Abstract
In the paper, a novel image encryption algorithm based on DNA sequence operations and chaotic systems is proposed. The encryption architecture of permutation and diffusion is adopted. Firstly, 256-bit hash value of the plain image is gotten to calculate the initial values and system parameters of the 2D Logistic-adjusted-Sine map (2D-LASM) and a new 1D chaotic system; thus, the encryption scheme highly depends on the original image. Next, the chaotic sequences from 2D-LASM are used to produce the DNA encoding/decoding rule matrix, and the plain image is encoded into a DNA matrix according to it. Thirdly, DNA level row permutation and column permutation are performed on the DNA matrix of the original image, inter-DNA-plane permutation and intra-DNA-plane permutation can be attained simultaneously, and then, DNA XOR operation is performed on the permutated DNA matrix using a DNA key matrix, and the key matrix is produced by the combination of two 1D chaotic systems. Finally, after decoding the confused DNA matrix, the cipher image is obtained. Experimental results and security analyses demonstrate that the proposed scheme not only has good encryption effect, but also is secure enough to resist against the known attacks.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
With the fast development and extensive applications of multimedia technology and Internet, more and more digital images carrying all kinds of information are produced and transmitted over the network, and security of images becomes more and more important [1–3]. Chaotic systems have many soaring and eminent characteristics, such as highly sensitivity to initial conditions and control parameters, ergodic behavior, deterministic in nature, pseudo randomness, non-periodicity, boundedness, and topological transitivity, those are similar to the counterparts in cryptography, and many image encryption algorithms based on chaotic systems are introduced to protect the images by transforming a meaningful original image into an unrecognizable and noise-like cipher images [4–14]. However, some of them have been broken and found insecure from the modern cryptographical point of view [15–24]. For example, aiming at a cryptosystem based on a spatiotemporal chaos with a dynamic keystream generator, Rabei Bechikh et al. [23] found that it was not sufficiently secure against chosen plaintext attack, and Li et al. [24] had broken a novel image encryption scheme based on improved hyperchaotic sequences with only one known-plaintext attack. Therefore, new and secure cryptosystems should be introduced to ensure information security.
Due to the massive parallelism, huge storage, and ultra-low-power consumption of DNA molecules, DNA computing has entered into the field of cryptography [25–27]. A number of image encryption schemes combining chaos and DNA computing have been presented [27–40]. The kernel of these algorithms is DNA encoding/decoding and DNA computing, and it consists of some algebra operations and biological operations, such as the complementary rule of bases, DNA addition operation, DNA subtraction operation, and DNA XOR operation. A novel image encryption algorithm was presented in ref. [28]; Lorenz and Chen chaotic systems were used to give the chaotic sequences; and DNA elongation operation, truncation operation, deletion operation, and addition operation were employed. Guesmi et al. [29] introduced a novel chaos-based image encryption scheme using DNA sequence operation and SHA-2 hash function, three-dimensional Lorenz system was used to generate the chaotic sequences, and DNA XOR operation was for diffusion. Wang et al. [30] proposed a novel image encryption method based on DNA sequence operations and coupled map lattice, extended hamming distance was employed to produce the initial values of the coupled map lattice (CML) system, and DNA addition operation and DNA subtraction operation are utilized in DNA level diffusion process.
However, recent cryptanalysis results about DNA-based image encryption algorithms have shown that some algorithms have security flaws. For example, Liu et al. [31] gave a RGB image encryption algorithm using DNA encoding and 1D Logistic map, Ozkaynak et al. [32] broke it using chosen-plaintext attack and obtained the secret key by four chosen plain images, and subsequently, the security of this algorithm was reevaluated by Liu et al. [33], and it has two shortcomings: one is that the encryption scheme has no sensitivity with the plain images, and the other is that it cannot resist against the known-plaintext and chosen-plaintext attacks. Recently, Hermassi et al. [34] analyzed an image encryption algorithm based on DNA addition and chaotic maps in ref. [35] and found that firstly, it is non-invertible, and the receiver cannot recover the plain image even if he gets the secret key, and secondly, it may not withstand chosen-plaintext attack. In addition, the previous DNA-based encryption schemes have demonstrated that the DNA encoding/decoding rules of the plain image and key image are set and the same for all the pixels [27, 28, 36, 37], it has nothing to do with the original image, which means that there are the same encoding rules for different original images, and at many times, the rules are secret keys and varying from 1 to 8 [29–31, 38–40]. This will downgrade the ability of the algorithm to resist against brute force attacks and chosen-plaintext attack.
In order to overcome the previous shortcomings from the image encryption algorithms based on chaotic maps and DNA computing, we present a novel image encryption scheme in the paper. The contributions of the proposed encryption scheme are described as follows. Firstly, a 2D Logistic-adjusted-Sine map (2D-LASM) and a new 1D chaotic system are used in the paper; their initial values and system parameters are computed from the SHA 256 hash value of the plain image, so the scheme is highly sensitive to the original image. Furthermore, a DNA encoding rule matrix is obtained from the chaotic sequences generated by 2D-LASM, and it is used to encode each pixel in the plain image; that is to say, every pixel has its own encoding rule and the rule depends on the original image; therefore, our algorithm may effectively resist the known-plaintext, chosen-plaintext and statistical attacks. Besides, DNA level row permutation and column permutation scheme are adopted, inter-DNA-plane permutation and intra-DNA-plane permutation schemes can make the DNA base elements of the plain image move to the same and other DNA planes, this can improve the encryption effect, and chaotic sequences used in confusion process are gotten from DNA encoding process, which can make full use of chaotic sequences and save encryption time. Lastly, in the diffusion process, highly random chaotic sequences are generated from two new 1D chaotic systems, the key matrix is produced by the DNA encoding rule in many existing methods, whereas the DNA base elements of the key matrix are gotten according to the sequence values in our method, which may shorten the encryption time, and we get the cipher image from the DNA XOR operation of the key matrix and the permutated original image.
The remaining of the paper is organized as follows: Sect. 2 describes the preliminary works, including two chaotic systems and DNA sequence operations. The detailed encryption scheme is presented in Sect. 3. Section 4 discusses the advantages of the encryption algorithm and Section 5 gives simulation results, the security analyses have been discussed in Sect. 6, and conclusions are drawn in the last section.
