Abstract
Background
Although optimal treatment of superficial esophageal squamous cell carcinoma (SCC) requires accurate evaluation of cancer invasion depth, the current process is rather subjective and may vary by observer. We, therefore, aimed to develop an AI system to calculate cancer invasion depth.
Methods
We gathered and selected 23,977 images (6857 WLI and 17,120 NBI/BLI images) of pathologically proven superficial esophageal SCC from endoscopic videos and still images of superficial esophageal SCC taken in our facility, to use as a learning dataset. We annotated the images with information [such as magnified endoscopy (ME) or non-ME, pEP-LPM, pMM, pSM1, and pSM2-3 cancers] based on pathologic diagnosis of the resected specimens. We created a model using a convolutional neural network. Performance of the AI system was compared with that of invited experts who used the same validation video set, independent of the learning dataset.
Results
Accuracy, sensitivity, and specificity with non-magnified endoscopy (ME) were 87%, 50%, and 99% for the AI system and 85%, 45%, 97% for the experts. Accuracy, sensitivity, and specificity with ME were 89%, 71%, and 95% for the AI system and 84%, 42%, 97% for the experts.
Conclusions
Most diagnostic parameters were higher when done by the AI system than by the experts. These results suggest that our AI system could potentially provide useful support during endoscopies.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Worldwide, esophageal cancer (EC) is the seventh most commonly diagnosed cancer, and has the sixth highest mortality rate, with an estimated 572,000 new cases and 509,000 deaths in 2018 [1]. Of its two subtypes, squamous cell carcinoma (SCC) and adenocarcinoma, SCC accounts for 80% of EC [1]. Superficial EC is asymptomatic, shows subtle mucosal change on endoscopic examination and is often missed until it has advanced to an advanced stage. The mainstay of treatment for advanced EC is esophagectomy [2,3,4,5,6,7,8,9,10], which has relatively high mortality rate. However, if detected as a superficial cancer, EC can be treated by less invasive treatment [2, 3, 6,7,8, 11,12,13].
In superficial EC, accurate assessment of invasion depth is particularly important for deciding treatment strategy [14,15,16,17]. Diagnosis by non-magnified endoscopy (ME) is based on macroscopic findings such as protrusion, depression, and hardness of the cancer. When ME is available, it usually follows non-ME, to observe blood vessel patterns, using narrow-band imaging (NBI) or blue laser imaging (BLI). Epithelium (EP)/lamina propria mucosa (LPM) and muscularis mucosa (MM)/submucosal cancers that invade up to 200 μm (SM1) are candidates for endoscopic resection (ER) because of their relatively low (< 10%) risk of metastasis [11,12,13,14, 16], whereas esophagectomy is mainly indicated for SM2-3 cancers, for which risk of metastasis exceeds 25% [11, 14, 15]. Thus differentiating EP-SM1 from SM2-3 is important, but these assessments are often unsatisfactory and subject to inter-observer variability [18].
A potential solution to improve both accuracy and variability of endoscopic diagnosis is using an artificial intelligence (AI) diagnostic system. Deep learning, the mainstay of AI systems, is typically based on convolutional neural networks (CNN), and has demonstrated good performances for visual tasks. This technology has been used to diagnose gastrointestinal cancers, including esophageal SCC [19,20,21,22]. Although various studies of AI systems to detect EC have been published, to our knowledge, the only report for assessing invasion depth was based on still images [23]. Studies that use still images may have some bias because usually the best images from the best areas are selected for the analysis. Validating a comprehensive diagnostic process with video images allows a more realistic assessment of the AI system.
Recent developments in deep learning-based techniques have included educating the system using images derived from video and free-hand marking with precise delineation of lesions. We therefore aimed to develop an AI system that used these new deep learning techniques, and to estimate its performance using video images by comparing the conclusions with those of experts in the field.
