Abstract
The analysis of genetic materials in the post-human genome project era has become an ever-expanding branch of research and thus routinely employed in majority of biochemical laboratories. Most of the diagnostic research area emphasizes on polymerase chain reaction for detecting pathogenic organisms. However, the conventional polymerase chain reaction requires expensive and sophisticated thermal cycler and is not handy owing to its large dimensions. Therefore, we fabricated a continuous-flow polymerase chain reaction chip on a PDMS based microfluidic platform to ease the hardship of the conventional system. Temperature being the most crucial factor in polymerase chain reaction, was monitored and regulated by thermostatic action using an on-line computer system. Indium tin oxide coated glass platform was used for heating as it is transparent and has good thermal conductivity under the influence of DC bias. The heating circuit used an ATMega 128 MCU to control the temperature. As a result, a precise and quick heating environment was maintained on the microfluidic chip to amplify the target DNA. We successfully amplified Lambda phage and Escherichia coli DNA on our chip to prove the practicality of the device.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Polymerase chain reaction (PCR) is used in a variety of applications such as genotyping, taxonomy, forensic investigation, cloning and molecular biology techniques, DNA sequencing, pathogen detection, drug discovery as well as environmental applications. During various stages of the disease diagnosis, genetic analysis has become a core application. In particular, the PCR is used as a technique to screen and amplify a specific DNA fragment to generate millions of copies of it. The conventional technique for performing PCR is time consuming and laborious. Another major drawback of a conventional thermal cycler is lack of portability due to its bulky size. Therefore, a miniaturized PCR system is highly in demand for a hand held on-site use. Owing to its widespread use and importance, a number of groups have tried to miniaturize PCR on a chip (Cho et al. 2006; Lee et al. 2008; Park et al. 2011). Miniaturized platform also offers a great potential of integrating different sample processing and detecting units on a single microfluidic chip, which are the essential objectives for the realization of a lab-on-a-chip device (Christensen et al. 2008; Jha et al. 2012; Kim et al. 2005).
Although a much has been done in minimizing the chip size, one of the major afflictions in realizing a portable device is the accurate control of temperature. Technically, PCR relies on thermal cycling; consisting of cycles of repeated heating and cooling of the reaction mixture for DNA melting, enzymatic replication, and re-annealing. A specific set of temperature range is used for each reaction depending on the type of DNA polymerase and the primer sequence used (Schneegaß and Köhler 2001). A high degree of temperature specificity is required for a successful amplification of DNA. If the temperature is not controlled in adequate; nonspecific amplification will increase due to mismatch annealing of primers. Aberration in temperature can also bar activation of enzyme or the melting of DNA. Therefore, integration of high performance temperature control system is a vital feature for the miniaturization of monolithic chip. Some of the reported continuous flowing PCR chips employed metal heating block and thin strips of metal to heat the reaction mixture (Kim et al. 2006; Schneegaß et al. 2001). But, the use of these blocks restricts the aim of miniaturization and lacked a precise temperature control system.
Therefore, in these regards, we fabricated a real-time temperature monitoring and control system based on the microcontroller unit (MCU) for the precise and online temperature control. The advent of personal computer enables large scale simulation and computing which was formerly not possible. A MCU is a specialized type of digital computer used to provide automatic sequencing or control of a system. An instruction programmed into the microcontroller’s memory tells it how to react to input conditions, and what types of signals to send to the outputs.
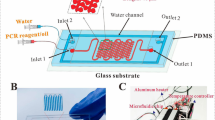
The microfluidic PCR chip was fabricated on ITO coated glass substrate while microchannels were engraved in polydimethylsiloxane (PDMS). The ITO microheater, along with PDMS based fluidic network and MCU system led to a miniaturized PCR system which could easily be developed into a handheld system. The final microfluidic PCR chip was used to amplify a target Lambda phage and Escherichia coli DNA.
2 Materials and methods
2.1 Device fabrication
The microfluidic PCR chip was made using conventional photolithographic techniques (Joung et al. 2008). The microchannels used in this study were molded in PDMS polymer using the negative molding method. For this purpose, 120 μm thick negative photoresist (PR) (SU-8 2075, Micro-Chem, USA) was spin-coated and patterned on a silicon wafer. A degassed mixture of Sylgard 184 (Dow corning, USA) silicone elastomer and curing agent (in 10:1 v/v ratio) was poured on the SU-8 patterned wafer and cured for 4 h at 72 °C. The PDMS mold thus formed was peeled off manually and drilled to make access holes of 3 mm diameter each. The width and depth of the microchannel created were 250 and 120 μm respectively with a total length of 2,790 mm for 30 cycles. ITO heaters were fabricated using photolithography and wet etch process. Positive PR (AZ-1512, Micro-Chem, USA) was spin-coated on ITO deposited glass (Samsung Corning) and was patterned by exposing UV light through a designed photomask. The exposed ITO film was then etched using the FeCl3/HCl solution for 1 h and the remaining PR was removed with acetone. Finally, the PDMS microchannel was bonded to the non-ITO side of glass wafer using UV ozone activation.