2 Preliminary works
2.1 Chaotic systems
2.1.1 2D Logistic-adjusted-Sine map
Employing the 1D Logistic map to adjust the input of the Sine map, a new 2D chaotic map named 2D Logistic-adjusted-Sine map (2D-LASM) is proposed [41], and it is defined as
where μ is the system parameter, μ ∈ [0, 1], and x and y are state variables, x , y ∈ (0, 1).
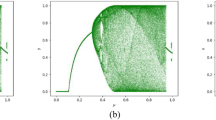
The bifurcation diagrams of the chaotic system are shown in Fig. 1, with the initial values x0 = 0.8 and y0 = 0.5. Compared to Logistic map and Sine map, 2D-LASM has a wider chaotic range and better ergodicity and unpredictability and is suitable for image encryption. From the figure, we can see that the system has good chaotic characteristics and the Lyapunov exponents are 0.8157 and 0.9787 with μ = 0.8.
2.1.2 1D chaotic map
A new 1D chaotic map [42] is used in the diffusion process, and it can be described by Eqs. (2) and (3):
where the system parameter r ∈ [1, 10] and the state variable z ∈ [−1, 1].
Its bifurcation diagram is shown in Fig. 2. From the figure, it can be known that the chaotic map has a dense set of periodic orbits, and its period is in the [−1, 1] interval with r > π/2.
In order to obtain highly random chaotic sequence, the combination of two 1D chaotic maps is employed, and it can be shown in the following equations:
Here, the system parameters r1 , r2 ∈ [1, 10], the initial values of the chaotic system\( {z}_0^1,{z}_0^2\in \left[-1,1\right] \), and the values produced by Eq. (6) are used to design the diffusion matrix in the diffusion process.
2.2 DNA sequence operations
2.2.1 DNA encoding and decoding rules
Each DNA sequence contains four nucleic acid bases, which are A (adenine), C (cytosine), G (guanine), and T (thymine), where A and T, G, and C are complementary pairs. Because 0 and 1 are complementary in the binary, so 00 and 11, 01, and 10 are also complementary. By using four bases A, C, G, and T to encode 00, 01, 10, and 11, there are 24 kinds of encoding rules. But there are only 8 kinds of them satisfying the Watson-Crick complementary rule [43], which are shown in Table 1. DNA decoding rules are the reverse operation of DNA encoding rules.
In this paper, we employ the DNA encoding rules to encode the plain image. For an 8-bit grayscale image, each pixel can be expressed as a DNA sequence whose length is 4. For example, when the pixel value is 174, its binary sequence is 10101110, and the DNA sequence is GGTG using the DNA encoding rule 1 to encode it, whereas using DNA encoding rule 1 to decode this DNA sequence, we can get a binary sequence 10101110, but if we utilize rule 2 to decode it, we obtain another binary sequence 01011101. Obviously, it is a simple way of encryption.
2.2.2 DNA XOR algebraic operation
With the rapid development of DNA computing, some biology operations and algebraic operation based on DNA sequence are presented, such as XOR operation. XOR operation of DNA sequences is manipulated according to traditional XOR in the binary. For there are eight kinds of DNA encoding rules, there exist eight types of DNA XOR rules, and one type of DNA XOR is listed in Table 2.
In the paper, the DNA XOR operation is used to fuse the plain image and the key image. For instance, there are two DNA sequences ACGT and CTAG; the XOR operation result of them is CGGC according to Table 2. From Table 2, it is obvious that every DNA base in every row or column is unique; therefore, the result of XOR operation is one and only.
3 The proposed encryption scheme
3.1 Generating the parameters and initial values of the chaotic systems
In the proposed encryption method, SHA 256 hash value of the plain image is employed to produce the system parameters and initial values of the chaotic systems. Even if there is only 1 bit between two plain images, their hash values will be completely different. So our encryption scheme highly depends on the original image. We firstly compute the SHA 256 hash value K of the original image and then divide the 256-bit secure key K into 8-bit blocks, which can be expressed as follows:
where in ki , j, i denotes the character number and j is the bit number in ki.
The intermediate parameters ti can be calculated as follows:
where li(i = 1, 2, 3, 4, 5, 6, 7) are the initial given parameters and values of the 2D-LASM and the 1D chaotic system, and they are μ0, \( {x}_0^{\prime } \), and \( {y}_0^{\prime } \) and \( {r}_1^{\prime } \), \( {r}_2^{\prime } \), \( {\left({z}_0^1\right)}^{\prime } \), and \( {\left({z}_0^2\right)}^{\prime } \), respectively. And ti is the processed value, and mod is the modular operator.
In view of the boundedness of parameters, the parameters and initial values of the chaotic systems can be given by
Here, μ, x0, and y0 are the parameter and initial values of the 2D-LASM, and r1, r2, \( {z}_0^1 \), and \( {z}_0^2 \) are the parameter and initial values of the 1D chaotic systems of Eqs. (4) and (5).
3.2 DNA encoding process of the plain image
Suppose the plain image P with the size of m × n, then convert every pixel of the image to its binary form, and get a matrix P1 of m × 8n. The detailed DNA encoding steps of the original image are as follows:
Step 1: Use μ, x0, and y0 produced in Sect. 3.1 to iterate the 2D-LASM 1000 times to avoid the transient effect.
Step 2: Continue to iterate the chaotic system s = max {m, 4n} times to obtain two chaotic sequences X and Y, then pick out the former m values from X to form a matrix X1 with the size of m × 1, next choose the former 4n values from Y to obtain a matrix Y1 of 1 × 4n, and finally get a matrix M of m × 4n by manipulating the matrix multiplying operation of X1 and Y1.
Step 3: Every element of the original image has different DNA encoding rule, considering that the elements of M fall in the ranges of 0 and 1, and then, the encoding rule matrix IT can be obtained as Eq. (16).
where M(i, j) and IT(i, j) denote the elements of M and IT located at the ith row and jth column, respectively, and 1 ≤ i ≤ m, 1 ≤ j ≤ 4n.