Methods
Preparation of learning dataset
This study was conducted at Osaka International Cancer Institute. Training of the system was conducted using endoscopic videos and images taken during daily endoscopic examinations. The endoscopic examination were carried out using the endoscopes GIF-RQ260Z, GIF-FQ260Z, GIF-Q240Z, GIF-H290Z, GIF-HQ290, GIF-H260Z, GIF-XP290N, GIF-Q260J, or GIF-H290 (Olympus, Tokyo, Japan) and video processors CV-260 (Olympus Co.) or EVIS LUCERA CV-260/CLV-260 and EVIS LUCERA ELITE CV-290/CLV-290SL (Olympus Medical Systems), or the endoscopes EG-L590ZW, EG-L600ZW, or EG-L600ZW7 (Fujifilm Co., Tokyo, Japan) and the video endoscopic system LASEREO (Fujifilm Co.). For observations that used the LASEREO system, white-light imaging (WLI) and blue laser imaging (BLI) which provide images similar to NBI, were used. A black soft hood was equipped on the tip of the endoscope to keep an appropriate distance between the tip of the endoscope and esophageal wall during magnifying observations. B-mode level 8 for NBI and level 5–6 for BLI were used for the structure enhancement function.
Initial routine inspections with non-ME with WLI were usually carried out to evaluate macroscopic type, protrusion, depression, and hardness of the lesions. ME was then conducted, with NBI or BLI, to evaluate the appearance of the superficial vascular architecture, especially changes in intrapapillary capillary loops. Finally, iodine staining was used to delineate the cancer spread. Pathological assessments of biopsies and resected specimens were conducted according to the Japanese classification of esophageal cancer [24, 25].
Creating the learning dataset
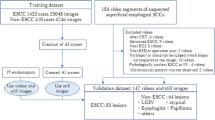
We gathered endoscopic videos taken between November 2015 and June 2019, and still images taken between December 2005 and April 2018, of superficial esophageal SCC treated in our facility. We included patients with (a) superficial esophageal SCC treated with ER or esophagectomy, and (b) with pathologic proof of cancer invasion depth. We excluded patients with severe esophagitis, those with histories of chemotherapy or radiation to the esophagus, lesions adjacent to ulcers or ulcer scars, and poor-quality images resulting from inadequate insufflation, bleeding, halation, blurring, poor focus, or mucus. We also excluded cancers diagnosed as EP-SM1 by surgical specimens. As surgically resected specimens are usually cut as much wider slices (5- to 7-mm) than are endoscopically resected specimens (2 mm), we speculated that deeper invasion cannot be excluded for EP-SM1 cancers diagnosed from surgically resected specimens.
Video images, which are composed of rapid successions of sequential images, were each divided into a series of still images. From this large volume of sequential still images (30 images per second of video), 1 out of every 30 still images was extracted and included in the learning dataset. Extracting still images from video images diversifies the shooting conditions of images, e.g., by distance, angle, and focus. The learning images included WLI, NBI, and BLI images (Fig. 1.)
Development of the deep learning-based artificial intelligence system for the diagnosis of cancer invasion depth. Video images were divided into a series of still images. Extracting still images from video images. These images were marked manually by precisely delineating cancers (Free-hand marking)
After selection, we had collected 23,977 images (6857 WLI and 17,120 NBI/BLI images) from 909 patients with pathologically proven superficial esophageal SCC as the learning dataset. We annotated the images with information such as non-ME or ME, use of irradiated light (WLI, NBI, BLI) and pathologic (p) EP-LPM, pMM, pSM1, and pSM2-3 cancers, based on pathologic diagnoses of the resected specimens. These images were marked manually by precisely delineating the boundaries. We marked the whole cancer area for pEP-MM cancers, but only the pSM invasion part for SM cancers (Fig. 2). Marking was conducted by eight endoscopists, and reconfirmed by a board-certified trainer (RI) at the Japan Gastroenterological Endoscopy Society.
