Abstract
Polymerase chain reaction (PCR) has been considered as the gold standard for detecting nucleic acids. The simple PCR system is of great significance for medical applications in remote areas, especially for the developing countries. Herein, we proposed a low-cost self-assembled platform for microchamber PCR. The working principle is rotating the chamber PCR microfluidic chip between two heaters with fixed temperature to solve the problem of low temperature variation rate. The system consists of two temperature controllers, a screw slide rail, a chamber array microfluidic chip and a self-built software. Such a system can be constructed at a cost of about US$60. The micro chamber PCR can be finished by rotating the microfluidic chip between two heaters with fixed temperature. Results demonstrated that the sensitivity of the temperature controller is 0.1℃. The relative error of the duration for the microfluidic chip was 0.02 s. Finally, we successfully finished amplification of the target gene of Porphyromonas gingivalis in the chamber PCR microfluidic chip within 35 min and on-site detection of its PCR products by fluorescence. The chip consisted of 3200 cylindrical chambers. The volume of reagent in each volume is as low as 0.628 nL. This work provides an effective method to reduce the amplification time required for micro chamber PCR.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Polymerase chain reaction (PCR) is a molecular biology that can produce an initially small amount of genetic material to substantial quantity of DNA. Because of its high sensitivity and specificity, PCR is widely applied in the field of forensics (Cavanaugh and Bathrick 2018), pathogen detection (Lee et al. 2019) and etc. Compared with the traditional method based on culture and morphology technology, PCR takes less time and fewer instruments (Gouriet et al. 2005; Liu et al. 2023). The main process of PCR is repeated thermal cycling, which offers the temperatures for denaturation, annealing, and extension. However, the temperature variation rate for conventional PCR thermal cyclers is about 4–6 °C (Chen et al. 2022). Consequently, it takes more than 50 min to complete 35 PCR cycles.
To simplify the heating process, various PCR techniques have been reported in the literature. For example, Krishnan’s group designed a device for PCR based on Rayleigh-Bénard convection (Krishnan et al. 2002). They successfully amplified a 295 bp fragment of single-copy β-Actin gene by maintaining the bottom and top of the reaction cell at 97 °C and 61 °C, respectively. However, we found that the circulatory flow inside the PCR solution was greatly dependent on the geometrical size of the cell, otherwise it may create weird circulatory flow, which will not bring the PCR mixture to the right temperature for amplification (Li et al. 2016). Notomi et al. developed the loop-mediated isothermal amplification, and the reaction was carried out at 65 °C for 1 h (Notomi et al. 2000). Although the heating process is quite simple, it requires four specially designed primers, which may induce false positives caused by cross-reaction. Furthermore, the aerosol contamination is easy to be induced when the lid is opened, which is prone to false positives. Continuous flow PCR proposed by Manz et al. was based continuously running the reagent into the micro channels placed on the heaters with fixed temperature (Kopp et al. 1998). We have also carried out series research of rapid detection of bacterial based on CF-PCR (Li et al. 2019, 2021; Yang et al. 2022). Although it can reduce the amplification time, the bubbles were easy to be generated in the microchannel, thus it may reduce the PCR efficiency. To increase the temperature variation rate, Wittwer’s team developed a rapid PCR prototype by rotating the reaction tube between two water-bath, which separately offered the annealing/expansion temperature (62–76 °C) and the continuous temperature (85–92 °C) (Farrar and Wittwer 2015). It realized amplification of 45- to 102-bp targets in 15–60 s, although the system is quite cumbersome. Most micro chamber PCR was performed on the flat PCR system (Cui et al. 2020; Zhu et al. 2014; Zhu et al. 2017; Song et al. 2015), and it takes more than 1 h to finish PCR because of the low temperature variation rate. Moreover, chamber-based digital PCR (cdPCR) and droplet-based dPCR (ddPCR) were also proposed for the accurate quantification of template nucleic acid (Huang et al. 2023; Ning et al. 2019; Suo et al. 2020), although the cost for instrument and consumables in dPCR was high. To solve this problem, we designed a novel ddPCR system for the rapid detection of bacterial based on CF-PCR (Li et al. 2023). There is also sophisticated new technology reported for the droplet manipulation, which may be applied in ddPCR (Wang et al. 2021). So far, most of the cdPCR systems depend on in situ PCR instrument. To reduce the cost, Hosokawa improved the structure of PDMS cdPCR microfluidic chip and successfully performed the experiment in a common PCR thermal cycler with well-type heat blocks designed for PCR tubes, but the protocol seems cumbersome (Hosokawa and Ohmori 2023).