Step 4: Group every two elements of matrix P1 from left to right and from top to bottom, then encode every element according to the corresponding DNA rule number of matrix IT, and finally obtain the encoded DNA matrix P2(m × 4n).
From the DNA encoding steps of the original image, we can see that the dynamical DNA encoding process is achieved and it is highly sensitive to the plain image. Encoding rule matrix is generated from the chaotic sequences, different original image has different encoding rule matrix, and every pixel has its own encoding rule. Therefore, the proposed scheme is highly sensitive with the plain image.
3.3 DNA level permutation
The DNA level permutation process is composed of row permutation and column permutation. The detailed steps are as follows:
Step 1: In order to save the encryption time and fully utilize the chaotic sequences, the sequences X and Y generated in Sect. 3.2 are used, X = {x1, x2, ⋯ , xs}, Y = {y1, y2, ⋯ , ys}.
Step 2: Two matrices X′ and Y′ are gotten through modifying the elements of the X and Y according to the following equation:
And here, xi and yi are the corresponding elements of X and Y, and after finishing Step 2, we may obtain two sequences, and they are \( {X}^{\prime }=\left\{{x}_1^{\prime },{x}_2^{\prime },\cdots, {x}_s^{\prime}\right\} \), \( {Y}^{\prime }=\left\{{y}_1^{\prime },{y}_2^{\prime },\cdots, {y}_s^{\prime}\right\} \).
Step 3: Manipulate the row permutation to the ith (1 ≤ i ≤ m) row of the encoded DNA matrix P2(m × 4n) gotten from Sect. 3.2 by the following rules:
-
Case 1:
Perform left cyclic shift on the ith row \( {x}_i^{\prime } \) times with i % 2 ≠ 0.
-
Case 2:
Implement right cyclic shift on the ith row \( {x}_i^{\prime } \) times with i % 2 = = 0.
After all rows have been permutated, a new matrix P3 is obtained.
Step 4: Implement column permutation to the jth (1 ≤ j ≤ 4n) column of the matrix P3 according to the following rules:
-
Case 1:
Perform up cyclic shift on the jth column \( {y}_j^{\prime } \) times with j % 2 ≠ 0.
-
Case 2:
Implement down cyclic shift on the jth column \( {y}_j^{\prime } \) times with j % 2 = = 0.
After every column has been permutated, the matrix P4 is obtained and the permutation process is finished.
3.4 DNA level diffusion
In this section, a key DNA matrix is firstly produced by the chaotic system, and then, it is used to diffuse the permutated DNA plain image and upgrade the encryption effect. The detailed steps can be described as follows:
Step 1: Compute the parameters r1, r2, \( {z}_0^1 \), and \( {z}_0^2 \) as described in Sect. 3.1, and next, iterate the 1D chaotic system shown in Eq. (4) with r1 and\( {z}_0^1 \) (1000 + 4mn) times, iterate the 1D chaotic system shown in Eq. (5) with r2 and \( {z}_0^2 \) (1000 + 4mn) times, discard the former 1000 values to avoid the transient effect, and obtain sequences Z1 and Z2.
Step 2: Sequence U is gotten by importing the corresponding elements of Z1 and Z2 into Eq. (5), and U = {u1, u2, ⋯ , u4mn} is obtained, where ui is its ith element, 1 ≤ i ≤ 4mn.
Step 3: In order to modify the elements of the plain image, we need a key matrix. Then, U is employed to generate the DNA key matrix Q by the following equation. Without the DNA encoding process, we can directly obtain the DNA base elements of the key matrix through Eq. (18), which can reduce the computation complexity of the algorithm and improve the encryption speed.
And here, Q = {q1, q2, ⋯ , q4mn}, qidenotes its ith elements, 1 ≤ i ≤ 4mn.
Next, transform the Q to a key matrix KK with the size of m × 4n.
Step 4: Manipulate DNA XOR operation to the permutated matrix P4 and key matrix KK through Eq. (19).
where C is the cipher DNA matrix and C(i, j) is its element located at ith row, jth column.
The decryption process of this part is to perform the inverse operation, and it is defined by
Subsequently, decode the matrix C according to the rule matrix IT that is obtained in Sect. 3.2; we may obtain the cipher image.
3.5 The complete encryption steps
The flowchart of our proposed encryption algorithm is illustrated in Fig. 3. The complete encryption steps are shown as follows.
-
Step 1:
Compute the external key K through the SHA 256 hash function of the plain image, and then, obtain the system parameters and initial values of the chaotic systems through Eqs. (7)–(15).
-
Step 2:
Implement DNA encoding process to the plain image P (m × n) as described in Sect. 3.2, and obtain the DNA matrix P2(m × 4n).
-
Step 3:
Perform DNA level permutation to matrix P2 as shown in Sect. 3.3, and get permutated matrix P4(m × 4n).
-
Step 4:
Manipulate DNA level diffusion to matrix P4 as illustrated in Sect. 3.4, and obtain the cipher DNA matrix C.
-
Step 5:
Decode matrix C through the rule matrix IT gotten in Sect. 3.2, and the cipher image is obtained.
The decryption process is the reverse operations of the encryption process.
4 Discussion
The proposed encryption scheme has some advantages as follows. Firstly, two chaotic systems, that is 2D-LASM and a new 1D chaotic system, are utilized; the system parameters and initial values are calculated by the SHA 256 hash value of the plain image and the given values. When the plain image has a little change, the 256-bit hash value is completely different. Even for the same original image, the parameters and the initial values are different, when we modify the given values, and then, a completely different cipher image is obtained. Therefore, the algorithm highly depends on the original image, and it may make known-plaintext attack invalid.
Secondly, in the DNA encoding process of the plain image, different from the past encoding scheme, where there is an encoding rule for all the pixels, a dynamical DNA encoding scheme is adopted in the paper. A new chaotic system named as 2D-LASM is used to produce the chaotic sequences; subsequently, an encoding rule matrix is obtained from them, and then, every pixel of the original image is encoded by the corresponding element in the rule matrix, which means that different pixels have different encoding rules and this may improve the ability of the proposed encryption scheme to withstand statistical attack.