Constructing the AI system
Our CNN model was a type of artificial neural network used in deep learning, that has been used to analyze visual imagery. Supervised learning was used to train the CNN in this study. With supervised learning, the training set is submitted as inputs to the system during the training phase. Each input is labeled with a desired output value (a correct diagnosis), which teaches the system to know what output is needed for a specific input. A large set of training endoscopic images with correct diagnostic information is required to create this function. The CNN learns filters that were previously hand-engineered in more traditional algorithms. This independence from prior knowledge represents a significant advantage of CNN models over other types of machine learning. The mathematical algorithm used was a Single Shot MultiBox Detector (https://arxiv.org/abs/1512.02325) and the base CNN was a Visual Geometry Group network consisting of 16 layers. All of the CNN layers were fine-tuned from weights of ImageNet using stochastic gradient descent as a back-propagation method with a global learning rate of 0.0001, 200 epochs, and a batch size of 24. Stochastic gradient descent can attenuate the risk of overfitting.
The CNN was trained by the dataset and validated using a PyTorch deep learning framework, which was developed by Facebook’s artificial intelligence research group. For the learning dataset, we included endoscopic images with various shooting conditions and resolutions, in consideration of the generalizability of the system. Each image was resized to 300 × 300 pixels for optimal CNN analysis. We determined appropriate values for hyperparameters (such as learning rate and weight decay) by repeated trial and error.
Evaluation of the AI system
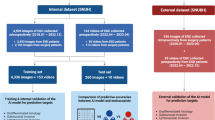
The AI system was evaluated using an independent validation dataset of 102 video images of superficial esophageal SCC taken at our hospital between January 2017 and January 2019. Of 102 videos, 87 (85.3%) were taken by endoscopists with 5–14 years’ expertise as clinicians. The selection criteria and exclusion criteria were the same as those of the learning dataset. Besides, in each video image, we extracted non-ME with WLI capable of recognizing macroscopic findings and the overall image of the lesion and ME with NBI/BLI capable of recognizing blood vessel pattern as the information necessary for the diagnosis of invasion depth. As a result, we prepared two types of videos—non-ME with WLI, and ME with NBI/BLI of 4–12 s.
The trained neural network generated diagnoses of EP- SM1 or SM2-3 cancer and probability scores between 0 and 1, corresponding to the probability of that diagnosis. Each frame was judged as SM2-3 cancer when probability score for SM2-3 cancer was 0.45 or greater in that frame (Figs. 3 and 4); the others (probability sore for SM2-3 < 0.45 and any probability score for EP-SM1) were judged as EP-SM1 cancer. Conclusive diagnosis of SM2-3 cancer by non-ME was conducted when ten times of “three almost serial appearance of positive images without interruption by 0.5 s” were confirmed in each video. This condition approximates frequent appearance of positive images for one second. Conclusive diagnosis of SM2-3 cancer by ME was conducted when ten times of “7 almost serial appearance of positive images without interruption by 0.2 s” were confirmed in each video. This condition approximates the frequent appearance of positive images for two to three seconds.
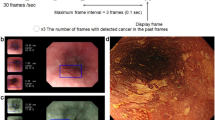
a A case of LPM cancer in the mid-esophagus, b with non-magnified white-light imaging. c The AI system correctly diagnosed the lesion by indicating it with a blue square frame as M_WLI (i.e., EP-SM1). d Same case with magnified narrow-band imaging. The AI system correctly diagnosed the lesion by indicating it with a blue square frame as M_NBI (i.e., EP-SM1)
a A case of SM3 cancer in the lower esophagus, b with non-magnified white-light imaging. c The AI system correctly diagnosed the lesion by indicating it with a blue square frame as SM2_WLI (i.e., SM2-3). d Same case with magnified narrow-band imaging. The AI system correctly diagnosed the lesion by indicating it with a blue square frame as SM2_NBI (i.e., SM2-3)
Evaluation by the expert
The performance of the AI system was compared with that of an invited group of 14 board-certified specialists (expert group) at the Japan Gastroenterological Endoscopy Society. Each of the experts had 7–25 years’ expertise as clinicians, had conducted 3000–20,000 endoscopic examinations, and routinely diagnosed gastrointestinal cancers. They were asked to observe the same videos as the AI system and diagnose cancer invasion depths as EP-SM1 or SM2-3.