2.2 Materials for polymerase chain reaction
In this study, two target genes were amplified for verifying the practicality of proposed microfluidic PCR chip. One of genes is from Lambda phage where the forward: GCA-AGT-ATC-GTT-TCC-ACC-GT and reverse: TTA-TAA-GTC-TAA-TGA-AGA-CAA-ATC-CC primers led to a product size of 100 bp while another one is from E. coli where the forward: ACT-TCA-GTC-GGA-TCC-GCT-GAT and reverse: TGT-AAA-CTC-GAG-TTC-CGC-ACT primers coded a product of 840 bp in length.
To perform the PCR, a pre-PCR mixture (Bionics, Korea) containing dNTPs, DNA polymerase, reaction buffer, and MgCl2 along with primers and template DNA (50 μL total volume) was injected in the microfluidic chip using syringe pump.
2.3 Temperature control system
Figure 1 shows the overall setup of microfluidic PCR along with the temperature sensor. The three different temperature zones namely denaturation, annealing, and extension for the PCR reaction were monitored through an MCU aided computer interface in real-time. As it was essential to precisely control the three temperature zones on the microfluidic PCR, we attached temperature sensors (LM35Z) on ITO using conducting glue. A fixed potential was supplied to the MCU which distributed it to the different temperature zones. For calibration, different voltages were given to MCU in steps. Voltage providing most stable temperature was selected for further experiment. The computer system operated the 8-bit microcontroller (ATmega128). It gathered the temperature signal from the sensors, processed and analyzed it, and finally controlled the flow of current through switching On/Off, MOSFET (metal–oxide–semiconductor field-effect transistor, IRF3205) thereby providing a means to control the heating efficiency of the microheater. A schematic diagram of this ASIC control system is shown in Fig. 2. The temperature inside the microchannel was also monitored in initial few devices by placing a thermocouple within the channel.
3 Results and discussion
3.1 Automatic real-time temperature control system
This control system offers an effective and automatic way to monitor and control the heating efficiency of the ITO microheater on the proposed on-chip PCR system. ITO was used as the microheater because of its transparency and it offers a linear calibration range for temperature upon application of DC power. The temperature of the PCR system can be changed online to make it a universal PCR and so can be used for amplification of any target sample. To show the potency of ITO electrodes as the microheater, a gradually increasing DC power was applied to the electrodes. Temperature on the other side of glass was measured using thermocouple. Linear temperature curve with respect to applied power was obtained which is shown in the Fig. 3a.
Calibration of temperature using MCU controlled ITO microheater. a Linear temeprature response of ITO electrodes with respect to applied power b feedback control calibration curve of tempertature for Lambda phage. Temperature range for on-chip PCR of Lambda phage DNA: denaturation, 92 °C; annealing, 55 °C and extension, 72 °C. c Calibration curve of temperature for E. coli. Temperature range for on-chip PCR of E. coli DNA: denaturation, 95 °C; annealing, 50 °C and extension, 72 °C
Calibration of voltage for MCU was done by applying 5, 10, 15, 17, and 18 V to it, while the DI water was flowing inside the microchannel at a rate of 4 μl/min. The temperature of each zone after 10 min of voltage application is summarized in Table 1. Though the required temperature was reached by applying 17, 18 V showed most stable and faster results. It took a mere 4 min to obtain a stable desired temperature using 18 V (Fig. 3b, c). Thermocouple placed inside the microchannel showed no noticeable difference in the temperature of the two sides of glass. The MCU prioritized heating of denaturation zone first followed by elongation and annealing. The denaturation, annealing and elongation temperature for Lambda phage and E. coli were 92, 55, 72 °C and 95, 50, 72 °C respectively (Fig. 3b, c).
Previously Huang et al. performed successful amplification of DNA in low-volume droplet, on their device which has a temperature controller. However, their device is as big as conventional thermocycler. Even if it achieves rapid micro-PCR, large scaled device cannot be used for hand-held application (Huang et al. 2011). Recently, Wang et al. and Zhu et al. used microcontroller unit (MCU) and LabVIEW based algorithm to monitor and control the temperature (Wang et al. 2011; Zhu et al. 2013). They used a Peltier thermoelectric cooler to cycle the temperature. Also, Peham et al. reported the use of Peltier devices attached with metal blocks to fabricate PCR. Such types of device are rather far from miniaturization, and make the process tedious and bulky (Peham et al. 2011). In contrast, patterned ITO heater which we fabricated enables realization of a hand-held device on a monolithic chip. Similarly, Xue et al. employed a NiCr film to heat the sample, which proved to be advantageous (Xue and Yan 2012).