Thirdly, in the confusion process, DNA level row permutation and column permutation are utilized, and DNA matrix elements of the original image can move across the DNA planes according to the chaotic sequences from the 2D-LASM. An image with the size of m × n has 8-bit planes, every 2 bit at the adjacent planes is encoded to DNA bases, and four DNA planes are gotten. When the chaotic sequence values are multiple of 4, the row permutation is to realize the move of the DNA base elements at their own DNA planes, and we call it as inter-DNA-plane permutation. Otherwise, the row permutation can make the DNA base elements move to other DNA planes, and we name it as intra-DNA-plane permutation. Therefore, the proposed DNA level row permutation can attain inter-DNA-plane permutation and intra-DNA-plane permutation at the same time. Besides, the subsequent column permutation can make the DNA base elements shift at their own planes and further upgrade the confusion effect.
Lastly, in the DNA level diffusion procedure, the combination of two new 1D chaotic systems is used to produce chaotic sequence; then, DNA base elements of the DNA key matrix may be easily gotten by the range of the chaotic sequence values. But in the previous studies, the DNA key matrix is obtained through encoding the key matrix using a certain DNA encoding rule. Compared with it, our method is easy to manipulate. In addition, DNA XOR operations of the key matrix and the permutated plain image matrix are employed to obtain the cipher image, and this can save much time and reduce the computational complexity of the presented algorithm. The diffusion process of the current plain image pixel is related to the key matrix and the previous cipher pixel, and so, the proposed algorithm has good diffusion effect.
The diffusion effect of the proposed algorithm in the encryption and decryption process can be plotted in Figs. 4 and 5. In these two figures, KK is the key matrix; P is the plain matrix; C is the cipher matrix; ⊕ denotes the DNA XOR operation; and KK(i), P(i), and C(i) are their corresponding ith element, 1 ≤ i ≤ 4mn. As can be seen from Fig. 4, when one current element of the plain image is modified, the current cipher element will change accordingly, and all the cipher elements after it will change; therefore, the confusion process has high sensitivity to the plain image. In Fig. 5, it is clear that the modification of the current cipher element will affect the current recovered element and all the recovered elements after it. So, the proposed confusion scheme has a good confusion effect, and our encryption algorithm has high security level.
5 Simulation results
We have employed Matlab 2014a to verify the encryption and decryption effect of the proposed algorithm in a personal computer with CPU 2.5 GHz and memory 4 GB, and the operating system is Microsoft Windows 7. The four different 512 × 512 Lena image, Baboon image, satellite image, and medical image are used as the plain images, and the simulation results are shown in Fig. 6. The parameters we used are as follows: μ0 = 0.5673, \( {x}_0^{\prime }=0.3791 \), \( {y}_0^{\prime }=0.7438 \), \( {r}_1^{\prime }=3.4586 \), \( {r}_2^{\prime }=6.1289 \), \( {\left({z}_0^1\right)}^{\prime }=0.1357 \), and \( {\left({z}_0^2\right)}^{\prime }=-0.4895 \). Figure 6a, d, g, j are the plain images, Fig. 6b, e, h, k are their corresponding cipher images, and the decrypted images are shown in Fig. 6c, f, i, l.
Simulation results. a Plain image of Lena. b Corresponding cipher image. c Decrypted image. d Plain image of Baboon. e Corresponding cipher image. f Decrypted image. g Plain image of city. h Corresponding cipher image. i Decrypted image. j Plain image of Brone. k Corresponding cipher image. l Decrypted image
From the figures, it is clear that the cipher images are similar to random noisy images, and no useful information can be found from them, which means that our algorithm can realize the encryption of many kinds of images, such as natural images, remote sensing images, and medical images. And thus, it has wide application fields. Besides, the decrypted images using the right decryption keys are the same with the plain images from the visual point. Thus, the proposed encryption methods can encrypt the original images and protect the private information effectively, and when the cipher images are transmitted to the authorized receivers, the right plain images can be easily recovered.
6 Security analyses
6.1 Histogram analysis
Histogram is employed to reflect the pixel distribution of an image. If the histogram of a cipher image is uniform, that is to say, that each gray level has equal probability, then the encryption scheme is more robust against statistical attack. The histogram analysis results by our algorithm are illustrated in Fig. 7. Figure 7a, c, e, g are the histograms of the plain images, and those of the corresponding cipher images are shown in Fig. 7b, d, f, h. It is clear that the gray value distribution is fairly uniform and significant from the original images. Hence, our algorithm can withstand statistical attack.
Furthermore, χ2 test and variances of histograms are used to evaluate the uniformity of the pixel-value distribution. The χ2 value and variances of histograms of an image with 256-Gy levels can be calculated as follows:
where ni is the occurrence frequency of gray level i, n/256 is the expected occurrence frequency of each gray level, and n is the number of all the pixels. Here, in Eq. (22), Z = {z1, z2, ⋯ , z256} is the vector of the histogram values, and zi and zj are the numbers of pixels which gray values are equal to i and j, respectively.
The quantitatively results for different images are listed in Table 3. When the significant level is 0.05, the corresponding χ2(0.05, 255) is 293.25 [44]. The lower the variance is, the higher the uniformity of the image is [45]. From the table, we can come to two conclusions: firstly, the χ2 results of the cipher images are less than 293.25, which means that the proposed algorithm has passed the χ2 test and it has enough high security level; secondly, the variances of the cipher images are lesser than those of the plain images, especially for Brone (512 × 512); the variance of the plain image is about 2.8 × 107, whereas that of the cipher image decreases to around 8.8 × 102, there has a large decrease, and these indicate that the histogram distributions of the cipher images are uniformly distributed, and our algorithm is highly secure.
6.2 Information entropy
6.2.1 Information entropy
Information entropy is an important parameter to describe the degree of disorder of a system. It has been applied in many fields, such as lossless data compression, statistical inference, cryptography, and machine learning. In general, the more uncertain or random the information source is, the more information entropy that it will contain. So, it is very useful for analyzing the randomness of an encryption algorithm.
