée F, Voyer H (1998) Effect of nonionic detergents on amplification of human papillomavirus DNA with consensus primers MY09 and MY11. J Clin Microbiol 36(4):1164
Erickson D, Liu X, Venditti R, Li D, Krull UJ (2005) Electrokinetically based approach for single-nucleotide polymorphism discrimination using a microfluidic device. Anal Chem 77(13):4000–4007
Felbel J, Bieber I, Pipper J, Köhler JM (2004) Investigations on the compatibility of chemically oxidized Silicon (SiOx)-Surfaces for applications towards chip-based polymerase chain reaction. Chem Eng J 101(1–3):333–338
Felbel J, Reichert A, Kielpinski M, Urban M, Henkel T, Häfner N, Dürst M, Weber J (2008) Reverse transcription-polymerase chain reaction (RT-PCR) in Flow-through Micro-reactors: thermal and fluidic concepts. Chem Eng J 135:S298–S302
Fukuba T, Yamamoto T, Naganuma T, Fujii T (2004) Microfabricated Flow-through device for DNA amplification—towards in Situ Gene Analysis. Chem Eng J 101(1–3):151–156
Han D, Jang YC, Oh SN, Chand R, Lim KT, Kim KI, Kim YS (2014) MCU Based Real-Time Temperature Control System for Universal Microfluidic PCR Chip. Microsyst Technol 20:471–476
Hsieh HY, Chang R, Huang YY, Juan PH, Tahara H, Lee KY, Vo DNK, Tsai MH, Wei PK, Sheen HJ (2022) Fan, Continuous Polymerase Chain Reaction Microfluidics Integrated with a gold-capped Nanoslit sensing chip for Epstein-Barr Virus Detection. Biosens Bioelectron 195:113672
Jiang X, Shao N, Jing W, Tao S, Liu S, Sui G (2014) Microfluidic chip integrating high Throughput continuous-Flow PCR and DNA hybridization for Bacteria analysis. Talanta 122:246–250
Johnson LM, Gao L, Shields IV CW, Smith M, Efimenko K, Cushing K, Genzer J, López GP (2013) Elastomeric microparticles for acoustic mediated bioseparations. J Nanobiotechnol 11:1–8
Joung SR, Kang CJ, Kim YS (2008) Series DNA amplification using the continuous-Flow polymerase chain reaction chip. Jpn J Appl Phys 47(2S):1342
Kaprou GD, Papadopoulos V, Papageorgiou DP, Kefala I, Papadakis G, Gizeli E, Chatzandroulis S, Kokkoris G, Tserepi A (2019) Ultrafast, Low-Power, PCB Manufacturable, continuous-Flow Microdevice for DNA amplification. Anal Bioanal Chem 411:5297–5307
Kim JA, Lee JY, Seong S, Cha SH, Lee SH, Kim JJ, Park TH (2006) Fabrication and characterization of a PDMS–Glass hybrid continuous-Flow PCR chip. Biochem Eng J 29(1–2):91–97
Kim HE, Schuck A, Kim WY, Jung EK, Hong YH, Kim YS (2022) PID temperature control system-based microfluidic PCR chip for genetic analysis. J Electr Eng Technol 17:495–501
Kopparthy VL, Crews ND (2020) Oscillating-Flow Microfluidic PCR system utilizing a thermal gradient for nucleic acid analysis. Biotechnol Bioeng 117(5):1525–1532
Kramer MF, Coen DM (2001) Enzymatic amplification of DNA by PCR: standard procedures and optimization. Curr Protoc Mol Bio 56(1):15–11
Kuan I, Gu W, Wu J, Wei C, Chen K, Yu C (2008) Effects of Grafting Poly (Ethylene Oxide) on the amplification efficiency of a poly (Dimethylsiloxane)-Based Flow-through PCR device. Chem Eng J 143(1–3):326–330
Kulkarni MB, Goyal S, Dhar A, Sriram D, Goel S (2021) Miniaturized and IoT enabled continuous-Flow-based microfluidic PCR device for DNA amplification. IEEE Trans Nanobiosci 21(1):97–104
