Abstract
Genetic programming (GP) is a popular and powerful optimization algorithm that has a wide range of applications, such as time series prediction, classification, data mining, and knowledge discovery. Despite the great success it enjoyed, selecting the proper primitives from high-dimension primitive set for GP to construct solutions is still a time-consuming and challenging issue that limits the efficacy of GP in real-world applications. In this paper, we propose a multi-population GP framework with adaptively weighted primitives to address the above issues. In the proposed framework, the entire population consists of several sub-populations and each has a different vector of primitive weights to determine the probability of using the corresponding primitives in a sub-population. By adaptively adjusting the weights of the primitives and periodically sharing information between sub-populations, the proposed framework can efficiently identify important primitives to assist the search. Furthermore, based on the proposed framework and the graphics processing unit computing technique, a high-performance self-learning gene expression programming algorithm (HSL-GEP) is developed. The HSL-GEP is tested on fifteen problems, including four real-world problems. The experimental results have demonstrated that the proposed HSL-GEP outperforms several state-of-the-art GPs, in terms of both solution quality and search efficiency.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Genetic programming (GP) is a powerful population-based search algorithm that solves optimization problems by evolving computer programs (Koza and Poli 2005). Over the past decades, GP has undergone a rapid development. A number of enhanced GP variants have been proposed, such as grammatical evolution (GE) (O’Neill and Ryan 2001), gene expression programming (GEP) (Ferreira 2006), Cartesian genetic programming (CGP) (Miller and Thomson 2000), linear genetic programming (LGP) (Brameier and Banzhaf 2007), and semantic genetic programming (SGP) (Moraglio et al. 2012; Ffrancon and Schoenauer 2015). The applications of GP are also multiplying fast, including time series prediction, classification, scheduling optimization, and others (Zhou et al. 2003; Schmidt and Lipson 2009; Espejo et al. 2010; Zhong et al. 2017a, b).
Despite its great success, GP has encountered a challenging issue in real-world applications (i.e., how to select proper primitives such as terminals (e.g., variable x) and functions (e.g., \(+\) and \(\sin \)) to construct solutions efficiently for a given problem). Traditionally, the primitive set is defined in an ad hoc manner, relying heavily on experts’ domain knowledge and experience. In many real-world applications where little or even no domain knowledge is available, we thus have to design as many primitives as possible to increase the probability that high-quality solutions are involved in the corresponding search space. However, this leads to an exponential increase in the range of the search space, which turns out to slow down the search speed seriously and makes GP get trapped into local optima easily. In the literature, several methods have been proposed to tackle this problem by using feature selection techniques (Chen et al. 2016, 2017; Harvey and Todd 2015). Nevertheless, these methods only focus on the terminal selection. Designing a more effective mechanism to select both important terminals and functions is still a challenging research topic remained unexplored in GP community.
To address the above issues, this paper proposes a multi-population GP framework with adaptively weighted primitives. In the proposed framework, the whole population is divided into a number of sub-populations. Each sub-population is assigned with primitives (i.e., terminals and functions) having different weights. The sub-populations are evolved independently using their own important primitives (i.e., those with larger weights) to construct solutions. By periodically sharing promising individuals among sub-populations and adaptively adjusting the weights of primitives based on the statistic information of surviving individuals, the proposed framework can gradually identify both important terminals and functions during the evolution processes online. Furthermore, based on the proposed multi-population framework, a high-performance self-learning gene expression programming algorithm named HSL-GEP is developed. The developed HSL-GEP contains a new way to cooperate different computing resources (e.g., CPU and GPU), and it adopts a recently published GP variant, self-learning gene expression programming (SL-GEP Zhong et al. 2016) as the GP solver for each sub-population in the proposed framework. In summary, the major contributions of the paper are as follows:
- (1)
A fast multi-population GP framework with adaptively weighted primitives is proposed. Unlike the existing methods that only identify important terminals, the proposed framework is capable of identifying both important terminals and functions efficiently to improve the search efficiency.
- (2)
An efficient algorithm, HSL-GEP, is developed based on the proposed multi-population framework and GPU computing technique. Unlike existing methods that only consider homogeneous computation resources, the proposed HSL-GEP can fully utilize both the heterogeneous hardware computing resources (i.e., GPU and multi-core CPU) and the characteristics of the algorithm architecture to accelerate the search.
- (3)
Comprehensive experiments on both benchmark and real-world problems are conducted to validate the proposed algorithm and to facilitate the real-world applications of the proposed method.
The rest of the paper is organized as follows. The related works of feature selection in GPs and GPU-based GPs are introduced in Sect. 2. The proposed multi-population GP framework is introduced in Sect. 3. Section 4 introduces the details of HSL-GEP. Section 5 presents the experimental studies, and Sect. 6 draws the conclusion.