Information entropy can be calculated by the following equation:
where p(mi) denotes the probability of symbol mi. For a random image with 256-Gy levels, the entropy should ideally be 8 [46]. If the entropy of the cipher image is less than 8, there is a possibility of predictability, and this is a threat to the algorithm security.
We test the information entropy of seven different images, and the results are listed in Table 4. Note that the results are very close to the theoretical value 8, which demonstrates that the cipher images are almost close to random sources and the information leakage in the encryption process is negligible. Compared with three encryption algorithms, it can be found that the results gotten from our proposed algorithm are the same with those in ref. [47, 48] and closer to the theoretical value of 8 than the algorithm in ref. [49], so our algorithm has a better property of information entropy.
6.2.2 Local Shannon entropy
Local Shannon entropy [50] is also used to test the local randomness of the plain image and the cipher image. The (k, TB)-local Shannon entropy may be defined using the following method:
-
Step 1:
Randomly choose non-overlapping image blocks S1, S2,…, Sk with TB pixels for a test image S with L intensity scales.
-
Step 2:
For all image blocks, calculate Shannon entropy H(Si) via Eq. (23).
-
Step 3:
Compute the mean of Shannon entropy over these k image blocks S1, S2,…, Sk as local Shannon entropy according to the following equation:
In the experiment, we choose k = 32 and TB = 1936 for the test images with L = 256, and the test results are shown in Table 5. The results indicate that the local Shannon entropies of the cipher images are greater than 7.90, and the cipher images generated by our encryption scheme have good local randomness.
6.3 Correlation analysis
We randomly choose 5000 pairs of pixels in horizontal, vertical, and diagonal directions from the plain image and cipher image and calculate the correlation coefficients according to the following equations:
where x and y denote gray level values of two adjacent pixels of the image, N is the total number of pixels chosen from the image, and E(x) and D(x) are the expectation and variance of variable x, respectively.
For the plain image Lena (512 × 512), we draw the distribution diagram of two horizontally, vertically, and diagonally adjacent pixels of the plain image and the cipher image in Fig. 8. From the figure, we can see that there is a strong correlation among adjacent pixels in the plain image, the correlation coefficients are close to 1, whereas the pixels of the cipher image are scattered over the complete plane and the strong correlations of adjacent pixels in the original image have been reduced by the proposed encryption scheme.
Correlation of two adjacent pixels of the plain image Lena (512 × 512) and its cipher image. a Horizontal direction in plain image. b Horizontal direction in cipher image. c Vertical direction in plain image. d Vertical direction in cipher image. e Diagonal direction in plain image. f Diagonal direction in cipher image
Next, the correlation coefficients of two horizontally adjacent pixels, vertically adjacent pixels, and diagonally adjacent pixels for seven different plain and cipher images have been given in Table 6. In Table 6, the correlation coefficients of the cipher images are less than 0.03 and much smaller than that of the original images, so the proposed algorithm successfully eliminates the correlation between adjacent pixels in the plain image, and it can withstand the statistical attack.
6.4 Differential attack analysis
Number of pixel change rate (NPCR) and unified average changing intensity (UACI) are utilized to evaluate the resisting differential attack performance of the proposed encryption algorithm. NPCR stands for the number of pixels change rate while one pixel of plain image is modified. UACI denotes the average intensity of differences between the original image and cipher image.
For two gray cipher images, the NPCR and UACI can be obtained by the following:
where D(i, j) is defined as
where W and H denote the width and height of the image and C1 and C2 are, respectively, the cipher images before and after one pixel of the original image is changed. The expected NPCR and UACI values for a 256-Gy scale image are 99.6094 and 33.4635% [51], respectively. The closer the NPCR and UACI are, the more effective the cryptosystem in resisting differential attack is.
Seven different images are used as the test images, we get the other images by randomly changing one pixel, and the results are shown in Table 7. From Table 7, it is obvious that high efficiency may be obtained by having NPCR >99.5% and UACI >33.4% through changing one pixel value in the plain images, and the proposed scheme is secure enough to effectively resist against differential attack.
6.5 Known-plaintext and chosen-plaintext attack analyses
There are four classical types of attacks, ciphertext-only attack, known-plaintext attack, chosen-plaintext attack, and chosen-ciphertext attack. Among them, known-plaintext and chosen-plaintext attacks are the more powerful attacks, and if a cryptosystem can resist these two attacks, it can withstand other attacks.
In the paper, we employ 2D-LASM and a new 1D chaotic system, and their initial values and system parameters are computed by the SHA 256 hash value of the plain image. 2D-LASM is used in the DNA encoding steps and permutation process of the plain image, and 1D chaotic system is employed in the confusion process; thus, our encryption algorithm depends on the plain image, and it can resist known-plaintext and chosen-plaintext attacks.
In real attack atmosphere, the opponent usually utilizes all black or white images as special original images to attack the encryption algorithms, for the special images can make the permutation process invalid, and they may get the secret keys and make the algorithm invalid. We have made simulation experiments on all white and black, and the results are shown in Fig. 9 and Table 8. As can be seen from the figure and table, the cipher images are something like noise, no useful information can be found from them, the histogram distributions are fairly uniform, the entropy values of the cipher images are closer to 8, correlation coefficients of the cipher images are less than 0.08, and all these results mean that the proposed algorithm can effectively encrypt all white and black images and withstand known-plaintext and chosen-plaintext attacks effectively.
6.6 Key space
A good image encryption algorithm has a large key space to make brute-force attack ineffective. In the proposed scheme, the keys are the initial given parameters and values of the 2D-LASM and the 1D chaotic system, and they are μ0, \( {x}_0^{\prime } \), and \( {y}_0^{\prime } \) and \( {r}_1^{\prime } \), \( {r}_2^{\prime } \), \( {\left({z}_0^1\right)}^{\prime } \), and \( {\left({z}_0^2\right)}^{\prime } \). If the computational precision of the computer is about 10−14, the key space of the initial given parameters is 1098, which is larger than 2100 [52], so the proposed method has a sufficiently large key space to resist the brute-force attacks.