Outcome measures
The main outcome measures were diagnostic accuracy (correctly diagnosed lesions/total lesions), sensitivity (correctly diagnosed SM2-3 cancers/total SM2-3 cancers), specificity (correctly diagnosed non-SM2-3 cancers/total non-SM2-3 cancers), positive predictive value (PPV; correctly diagnosed SM2-3 cancers /total lesions diagnosed as SM2-3 cancers), negative predictive value (NPV; correctly diagnosed non-SM2-3 cancers/total lesions diagnosed as non-SM2-3 cancers), and diagnostic time. The results are shown as averages and 95% confidence intervals (CI). Inter-observer variation in the diagnosis of esophageal SCC was assessed using kappa statistics; κ > 0.8 denoted almost perfect agreement, 0.8–0.6, substantial agreement; 0.6–0.4, moderate agreement; 0.4–0.2, fair agreement; < 0.2, slight agreement; 0, agreement equal to chance; and < 0 suggested disagreement. All calculations were performed on a personal computer using EZR software, version 1.40 (Saitama Medical Center, Jichi Medical University, Japan).
Ethics
This study was approved by the Institutional Review Board of Osaka International Cancer Institute (2017–1710059178) and Japan Medical Association (JMA-IIA00283).
Results
Performance of AI system and expert SM2-3 diagnosis with video
Patient and lesion characteristics are summarized in Table 1. For non-ME videos, accuracy, sensitivity and specificity were 87.3%, 50.0% and 98.7% for the AI system, and 84.7%, 44.9% and 96.9% for the experts (Fig. 5, Table 2). The accuracy and specificity were each higher for the AI system than the upper 95% CI limits for the experts. For the ME videos, accuracy, sensitivity and specificity were 89.2%, 70.8% and 94.9% for the AI system and 84.4%, 42.3% and 97.3% for the experts (Fig. 5, Table 2). In the ME diagnosis, the accuracy and sensitivity for the AI system were higher than upper 95% CI limits for the experts; however, AI specificity was lower than the lower 95% CI limit for the experts.
Diagnosis time for AI was 30 fps on a high-speed computer, which can be adapted as real-time diagnosis. Experts averaged 165 min. Intra-observer agreement of 14 experts was moderate, with Fleiss’ κ 0.4–0.6 for both non-ME and ME (Table 3).
Discussion
Our AI system performed favorably in diagnosing invasion depth of superficial esophageal SCCs in video images, with accuracies of 87.3% and 89.2% for non-ME and ME, respectively. These values were comparable or even better than those of expert endoscopists with long experience.
To our knowledge, this is the first report of the AI system diagnosing EC invasion depth from video images. Although a previous report has shown an AI system effectively diagnosing cancer invasion depth based on still images [23], studies that use still images may have some bias because they usually select the best images from the best areas. In contrast, video images may include poor-quality frames that more accurately reflect real-time clinical images. As our AI showed good performance in diagnosing video images, we could confirm its robustness in diagnosing cancer invasion depth.
In this study, we developed an AI system to differentiate between EP-SM1 and SM2-3 cancers, because of the different treatment strategies for these two categories of cancers. However, we can also develop the AI system to differentiate M and SM cancers using the same dataset, simply by changing the training algorithm.
Endoscopic diagnosis of cancer invasion depth requires real-time endoscopic techniques and sufficient expertise to evaluate various endoscopic findings of EC, such as protrusion, hardness, and microvascular changes. Unsurprisingly, the diagnostic performance of experienced endoscopists with longer experience is reportedly better than that of those with less experience [23]. In the present study, the diagnostic accuracy of the AI system was comparable to that of experts. Notably, for the ME videos, the accuracy of the AI system exceeded the 95% CI of experts. Considering that our AI system had such good performance, this system could potentially provide valuable support for endoscopists.