To overcome this problem, we herein proposed a novel self-assembled platform for microchamber PCR. The working principle is rotating the chamber PCR microfluidic chip between two heaters with fixed temperature. Finally, we successfully amplified the target of Porphyromonas gingivalis (P.g) in the microfluidic chamber array, and we realized the on-site detection of positives by fluorescence. Such a system is of low cost and easy to assemble, and may offer a new way for the point-of-care testing application.
2 Materials and methods
2.1 Materials and reagents
The mineral oil was purchased from Aladdin (USA). The primers were synthesized by Sangon Biotech (Shanghai). Hydroxyethyl cellulose (HEC, 1300 k) was purchased from Sigma-Aldrich (Saint Louis, USA). SpeedSTAR HS DNA Polymerase and 100 bp DNA ladder were bought from Takara (Beijing, China). 2×SYBR Green PCR Mastermix (Solarbio, Beijing, China) was used for DNA amplification.
2.2 Construction of the chamber PCR microfluidic chip
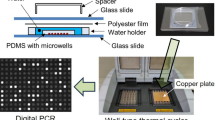
As shown in Fig. 1A, the chamber PCR microfluidic chip is mainly composed of three layers. The micropatterned polydimethylsiloxane (PDMS) consisted of 3200-cylinder microchambers (radius/depth = 50 μm/80 µm) was bonded on a 0.17 mm substrate glass, and another 0.17 mm glass slide was bonded to the top surface of PDMS to prevent reagent evaporation. The microchambers were connected by the main channel (width/depth = 50 μm /80 µm). Each microchamber was connected to the main channel by a branch channel (length/width/depth = 40 μm/ 30 μm/80 µm). A water channel surrounded the microfluidic chip was fabricated to prevent PCR reagent from evaporation. The photo of the microfluidic chip was shown in Fig. 1B.
2.3 Construction of the microchamber PCR system
The self-assembled platform is shown in Fig. 1C. It mainly consisted of two PID temperature controllers, two aluminum heaters, a slide screw module with a power supply, and a single chip microcomputer. The accuracy of the proportional-integral-differential temperature controllers (M101, SinoTimer, Shanghai, China) was 0.1 °C. The holder supporting the heaters was made of nylon material and it was fabricated by a 3D printer. The microfluidic chamber chip was fixed on the screw slide, and then it was rotated between two heaters (see Movie S1 in the supporting information). The process was controlled by a self-programmed software.
2.4 Fabrication of the microfluidic chip
The micropatterned PDMS structure was made by replica molding technology. The photomask was designed by CAD software. Firstly, SU8-2025 photoresist was spin-coated onto a silicon wafer (1000 rpm, 30 s). Next, the wafer was separately heated at 65 °C for 3 min and 95 °C for 9 min to enhance the adhesion. The photoresist was engraved by a mask aligner MJB4 (SUSS Microtec, Germany), and the mold was heated at (65 °C, 3 min) and (95 °C, 7 min), respectively. Then it was successively placed in the developer solution (Alfa Aesar, USA) to clean the channel, tested by isopropyl alcohol (Aladdin, Shanghai, China), dried at room temperature, and baked at 170 °C for 15 min.
Next, the patterned silicon was treated by trichloro (1 H, 1 H, 2 H, 2 H-perfluorooctyl) silane (Sigma-Aldrich China, Shanghai, China) vapor for 30 min to avoid adhesion to silicone rubber. PDMS mixed at ratio of 10:1 prepolymer and curing agent was degassed in a vacuum-drying oven (HASUC, Shanghai, China), and then it was poured into the mold and degassed for 1 h. The patterned PDMS was desquamated from the mold after solidification at 80 °C for 1 h. Finally, PDMS-glass bonding was achieved after plasma treatment, and the assembled microfluidic chip was placed at 100 °C for 30 min.
2.5 Experimental protocol
A piece of 2.5 mm thick PDMS layer was stuck onto the top of the chip, and then the chip was degassed in a vacuum-drying oven (-0.1 MPa, 5 min). The PCR reagent was loaded by pressure difference induced from the interior and exterior PDMS. Next, the chip was fixed on the spiral slide screw, and then the chip was continuously rotated between the two heaters (see Figure S1 in the Supporting Information). Its speed was controlled by a self-programmed software based on C + + in Microsoft Visual Studio 2019. Finally, the chip was observed by Ti2 inverted microscope (Nikon, Japan) after the thermal cycle.