6.7 Key sensitivity
Key sensitivity includes encryption key (EK) sensitivity and decryption key (DK) sensitivity. For a good encryption scheme, in the encryption process, when the EK has a trivial change, the corresponding encrypted image should have a complete difference, and the recovered image is different from the plain image with slightly changed DK.
In the paper, the secret keys consist of μ0, \( {x}_0^{\prime } \), \( {y}_0^{\prime } \), \( {r}_1^{\prime } \), \( {r}_2^{\prime } \), \( {\left({z}_0^1\right)}^{\prime } \), and \( {\left({z}_0^2\right)}^{\prime } \). The 512 × 512 Lena image (shown in Fig. 6a) is used as the test image, the cipher image is Fig. 6b with the right EK, and the recovered image is Fig. 6c with the right DK. Next, we test the key sensitivity in the encryption and decryption process.
6.7.1 Encryption process
First, we modify one of μ0, \( {x}_0^{\prime } \), \( {y}_0^{\prime } \), \( {r}_1^{\prime } \), \( {r}_2^{\prime } \), \( {\left({z}_0^1\right)}^{\prime } \), and \( {\left({z}_0^2\right)}^{\prime } \) with 10−14 and others are the same, and the corresponding cipher images and their differences with the Fig. 6b are illustrated in Fig. 10 and Table 9. From the figure and the table, it is clear that when the EK has a trivial change, the encrypted images have a complete change, and more than 99% pixels are modified compared with Fig. 6b, which means that the encryption process is quite sensitive to the EK.
Key sensitivity test results. a Cipher image when μ0 is changed. b Differential image between a and Fig. 6b. c Cipher image when \( {x}_0^{\prime } \) is changed. d Differential image between c and Fig. 6b. e Cipher image when \( {y}_0^{\prime } \) is changed. f Differential image between e and Fig. 6b. g Cipher image when \( {r}_1^{\prime } \) is changed. h Differential image between g and Fig. 6b. i Cipher image when \( {r}_2^{\prime } \) is changed. j Differential image between i and Fig. 6b. k Cipher image when \( {\left({z}_0^1\right)}^{\prime } \) is changed. l Differential image between k and Fig. 6b. m Cipher image when \( {\left({z}_0^2\right)}^{\prime } \) is changed. n Differential image between m and Fig. 6b
6.7.2 Decryption process
In this section, we use one of μ0, \( {x}_0^{\prime } \), \( {y}_0^{\prime } \), \( {r}_1^{\prime } \), and \( {r}_2^{\prime } \) with 10−14 change and others are the same to decrypt the cipher image shown in Fig. 6b, the recovered images are illustrated in Fig. 11, the images are something like noise, we cannot find any useful information from them, and the differences with the plain image(Fig. 6a) are more than 99% listed in Table 10. From the previous key sensitivity results, we can watch that the decryption process is also highly sensitive to the DK.
Based on the key sensitivity analyses in the encryption process and decryption process, we can conclude that the proposed encryption algorithm is quite sensitive to the secret key and may resist the differential attack.
6.8 Comparison with existing schemes
In this section, we firstly compare the proposed algorithm with several existing chaos-based encryption scheme and give some quantitative results. Lena (512 × 512) is used as the test image; we encrypt it using different algorithms and compute the histograms and the information entropies and the correlation coefficients of two adjacent pixels in the cipher images. The comparison results are listed in Table 11. It can be seen from the table that our algorithm is best than those in refs. [53] and [54] as for histogram and information entropy and has the competitive performance compared with the encryption schemes in refs. [47–49, 55, 56].
Next, we compare the security performance of the proposed algorithm with two encryption schemes based on DNA computing and chaotic systems. Liu et al. [31] presented an image encryption algorithm, and DNA algebraic operations, DNA binary encoding rules, and chaotic sequences from one-dimensional Logistic map are utilized in permutation-diffusion encryption architecture. Wang et al. [30] introduced a novel image encryption scheme based on DNA sequence operations and CML, and diffusion-permutation-diffusion encryption process was adopted; firstly, bitwise exclusive OR operation was performed on the plain image and chaotic sequences generated from the CML, then the diffused images are transformed to a DNA matrix by a kind of DNA encoding rule, next the DNA matrix was permutated and diffused, and finally, the cipher image was obtained using a kind of DNA decoding rule.
We compare our algorithm with them from the view of security, and the results are listed in Table 12. From the table, it is clear that the proposed algorithm has the largest key space, high sensitivity to the secret key and plain image, dependence on the plain image, uniform histogram, near-zero correlation, and close to 8 information entropy, and the DNA encoding and decoding rules are dependent on the original image. However, the algorithm in ref. [31] is not sensitive to the secret key and plain image, the encryption process has no relationship with the original image, DNA encoding/decoding rules have no dependence on the plain image, and it has been broken with only one known plaintext [33]. As for the encryption scheme in ref. [30], DNA encoding/decoding rules are regarded as a secret key, varying from 1 to 8, and not dependent on the plain image, and it can be attacked by known-plaintext and chosen-plaintext attacks. As a consequence, the proposed algorithm has the highest security level.
6.9 Computational and complexity analyses
We analyze the computation and complexity of our encryption scheme and compare it with ref. [30] from two points: one is the time-consuming part of generating chaotic sequences, and the other is the DNA part. In the proposed algorithm, we adopt the architecture of permutation and confusion. In the permutation process, 2D-LASM is utilized to produce the chaotic sequences for row permutation and column permutation, and in the diffusion steps, two 1D chaotic systems are used to generate the chaotic sequences. In ref. [30], there are three encryption steps, that is diffusion, permutation, and diffusion, and in order to overcome the shortcoming that the chaotic dynamics degrades rapidly, the complex CML spatiotemporal chaotic system is employed to obtain the chaotic sequences, whereas many low dimensional chaotic systems are adopted in our algorithm, and the low dimensional system is easy to implement and can run faster.
As for the DNA part, there are two differences between the two schemes in computational complexity. One is that the algorithm in ref. [30] uses the DNA encoding and decoding rules as two secret keys; however, we obtain the encoding rule matrix from the chaotic sequences in the paper, the generation of the chaotic sequences needs time, the same chaotic sequences are used in the permutation step, and thus, there is no additional time-consuming from the view of the whole encryption process. The other is that the scheme in ref. [30] uses the DNA addition operation and subtraction operation, and the proposed algorithm adopts DNA XOR operation, which is more efficient.