To confirm the temperature profile, we also tried to measure the temperature of device using infrared camera. Since the transmittance/emissivity of PDMS is not 1, we could not capture the exact thermal profile of the ITO heater. Supplementary data, Figure S1 and S2 shows the IR image of the device used for amplification of Lamba phage and E. coli DNA. We believe that some amount of light was absorbed by PDMS during measurement and thus the camera could not measure accurate temperatures. Nevertheless a good co-relation was seen between the actual temperature and the temperature measured using IR camera (Fig. S3).
3.2 Microfluidic PCR chip
The PDMS based microchannel included in the microfluidic PCR chip acted as a reservoir and helped with the flow of sample within different temperature zones. The patterned ITO electrode assisted in maintaining the temperature zones, which was controlled using an MCU based temperature sensor circuit. PCR mixture flew continuously inside the microchannel passing over different temperature zones. The sequential change in temperature resulted in amplified DNA product.
To prevent the binding of biomolecules on the PDMS surface, an acid/base solution treatment was done. HCl and NaOH solution was flown to neutralize the inner charge of PDMS surface. Bovine serum albumin (BSA) was then injected and incubated for 30 min inside the microchannel to carry out an efficient surface treatment (Fang et al. 2009; McDonald et al. 2000; Shin et al. 2006). After surface treatment, heater supply was started to initiate the PCR. At the desired temperature level, the PCR mixture containing template DNA, primers, dNTPs, DNA polymerase and PCR buffer was injected using a syringe pump at an optimized flow rate of 4 μl/min. Total times taken to perform 30 cycles was around 45 min.
3.3 Polymerase chain reaction of microfluidic chip
The fabricated microfluidic chip was used for an on-chip amplification of target genes from Lambda phage and E. coli genomic DNA. Continuous flow of reaction mixture in different temperature zones above ITO heater led to PCR of the target gene. The agarose gel electrophoresis of on-chip amplified PCR amplicon is shown in Fig. 4. Lane 1 and 2 of Fig. 4a and lane 1 of Fig. 4b represents the amplicon after 30 cycles in pre-treated microchannel, while lane 3 of Fig. 4a and lane 2 of Fig. 4b denotes the PCR product from the untreated microchannel. The results clearly demonstrate that the PCR amplification in untreated channels decreases the product concentration due to the binding of biomolecules to the PDMS surface. We also used negative control which is shown in lane 4 of Fig. 4a and lane 3 of Fig. 4b. Negative controls have only the PCR premix solution lacking the template DNA. These results validate the specific and successful amplification of target genes on our continuous flow microfluidic PCR chip integrated with a precise temperature control system. An unstable system temperature would have led to unspecific binding of primers to negative DNA samples, resulting in false positive result.
Gel electrophoresis image : a image of on-chip PCR product of Lambda phage. M Marker, lanes 1 and 2 amplicon from pre-treated microchannel, lane 3 untreated microchannel, lane 4 negative control, b image of on-chip PCR product of E. coli. M Marker, lane1 amplicon from pre-treated microchannel, lane 2 untreated microchannel, lane 3 negative control
4 Conclusion
We have demonstrated a microfluidic PCR system based on a real-time temperature monitoring. ITO based microheaters were used to heat the DNA sample, while the sample was contained in a PDMS based microchannel. A low DC bias was passed through the ITO electrodes which raised the temperature and was controlled using an MCU based temperature sensor. The temperature system worked using the thermostatic action with a real-time on-line computer system which can adapt to any type of the target gene. We could successfully amplify target genes of Lambda phage DNA and E. coli DNA and proved the practicality of our device.