Kwak BJ, Kim H, Park N, Hahn JH (2021) Microchip for continuous DNA analysis based on gel Electrophoresis coupled with co-injection of size markers and in-Channel staining. Anal Bioanal Chem 413(23):5685–5694
Li S, Fozdar DY, Ali MF, Li H, Shao D, Vykoukal DM, Floriano PN, Olsen M, McDevitt JT, Gascoyne PRC, Chen S (2006) A continuous-Flow polymerase chain reaction microchip with Regional Velocity Control. J Microelectromechanical Syst 15(1):223–236
Mullis KB, Faloona FA (1987) Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods in Enzymology, vol 155. Academic, pp 335–350
Nakayama T, Kurosawa Y, Furui S, Kerman K, Kobayashi M, Rao SR, Yonezawa Y, Nakano K, Hino A, Yamamura S, Takamura Y, Tamiya E (2006) Circumventing air bubbles in Microfluidic systems and quantitative continuous-Flow PCR applications. Anal Bioanal Chem 386(5):1327–1333
Peham JR, Grienauer W, Steiner H, Heer R, Vellekoop MJ, Nöhammer C (2011) Wiesinger-Mayr, Long Target Droplet polymerase chain reaction with a Microfluidic device for high-throughput detection of pathogenic Bacteria at clinical sensitivity. Biomed Microdevices 13:463–473
Qin K, Lv X, Xing Q, Li R, Deng Y (2016) A BSA coated NOA81 PCR chip for gene amplification. Anal Methods 8(12):2584–2591
Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N (1985) Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of Sickle Cell Anemia. Science 230(4732):1350–1354
Schneegaß I, Bräutigam R, Köhler JM (2001) Miniaturized Flow-through PCR with different template types in a Silicon Chip Thermocycler. Lab Chip 1(1):42–49
Seetasang S, Xu Y (2022) Recent progress and perspectives in applications of 2-Methacryloyloxyethyl phosphorylcholine polymers in Biodevices at Small scales. J Mater Chem B 10:2323–2337
Trinh KTL, Lee NY (2022) Fabrication of wearable PDMS device for rapid detection of nucleic acids via recombinase polymerase amplification operated by human body heat. Biosensors 12(2):72
Wang JH, Chien LJ, Hsieh TM, Luo CH, Chou WP, Chen PH, Chen PJ, Lee DS, Lee GB (2009) A miniaturized quanti-tative polymerase chain reaction system for DNA amplification and detection. Sens. Actuators B Chem 141(1):329–337
Wu J, Guo W, Wang C, Yu K, Ma Y, Chen T, Li Y (2015) Research to improve the efficiency of double stereo PCR microfluidic chip by passivating the Inner Surface of Steel Capillary with NOA61. Cell Biochem Biophys 72:605–610
Xia YM, Hua ZS, Srivannavit O, Ozel AB, Gulari E (2007) Minimizing the Surface Effect of PDMS–Glass microchip on polymerase chain reaction by dynamic polymer passivation. J Chem Technol Biotechnol 82(1):33–38
Acknowledgements
The authors thank the National Science and Technology Council of the Republic of China for financially supporting this research under Contract No. MOST 110-2313-B-020-003-.
Author information
Authors and Affiliations
Contributions
J.J.C. and X.C.Q. conceptualized the design, and X.C.Q. implemented the platform. J.J.C. supervised the project. J.J.C. and X.C.Q. prepared the initial draft. J.J.C. got the funding. All the authors contributed to the manuscript finalization.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, J.J., Qiu, X.C. The effect of the surface passivation on polymerase chain reaction inside a continuous flow microfluidic chip. Microsyst Technol (2024). https://doi.org/10.1007/s00542-024-05675-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00542-024-05675-2