7 Conclusions
In the paper, a novel image encryption scheme is introduced using DNA sequence operations and chaotic systems. Permutation and diffusion are adopted. Two simple chaotic systems, 2D-LASM and a new 1D chaotic system, are used to generate the chaotic sequences, and their initial values and system parameters are calculated by the 256-bit hash value of the plain image. The DNA encoding/decoding rule matrix, the chaotic sequences employed in the permutation, and diffusion stages are both dependent on the original image; we can attain “one plain image, one secret key”; therefore, our algorithm can effectively resist the known-plaintext and chosen-plaintext attacks. Besides, different from the existing encryption algorithm based on DNA computing, the DNA encoding/decoding rule for every pixel in the original image is different and varies with the plain image in the paper, and this may upgrade the attacking ability to statistical attack.
Experiment results of four different types of original images illustrate that the proposed algorithm has good encryption results and may be applied for encrypting all kinds of images, such as medical images, remote sensing image, and others. In addition, we have made the security analyses on histogram, information entropy, correlation, differential attack, known-plaintext and chosen-plaintext attacks, key space, key sensitivity, comparison with existing schemes, and computational and complexity; the results show that our encryption scheme has large key space, high key sensitivity, and high security level to resist against common attacks. So the proposed algorithm can be applied in the image secure communication. But at present, in the proposed encryption algorithm, DNA computing operations may cost much time. In the future, the biocomputers may be designed and utilized, and biocomputers use systems of biologically derived molecules—such as DNA and proteins—for computational calculations involving storing, retrieving, and processing data. And then, the DNA computing operations may be done by biological molecules, and the proposed algorithm may be performed easily and efficiently.
References
Zhou YC, Hua ZY, Pun CM, Philip Chen CL (2015) Cascade chaotic system with applications. IEEE T Cybernetics 45(9):2001–2012
Chen JX, Zhu ZL, Fu C, Zhang LB, Zhang YS (2015) An efficient image encryption scheme using lookup table-based confusion and diffusion. Nonlinear Dyn 81:1151–1166
Wen W Y, Zhang Y S, Fang Y M, Fang Z J 2016. Image salient regions encryption for generating visually meaningful ciphertext image. Neural Comput & Applic; Doi: 10.1007/s00521–016–2490-6
Wu XJ, Wang DW, Kurths J, Kan HB (2016) A novel lossless color image encryption scheme using 2D DWT and 6D hyperchaotic system. Inf Sci 349-350:137–153
Tong XJ, Wang Z, Zhang M, Liu Y, Xu H, Ma J (2015) An image encryption algorithm based on the perturbed high-dimensional chaotic map. Nonlinear Dyn 80:1493–1508
Assad SEI, Farajallah M (2016) A new chaos-based image encryption system. Signal Process: Image 41:144–157
Zhang YQ, Wang XY (2015) A new image encryption algorithm based on non-adjacent coupled map lattices. Appl Soft Comput 26:10–20
Diaconu A-V (2015) Circular inter-intra pixels bit-level permutation and chaos-based image encryption. Inf Sci 3:1–14
Hsiao H-I, Lee J (2015) Color image encryption using chaotic nonlinear adaptive filter. Signal Process 117:281–309
Xu L, Li Z, Li J, Hua W (2016) A novel bit-level image encryption algorithm based on chaotic maps. Opt Laser Eng 78:17–25
Zhou NR, Pan SM, Chen S, Zhou ZH (2016) Image compression-encryption based on hyper-chaotic system and 2D compressive sensing. Opt Laser Technol 82:121–133
Ye GD, Huang XL (2016) A secure image encryption algorithm based on chaotic maps and SHA-3. Secur Commun Netw 9:2015–2023
Chai XL, Gan ZH, Chen YR, Zhang YS (2017) A visually secure image encryption scheme based on compressive sensing. Signal Process 134:35–51
Chai XL, Gan ZH, Yang K, Chen YR, Liu XX (2017) An image encryption algorithm based on the memristive hyperchaotic system, cellular automata and DNA sequence operations. Signal Process: Image 52:6–19
Özkaynak F, Yavuz S (2014) Analysis and improvement of a novel image fusion encryption algorithm based on DNA sequence operation and hyper-chaotic system. Nonlinear Dyn 78(2):1311–1320
Li CQ, Lo KT (2011) Optimal quantitative cryptanalysis of permutation-only multimedia ciphers against plaintext attacks. Signal Process 91(4):949–954
Solak E, Cokal C, Yildiz OT, Biyikiglu T (2010) Cryptanalysis of Fridrich’s chaotic image encryption. Int J Bifurc Chaos 20:1405–1413
Zhang Y, Li C, Li Q, Zhang D, Shu S (2012) Breaking a chaotic image encryption algorithm based on perceptron model. Nonlinear Dyn 69:1091–1096
Li C, Zhang Y, Ou R, Wong KW (2012) Breaking a novel colour image encryption algorithm based on chaos. Nonlinear Dyn 70:2383–2388
Ozkaynak F, Ozer AB (2016) Cryptanalysis of a new image encryption algorithm based on chaos. Optik 127:5190–5192
Li CQ (2016) Cracking a hierarchical chaotic image encryption algorithm based on permutation. Signal Process 118:203–210
Eric Xie Y, Li CQ, Yu SM, Lü JH (2017) On the cryptanalysis of Fridrich’s chaotic image encryption scheme. Signal Process 132:150–154
Bechikh R, Hermassi H, Abd EI-Latif AA, Rhouma R, Belghith S (2015) Breaking an image encryption scheme based on spatiotemporal chaotic system. Signal Process: Image 39:151–158
Li CQ, Liu YS, Xie T, Michael Chen ZQ (2013) Breaking a novel image encryption scheme based on improved hyperchaotic sequences. Nonlinear Dyn 73:2083–2089