3 Results and discussion
3.1 Reagent self-priming compartmentalization
PDMS is a porous matrix with high gas solubility or gas permeability (Xu et al. 2015). This phenomenon is the same as the principle of solute dissolution in solvent, which can be explained by Henry’s Law (Merkel et al. 2000). Air concentration of PDMS decreases when the whole microfluidic chip is placed in a vacuum drying oven for degassing, thus the air diffuses out of PDMS blocks. Then it was removed from the vacuum-drying oven and placed under normal atmosphere. There will be pressure difference between the inside and outside of the PDMS, and the air will be slowly dissolved into the PDMS block to balance the pressure difference. Liquid can be withdrawn into the embedded dead-end microfluidic channels when degassed PDMS restore air inside it.
The process of cyan ink self-priming compartmentalization: (A) The solution was priming into the microchambers. (B) The micro chambers were gradually filled with solution. (C) Air was pushed into the main channel. (D) The mineral oil was pushed into the branch channel and main channel. All the microchambers were compartmentalized
We chose cyan ink as a substitute for PCR reagent to observe the sample loading process. First, the channel inlet and outlet were pasted with transparent tape, and the chip was degassed at -0.1 MPa for 5 min. 4.7 µL cyan ink was injected into the main channel from inlet 1 by a syringe, and the cyan ink was quickly inhaled into the branch microchannel and microchamber (Fig. 2A), then the microchamber was gradually filled with cyan ink (Fig. 2B). Next, we removed the transparent tape from the inlet and outlet, and injected air from inlet 1 to push the excess reagent from the main channel (Fig. 2C), which can reduce the resistance of the oil phase in the channel and forbid the reagent to be extruded from the microchamber due to excessive pressure. A constant pressure pump (Wenhao Co., Ltd, Suzhou, China) was applied to push mineral oil into the main channel. Finally, the main and branch channels were filled with mineral oil, and each microchamber was occupied by cyan ink (Fig. 2D).
3.2 The effect of PDMS thickness on sample loading
PDMS has excellent air permeability since air is mainly stored in porous PDMS. A negative pressure environment generated inside degassed PDMS can offer self-priming power to inhale the reagent into each microchamber, and thus we suppose that thicker PDMS has stronger ability to inhale the test solution. Therefore, we fabricated PDMS chips with thickness varied from 0.5 to 3 mm. Each microfluidic chip was degassed at -0.1 MPa for 5 min, and we employed a self-programmed software based on OpenCV 3.1.0 to count the microchambers filled with reagent. Data in Fig. 3A showed that the number of microchambers filled with reagent increased with the thickness of PDMS, and it is easy to fill all microchambers with reagent in 3 mm thick PDMS microfluidic chip.
However, we should notice that the thicker PDMS may lead to serious water evaporation during heating because PDMS is a porous material. It is better to make the micropatterned PDMS as thin as possible to reduce water evaporation. To overcome this problem, we separately pasted 0.5 mm, 1.5 mm, and 2.5 mm thick PDMS blank layers at the top of a 0.5 mm thick microfluidic chip as a vacuum-driven power source. Data in Fig. 3B revealed that the number of filled microchambers was not reduced if the thickness of the PDMS blank layer and the substrate was the same as the thickness of PDMS microfluidic chip in Fig. 3A. Finally, we selected 0.5 mm thick microchamber chip with a 2.5 mm thick PDMS blank layer as the surface, and placed it in a vacuum-drying oven for degassing.
3.3 The stability of the temperature and duration for heating
Temperature variation rate and stability of temperature play a nonnegligible role in PCR. We next evaluated the temperature of the glass slide by TCM-M207 temperature sensors (EasyShining Technology, Chengdu, China), this is because the chamber was in close contact with the surface of the glass slide. To test the temperature variation rate, we used a 0.17 mm thick glass slide to substitute for the chamber microfluidic chip. The glass slide was rotated between two heaters (95 °C and 65 °C). Curve in Fig. 4 revealed the temperature variation rate was about 1.36 °C/s and 1.49 °C/s for the temperature increase and decrease, respectively. Moreover, we measured the temperature distribution by an infrared thermal camera (Testo 865, Testo, Inc., Germany). Figure 4A showed that the temperature of the glass slide was slightly higher than the atmospheric temperature when it was initially placed on the heater (95 °C), and then it gradually reached a stable temperature (Fig. 4B), which corresponded ① in the curve. Figure 4C and D separately demonstrated the temperature distribution for the glass slide moved to another heater (65 °C), which corresponded to the ② and ③ in the curve. Finally, the temperature of the surface of glass slide was 95 ± 0.41 °C and 65 ± 1.16 °C, respectively.
We also tested the stability of rotating by measuring the duration of glass slide stayed in each heater. We have set 20 and 30 s for the glass slide to stay on each heater by software. Data in Fig. 5 revealed that the real duration for them was 19.97 ± 0.02 s and 29.99 ± 0.02 s, respectively.