Our AI system showed better performance with ME videos than non-ME videos. A possible explanation for this is that ME uses more objective endoscopic findings, such as loop vessels, non-loop vessels, and dilated vessels, which contributed to the education of our AI system [26]. Another possible explanation is that peristalsis of the esophagus in non-ME videos might cause misdiagnosis by interpreting protrusion during peristalsis as a sign of SM2-3 invasion. When this system is used in clinical practice, we may have to consider limiting the condition of AI diagnosis, i.e. without peristalsis or full-open lumen, to improve its accuracy.
Sensitivities for SM2-3 cancer were higher with the diagnoses by the AI system than the experts, while there was no tendency in specificities. Sensitivity and specificity show what proportion of SM2-3 and EP-SM1 cancers were correctly diagnosed. One possible explanation for lower sensitivities by experts is that they may sometimes hesitate to diagnose cancers as SM2-3. Because misdiagnosing EP-SM1 cancer as SM2-3 cancer may lead patients to receive unnecessary and invasive treatment.
There are some limitations in this study. First, the validation video set was comprised of short videos of 4–12 s, which do not represent real clinical practice. In real-time diagnosis, poor-quality images resulting from poor insufflation, bleeding, halation, blurring, bad focus, or mucus may be included. However, once detected, cancer invasion depth can be evaluated under selected good conditions. Accordingly, extracting short video images for validation may not cause much bias to the results. Another limitation is its retrospective style. This system should be evaluated in real-time situation in the near future. Furthermore, there were few cases of SM1 as the composition of the validation dataset. This is because surgically resected specimens were excluded. The evaluation of pathology is more accurate in endoscopically resected specimens than in surgically resected specimens, especially for SM1. Therefore, it could not be included as appropriate cases.
In conclusion, our study showed the effectiveness of an AI system in evaluating cancer invasion depth using validation videos. Most diagnostic parameters of the AI system were better than those of the experts. Based on these favorable results, our AI system could provide valuable support for many endoscopists.
References
Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–53.
Tachimori Y, Ozawa S, Numasaki H, et al. Comprehensive registry of esophageal cancer in Japan, 2012. Esophagus. 2019;16:221–45.
Kodama M, Kakegawa T. Treatment of superficial cancer of the esophagus: a summary of responses to a questionnaire on superficial cancer of the esophagus in Japan. Surgery. 1998;123:432–9.
Igaki H, Kato H, Tachimori Y, et al. Clinicopathologic characteristics and survival of patients with clinical Stage I squamous cell carcinomas of the thoracic esophagus treated with three-field lymph node dissection. Eur J Cardiothorac Surg. 2001;20:1089–94.
Yamamoto S, Ishihara R, Motoori M, et al. Comparison between definitive chemoradiotherapy and esophagectomy in patients with clinical stage I esophageal squamous cell carcinoma. Am J Gastroenterol. 2011;106:1048–54.
Birkmeyer JD, Siewers AE, Finlayson EV, et al. Hospital volume and surgical mortality in the United States. N Engl J Med. 2002;346:1128–37.
Chang AC, Ji H, Birkmeyer NJ, et al. Outcomes after transhiatal and transthoracic esophagectomy for cancer. Ann Thorac Surg. 2008;85:424–9.
Swisher SG, Deford L, Merriman KW, et al. Effect of operative volume on morbidity, mortality, and hospital use after esophagectomy for cancer. J Thorac Cardiovasc Surg. 2000;119:1126–32.
Begg CB, Cramer LD, Hoskins WJ, et al. Impact of hospital volume on operative mortality for major cancer surgery. JAMA. 1998;280:1747–51.
Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598–606.
Shimizu Y, Tsukagoshi H, Fujita M, et al. Long-term outcome after endoscopic mucosal resection in patients with esophageal squamous cell carcinoma invading the muscularis mucosae or deeper. Gastrointest Endosc. 2002;56:387–90.