References
Cho Y-K, Kim J, Lee Y, Kim Y-A, Namkoong K, Lim H, Oh KW, Kim S, Han J, Park C, Pak YE, Ki C-S, Choi JR, Myeong H-K, Ko C (2006) Clinical evaluation of micro-scale chip-based PCR system for rapid detection of hepatitis B virus. Biosens Bioelectron 21(11):2161–2169. doi:10.1016/j.bios.2005.10.005
Christensen TB, Bang DD, Wolff A (2008) Multiplex polymerase chain reaction (PCR) on a SU-8 chip. Microelectron Eng 85(5–6):1278–1281. doi:10.1016/j.mee.2008.01.066
Fang TH, Ramalingam N, Xian-Dui D, Ngin TS, Xianting Z, Lai Kuan AT, Peng Huat EY, Hai-Qing G (2009) Real-time PCR microfluidic devices with concurrent electrochemical detection. Biosens Bioelectron 24(7):2131–2136. doi:10.1016/j.bios.2008.11.009
Huang T-T, Wang J-S, Yu C-S, Hu Y-C (2011) A novel rapid-reaction nucleic-acid amplification device using micro-volume chips. Instrumentation and measurement technology conference (I2MTC), 2011 IEEE doi:10.1109/imtc.2011.5944229
Jha SK, Chand R, Han D, Jang Y-C, Ra G-S, Kim JS, Nahm B-H, Kim Y-S (2012) An integrated PCR microfluidic chip incorporating aseptic electrochemical cell lysis and capillary electrophoresis amperometric DNA detection for rapid and quantitative genetic analysis. Lab Chip. doi:10.1039/c2lc40727b
Joung S-R, Kang CJ, Kim Y-S (2008) Series DNA amplification using the continuous-flow polymerase chain reaction chip. Jpn J Appl Phys 47(2):1342–1345. doi:10.1143/jjap.47.1342
Kim J-H, Na K-H, Kang CJ, Kim Y-S (2005) A disposable thermopneumatic-actuated micropump stacked with PDMS layers and ITO-coated glass. Sens Actuators A 120(2):365–369. doi:10.1016/j.sna.2004.12.024
Kim JA, Lee JY, Seong S, Cha SH, Lee SH, Kim JJ, Park TH (2006) Fabrication and characterization of a PDMS: glass hybrid continuous-flow PCR chip. Biochem Eng J 29(1–2):91–97. doi:10.1016/j.bej.2005.02.032
Lee SH, Kim S-W, Kang JY, Ahn CH (2008) A polymer lab-on-a-chip for reverse transcription (RT)-PCR based point-of-care clinical diagnostics. Lab Chip 8(12):2121. doi:10.1039/b811131f
McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJ, Whitesides GM (2000) Fabrication of microfluidic systems in poly(dimethylsiloxane). Electrophoresis 21(1):27–40. doi:10.1002/(SICI)1522-2683(20000101)21:1<27:AID-ELPS27>3.0.CO;2-C
Park S, Zhang Y, Lin S, Wang T-H, Yang S (2011) Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol Adv 29(6):830–839. doi:10.1016/j.biotechadv.2011.06.017
Peham JR, Grienauer W, Steiner H, Heer R, Vellekoop MJ, Nohammer C, Wiesinger-Mayr H (2011) Long target droplet polymerase chain reaction with a microfluidic device for high-throughput detection of pathogenic bacteria at clinical sensitivity. Biomed Microdevices 13(3):463–473. doi:10.1007/s10544-011-9514-x
Schneegaß I, Köhler JM (2001) Flow-through polymerase chain reactions in chip thermocyclers. Rev Mol Biotechnol 82(2):101–121. doi:10.1016/s1389-0352(01)00033-2
Schneegaß I, Bräutigam R, Köhler JM (2001) Miniaturized flow-through PCR with different template types in a silicon chip thermocycler. Lab Chip 1(1):42. doi:10.1039/b103846j
Shin KS, Kim YH, Min JA, Kwak SM, Kim SK, Yang EG, Park JH, Ju BK, Kim TS, Kang JY (2006) Miniaturized fluorescence detection chip for capillary electrophoresis immunoassay of agricultural herbicide atrazine. Anal Chim Acta 573–574:164–171. doi:10.1016/j.aca.2006.05.099
Wang L, Song HX, Chen T, Wang ZH (2011) A design of dspic based microfluidic pcr chip temperature controlling system. Appl Mech Mater 108:206–211. doi:10.4028/www.scientific.net/AMM.108.206
Xue N, Yan W (2012) Glass-based continuous-flow pcr chip with a portable control system for DNA amplification. IEEE Sens J 12(6):1914–1918. doi:10.1109/jsen.2011.2182047
Zhu J, Palla M, Ronca S, Wapner R, Ju J, Lin Q (2013) A MEMS-based approach to single nucleotide polymorphism genotyping. Sens Actuators A 195:175–182. doi:10.1016/j.sna.2012.07.025
Acknowledgments
This paper was supported by Samsung Research Fund, Sungkyunkwan University, 2013.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Han, D., Jang, YC., Oh, SN. et al. MCU based real-time temperature control system for universal microfluidic PCR chip. Microsyst Technol 20, 471–476 (2014). https://doi.org/10.1007/s00542-013-1970-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00542-013-1970-1