3.4 Specificity and selectivity of the primers
Prior to performing the microchamber PCR, we performed the amplification of P.g in T100 thermal cycler (Bio-Rad). 50 µL PCR reagent contained 1.0 µL sample and 5.0 µL 10× Fast Buffer I, 4.0 µL dNTP mixture (2.5 mM), 2.0µL 200 nM primers (Forward: TGACCTTAAGCCTGTAG; Reverse, TCATCTCGTGTCCGATCACT), 0.25 µL SpeedSTAR HS DNA Polymerase and 37.75 µL ultra-pure water (see Table S1 in the supporting information). The cycling program consisted of 95 °C for 10 s (denaturation) and 64 °C for 30 s (annealing and extension) with an initial cycle of 95 °C for 2 min. The size of the amplicon was 169 bp. Negative control was conducted under the same PCR conditions. Then we carried out separation of the PCR product and 100 bp DNA ladders (Solarbio, China) in 0.5% hydroxyethyl cellulose on the self-built capillary electrophoresis system (Li et al. 2016). The background electrolyte was diluted 0.5× TBE (1× TBE = 89 mM Tris/89 mM boric acid/2 mM EDTA, pH 8.4). The effective length and the total length of the capillary were 12 and 15 cm, respectively. The sample was electrokinetically introduced into the capillary at 100 V/cm for 1.0 s. They were separated at 100 V/cm. Data in Fig. 6 revealed that the DNA ladders were well resolved and the linear relationship existed between size of DNA and the migration time (correlation coefficient R, 0.997), and thus we supposed the peak was from the positives of PCR. Moreover, the peak corresponding the PCR products was not observed in negative control (data not shown).
3.5 Detection of periodontal pathogen by microfluidic chamber chip
Finally, we amplified the target genes of P.g in the chamber PCR microfluidic chip based on the self-built platform. The PCR solution contained 5.0 µL template, 12.5 µL 2×SYBR Green PCR mix, 0.5 µL Primer1 (10 µM), 0.5 µL Primer2 (10 µM) and 6.5 µL ddH2O (see Table S2 in the supporting information). The size of the amplicon was 169 bp. The microfluidic chamber chip consisted of 3200 micro chambers. The depth and radius for the chamber were 80 μm and 50 μm, respectively. The capacity of a single microchamber was 0.628 nL. Figure 7A showed the image of the microchambers prior to be filled with PCR reagent. The self-assembled platform for PCR reaction was performed according to following thermal cycling profile: 95 °C for 2 min, 40 thermal cycles of 95 °C for 20 s and 64 °C for 30 s. For negative control, we performed the micro chamber PCR with reagent contained no DNA template (Fig. 7B). Figure 7C showed the microchambers filled with reagent prior to PCR. It showed that fluorescence was not observed in Fig. 7A-C, and fluorescence from the microchamber was observed (Fig. 7D) after PCR, indicating that PCR is feasible in the self-built platform. By calculating the grayscale, we found that the ratio of the fluorescence intensity corresponding to the bright (positive) microwells and the dim (negative) ones was about 1.5 (see Figure S2 in the supporting information). We also measured the limit of detection of the chip. The absolute quantification of the template DNA was carried out based on Eq. (1) described in Ref (Hosokawa and Ohmori 2023). Experiments demonstrated that the lowest concentration was about 1.05 × 101 copies/µL, which was close to our previous result (Shen et al. 2021).
4 Conclusions
In summary, we have built a novel self-built platform for chamber PCR microfluidic chip. We have proposed a method to improve reagent self-priming compartmentalization and avoid the reagent evaporation by placing a glass slide above the PDMS microfluidic chip. We also tested the temperature distribution and stability of the glass slide rotating between two heaters. Finally, we have realized the amplification of P.g in the micro chamber microfluidic chip within 35 min and on-site detection of its PCR products. The advantages of such system are mainly low-cost, easy to miniaturize, and light in weight. In the next step, we are trying to miniaturize the large field of view imaging system developed in our lab, and combine it with the system reported in this work, and make the low-cost microchamber PCR microfluidic chip more practical. We believe that it has potential application in the field of bacterial diagnosis.