Celland CT, Risca V, Bancroft C (1999) Hiding messages in DNA microdots. Nature 399:533–534
Enayatifar R, Sadaei HJ, Abdullah AH, Lee M, Isnin IF (2015) A novel chaotic based image encryption using a hybrid model of deoxyribonucleic acid and cellular automata. Opt Laser Eng 71:33–41
Babaei M (2013) A novel text and image encryption method based on chaos theory and DNA computing. Nat Comput 12:101–107
Zhang Q, Guo L, Wei X (2013) A novel image fusion encryption algorithm based on DNA sequence operation and hyper-chaotic system. Optik 124:3596–3600
Guesmi R, Farah MAB, Kachouri A, Samet M (2016) A novel chaos-based image encryption using DNA sequence operation and secure hash algorithm SHA-2. Nonlinear Dyn 83:1123–1136
Wang XY, Zhang YQ, Bao XM (2015) A novel chaotic image encryption scheme using DNA sequence operations. Opt Lasers Eng 73:53–61
Liu L, Zhang Q, Wei X (2012) A RGB image encryption algorithm based on DNA encoding and chaos map. Comput Electr Eng 38:1240–1248
Ozkaynak F, Ozer A, Yavuz S 2013. Security analysis of an image encryption algorithm based on chaos and DNA encoding. In: Signal processing and communications applications conference (SIU), pp.1–4
Liu Y, Tang J, Xie T (2014) Cryptanalyzing a RGB image encryption algorithm based on DNA encoding and chaos map. Opt Lasers Eng 60:111–115
Hermassi H, Belazi A, Rhouma R, Belghith SM (2014) Security analysis of an image encryption algorithm based on a DNA addition combining with chaotic maps. Multimed Tools Appl 72:2211–2224
Zhang Q, Guo L, Wei X 2010. Image encryption using DNA addition combining with chaotic maps. Math Comput Model. (11–12): 2028–2035
Huang X, Ye G (2014) An image encryption algorithm based on hyper-chaos and DNA sequence. Multimed Tools Appl 72:57–70
Zhang Q, Guo L, Wei XP (2010) Image encryption using DNA addition combining with chaotic maps. Math Comput Model 52:2028–2035
Zhang YQ, Wang XY, Liu J, Chi ZL (2016) An image encryption scheme based on the MLNCML system using DNA sequences. Opt Lasers Eng 82:95–103
Zhang Q, Liu L, Wei X (2014) Improved algorithm for image encryption based on DNA encoding and multi-chaotic maps. AEU-Int J Electron C 68:186–192
Liu H, Wang X, Abdurahman K (2012) Image encryption using DNA complementary rule and chaotic maps. Appl Soft Comput 12:1457–1466
Hua ZY, Zhou YC (2016) Image encryption using 2D Logistic-adjusted-Sine map. Inf Sci 339:237–253
Dascalescu A-C, Boriga RE, Diaconu A-V (2013) Study of a new chaotic dynamical system and its usage in a novel pseudorandom bit generator. Math Probl Eng 2013:769108
Watson JD, Crick FHC (1953) A structure for deoxyribose nucleic acid. Nature 171(4356):737–738
Zhang XP, Zhao ZM, Wang JY (2014) Chaotic image encryption based on circular substitution box and key stream buffer. Signal Process: Image 29:902–913
Zhang YQ, Wang XY (2014) A symmetric image encryption algorithm based on mixed linear- nonlinear coupled map lattice. Inf Sci 273:329–351
Mirzaei O, Yaghoobi M, Irani H (2012) A new image encryption method: parallel sub-image encryption with hyper chaos. Nonlinear Dyn 67:557–566
Ye GD (2014) A block image encryption algorithm based on wave transmission and chaotic systems. Nonlinear Dyn 75:417–427
Wang XY, Xu DH (2014) A novel image encryption scheme based on Brownian motion and PWLCM chaotic system. Nonlinear Dyn 75(1–2):345–353
Zhou YC, Cao WJ, Philip CCL (2014) Image encryption using binary bitplane. Signal Process 100:197–207
Wu Y, Zhou YC, George S, Sos A, Noonan Joseph P, Premkumar N (2013) Local Shannon entropy measure with statistical tests for image randomness. Inf Sci 222:323–342
Zhu C (2012) A novel image encryption scheme based on improved hyper-chaotic sequences. Opt Commun 285(1):29–37
Àlvarez G, Li S (2006) Some basic cryptographic requirements for chaos-based cryptosystems. Int J Bifur Chaos 16(8):2129–2151
Fouda JAE, Effa JY, Sabat M (2014) Ali. A fast chaotic block cipher for image encryption. Commun Nonlinear Sci Numer Simul 19(3):578–588
Zhang X, Zhao Z (2014) Chaos-based image encryption with total shuffling and bidirectional diffusion. Nonlinear Dyn 75(1–2):319–330
Chai X L, Yang K, Gan Z H 2016. A new chaos-based image encryption algorithm with dynamic key selection mechanisms. Multimed Tools Appl doi: 10.1007/s11042–016-3585-x
Chai XL (2017) An image encryption algorithm based on bit level Brownian motion and new chaotic systems. Multimed Tools Appl 76:1159–1175
Acknowledgements
All the authors are deeply grateful to the editors for smooth and fast handling of the manuscript. The authors would also like to thank the anonymous referees for their valuable suggestions to improve the quality of this paper. This work is supported by the National Natural Science Foundation of China (Grant No. 41571417 and U1604145), Natural Science Foundation of the United States (Grant No. CNS-1253424 and ECCS-1202225), Science and Technology Foundation of Henan Province of China (Grant No. 152102210048), Foundation and Frontier Project of Henan Province of China (Grant No. 162300410196), China Postdoctoral Science Foundation (Grant No. 2016M602235), Natural Science Foundation of Educational Committee of Henan Province of China (Grant No. 14A413015), and the Research Foundation of Henan University (Grant No. xxjc20140006).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Chai, X., Gan, Z., Yuan, K. et al. A novel image encryption scheme based on DNA sequence operations and chaotic systems. Neural Comput & Applic 31, 219–237 (2019). https://doi.org/10.1007/s00521-017-2993-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-017-2993-9