Katada C, Muto M, Momma K, et al. Clinical outcome after endoscopic mucosal resection for esophageal squamous cell carcinoma invading the muscularis mucosae–a multicenter retrospective cohort study. Endoscopy. 2007;39:779–83.
Yamashina T, Ishihara R, Nagai K, et al. Long-term outcome and metastatic risk after endoscopic resection of superficial esophageal squamous cell carcinoma. Am J Gastroenterol. 2013;108:544–51.
Akutsu Y, Uesato M, Shuto K, et al. The overall prevalence of metastasis in T1 esophageal squamous cell carcinoma: a retrospective analysis of 295 patients. Ann Surg. 2013;257:1032–8.
Holscher AH, Bollschweiler E, Schroder W, et al. Prognostic impact of upper, middle, and lower third mucosal or submucosal infiltration in early esophageal cancer. Ann Surg. 2011;254:802–7 (discussion 7-8).
Kitagawa Y, Uno T, Oyama T, et al. Esophageal cancer practice guidelines 2017 edited by the Japan Esophageal Society: part 1. Esophagus. 2019;16:1–24.
Pimentel-Nunes P, Dinis-Ribeiro M, Ponchon T, et al. Endoscopic submucosal dissection: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy. 2015;47:829–54.
Yoshida M, Momma K, Hanashi T. Endoscopic evaluation of depth of cancer invasion in cases with superficial esophageal cancer. Stomach Intestine. 2001;36:295–306.
Hirasawa T, Aoyama K, Tanimoto T, et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653–60.
Horie Y, Yoshio T, Aoyama K, et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25–322.
Luo H, Xu G, Li C, et al. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20:1645–54.
Ohmori M, Ishihara R, Aoyama K, et al. Endoscopic detection and differentiation of esophageal lesions using a deep neural network. Gastrointest Endosc. 2019;91(301–309):e1.
Nakagawa K, Ishihara R, Aoyama K, et al. Classification for invasion depth of esophageal squamous cell carcinoma using a deep neural network compared with experienced endoscopists. Gastrointest Endosc. 2019;90:407–14.
Japanese Classification of Esophageal Cancer, 11th edition: part I. Esophagus. 2017;14:1–36.
Japanese Classification of Esophageal Cancer, 11th edition: part II and III. Esophagus. 2017;14:37–65.
Oyama T, Inoue H, Arima M, et al. Prediction of the invasion depth of superficial squamous cell carcinoma based on microvessel morphology: magnifying endoscopic classification of the Japan Esophageal Society. Esophagus. 2017;14:105–12.
Acknowledgements
We thank Naoki Dan (Suita Municipal Hospital), Kengo Nagai (Suita Municipal Hospital), Yoshiki Onishi (Japan Community Health care Organization Osaka Hospital), Tomoyo Kanno (Japan Community Health care Organization Osaka Hospital), Satoshi Hiyama (Japan Community Health care Organization Osaka Hospital), Shoichiro Kawai (Osaka General Medical Center), Tadashi Kegasawa (Ikeda City Hospital), Yoshitaka Yamaguchi (Ikeda City Hospital), and Yasushi Matsumoto (Ikeda City Hospital) for acting as experienced endoscopists. We also thank Marla Brunker, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Study concept and design: YS, RI. Acquisition of data: YS, RI, AS, TI, KM, MM, KW, MK, HF. Analysis and interpretation of data: YS, RI, YK. Drafting of the manuscript: YS, RI. All authors have approved the final draft submitted.
Corresponding author
Ethics declarations
Conflict of interest
Tomohiro Tada is shareholder of AI Medical Service. All other authors declare that they have no conflict of interest.
Provenance and peer review
Not commissioned; externally peer reviewed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shimamoto, Y., Ishihara, R., Kato, Y. et al. Real-time assessment of video images for esophageal squamous cell carcinoma invasion depth using artificial intelligence. J Gastroenterol 55, 1037–1045 (2020). https://doi.org/10.1007/s00535-020-01716-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-020-01716-5