References
S.E. Cavanaugh, A.S. Bathrick, Forensic science international: Genetics. 32,40 – 9(2018)
S. Chen, Y. Sun, F. Fan, S. Chen, Y. Zhang, Y. Zhang et al., TRAC Trends Anal. Chem. 157,116737(2022)
X. Cui, L. Wu, Y. Wu, J.H. Zhang, Q. Zhao, F.X. Jing et al., Anal. Chim. Acta 1107,127 – 34(2020)
J.S. Farrar, C.T. Wittwer, Clin Chem. 61,145 – 53(2015)
F. Gouriet Fdr, Fenollar, J.-Y. Patrice, M. Drancourt, D. Raoult, J. Clin. Microbiol. 43, 4993–5002 (2005)
K. Hosokawa, H. Ohmori, Analytical Sciences. 39,2067-74(2023)
Y. Huang, Z. Gao, C. Ma, Y. Sun, Y. Huang, C. Jia et al., Analyst (2023)
M.U. Kopp, A.J. Mello, A. Manz, Science. 280, 1046–1048 (1998)
M. Krishnan, V.M. Ugaz, M.A. Burns, Science. 298, 793 (2002)
S.H. Lee, S.-Y. Ruan, S.-C. Pan, T.-F. Lee, J.-Y. Chien, P.-R. Hsueh, J. Microbiol. Immunol. Infect. 52, 920–928 (2019)
Z. Li, Y. Zhao, D. Zhang, S. Zhuang, Y. Yamaguchi, Sens. Actuators B 230,779 – 84(2016)
Z. Li, R. Ju, S. Sekine, D. Zhang, S. Zhuang, Y. Yamaguchi, Lab. On a Chip. 19, 2663–2668 (2019)
Z. Li, J. Liu, P. Wang, C. Tao, L. Zheng, S. Sekine et al., Lab. On a Chip. 21, 3159–3164 (2021)
Z. Li, Y. Wang, Z. Gao, S. Sekine, Q. You, S. Zhuang et al., Anal. Chim. Acta 1251,340995(2023)
C. Liu, W. Eschen, L. Loetgering, D.S. Penagos Molina, R. Klas, A. Iliou et al., PhotoniX. 4, 1–15 (2023)
T.C. Merkel, V.I. Bondar, K. Nagai, B.D. Freeman, I. Pinnau, J. Polym. Sci. Pol. Phys. 38,415 – 34(2000)
Y. Ning, X. Cui, C. Yang, F. Jing, X. Bian, L. Yi et al., Anal. Chim. Acta. 1055, 65–73 (2019)
T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino et al., Nucleic Acids Res. 28, e63 (2000)
J. Shen, J. Zheng, Z. Li, Y. Liu, F. Jing, X. Wan et al., Lab. On a Chip. 21, 3742–3747 (2021)
Q. Song, Y.B. Gao, Q.Y. Zhu, Q.C. Tian, B.W. Yu, B.F. Song et al., Biomed. Microdevices. 17, 64 (2015)
T. Suo, X. Liu, J. Feng, M. Guo, W. Hu, D. Guo et al., Emerg. Microbes Infections. 9, 1259–1268 (2020)
H. Wang, Y.-L. Zhang, D.-D. Han, W. Wang, H.-B. Sun, PhotoniX. 2, 1–13 (2021)
L. Xu, H. Lee, D. Jetta, K.W. Oh, Lab. On a Chip. 15, 3962–3979 (2015)
B. Yang, P. Wang, Z. Li, C. Tao, Q. You, S. Sekine et al., Lab. On a Chip. 22, 733–737 (2022)
Q.Y. Zhu, L. Qiu, B.W. Yu, Y.N. Xu, Y.B. Gao, T.T. Pan et al., Lab. On a Chip. 14, 1176–1185 (2014)
Q.Y. Zhu, Y.N. Xu, L. Qiu, C.C. Ma, B.W. Yu, Q. Song et al., Lab. On a Chip. 17, 1655–1665 (2017)
Acknowledgements
This work was supported by Science and Technology Commission of Shanghai Municipality, China (No.18441900400 and No.19ZR1477500). It was also supported by Excellent Young Talents support plan in Colleges of Anhui Province Grant (gxgnfx 2021167).
Author information
Authors and Affiliations
Contributions
Z. L: Project administration, Supervision, Funding acquisition, Writing; X. M, X. W, X. Y: Methodology, Data acquisition; Z. Z and B. Y: Methodology, Periodontal pathogen extraction; D. Z: Project administration, Supervision; Y. Y: Project administration, Supervision, Review &editing; J. Y and Y. Z: Data analysis, Review &editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have declared no conflict of interest.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Z., Ma, X., Zhang, Z. et al. A rapid and low-cost platform for detection of bacterial based on microchamber PCR microfluidic chip. Biomed Microdevices 26, 20 (2024). https://doi.org/10.1007/s10544-024-00699-x
Accepted:
Published:
DOI: https://doi.org/10.1007/s10544-024-00699-x