2 Related works
In this section, we review the related research works on: (1) improving GP using feature selection, and (2) improving GP using GPU parallel computing.
2.1 Improving GP using feature selection
Feature selection is a common technique to improve the performance of a learning algorithm (Xue et al. 2016; Zhai et al. 2014). Generally, the goal of feature selection is to select a subset of relevant features from the original redundant feature set, so that the selected features can effectively model the system. By removing redundant features, we can reduce the search space and training time, and enhance the generality of the obtained solutions.
Only several preliminary efforts have been made to apply feature selection methods to GP high-dimension regression problems in past few years (Chen et al. 2016, 2017), though various feature selection methods have been proposed on classification and clustering problems (Ahmad et al. 2015; Xue et al. 2013; Hancer et al. 2018; Gu et al. 2018; Neshatian and Zhang 2009; Sandin et al. 2012; Ahmed et al. 2013; Moore et al. 2013; Deng et al. 2019a). For example, Chen et al. (2016) assumed that the features appearing in the better individuals are more important. They calculated the frequency of distinct features in the top best individuals in each generation and selected those with higher frequency as the important features. To further improve the robustness and the efficiency of the feature selection, Chen et al. (2017) introduced the permutation importance metric in feature selection. In this method, all the distinct features in the best-of-run individuals are permutated into other features and these permutated individuals are re-evaluated. The difference in permutated fitness and the original fitness is regarded as the importance of the features. Besides, feature selection techniques have also been applied to improve GP to solve other real-world applications, such as job shop scheduling (Mei et al. 2016b; Riley et al. 2016; Mei et al. 2017) and oral bio-availability problem (Dick et al. 2015). The above methods have shown great potential for feature selection in improving GP. However, existing methods only focus on selecting terminals, which is not efficient enough, especially on cases where the function set is large and contains redundant primitives. In this paper, we propose a new framework that can efficiently select both terminal and function primitives.
2.2 Improving GP using GPU
Benefiting from the strong computing power, GPU has been adopted to develop various parallel GPs in the literature. The first programmable-GPU-based GP is perhaps proposed by Harding and Banzhaf (2007), where a SIMD interpreter (a GPU programming framework) is developed to evaluate the whole population of GP in parallel. Later, Harding, Banzhaf, and Langdon developed a series of parallel GPs (Langdon 2011; Harding and Banzhaf 2007; Banzhaf et al. 2008; Langdon 2010) based on the Compute Unified Device Architecture (CUDA/C++) and different GP variants. Though these works mainly focused on the parallelization of fitness evaluation, they have demonstrated the great potential of GPU in accelerating the search efficiency of GP.
Further, a number of efforts have been made to improve GPU-based GPs by properly utilizing the distinct characteristics (e.g., memory category and memory hierarchy) of GPU. From the view of memory category, Shao et al. (2012) decoded the chromosomes into post-order trees to make full use of the constant memory of GPU, while Cano and Ventura (2014) transferred the chromosomes into shared memory of GPU to utilize threads in GPU to decode the independent subtrees of the chromosomes. Besides, Chitty proposed two works (Chitty 2016a, b) to improve the performance of GPU-based GPs from the perspective of memory hierarchy. In Chitty (2016b), Chitty designed a preemptive parallel structure to schedule both the L1 cache and shared memory of GPU. In Chitty (2016a), Chitty further proposed a two-dimension stack to improve the data transfer efficiency between L1 cache and global memory of GPU. Both of these two works successfully accelerated the search efficacy. Besides, GPU-based GPs have also been applied to various real-world applications such as the concrete industry and the graph coloring problem (Gandomi et al. 2016; Chen et al. 2015; Rojas and Meza 2015).
Generally, in existing GPU-based GPs, the evolution process is divided into separated stages and GPU is utilized in certain stages such as fitness evaluation. Different stages are performed sequentially, which means the GPU and CPU have to wait for each other alternatively. Keep the above in mind, this paper proposes a hierarchical parallel GP framework which can keep GPU and CPU busy throughout the evolution process. When some individuals are evaluated in GPU threads, genetic operations of other individuals can be performed simultaneously in CPU threads. By this mean, the computing resources of multi-core CPU and GPU can be utilized in a more flexible and efficient manner.
3 Proposed environment-vector-based multi-population GP framework
3.1 The proposed framework
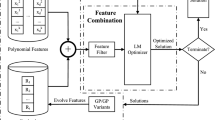
Since the multi-population is an effective way to solve the large-scale optimization problems (Deng et al. 2017, 2019b; Mei et al. 2016a; Antonio and Coello 2018; Yang et al. 2008), the idea of multi-population is also adopted in our proposed framework. In this section, the environment-vector-based multi-population GP framework (EMGP) is presented. In EMGP, the whole population is divided into multiple sub-populations, and the searching direction of each sub-population is guided by an environment vector (EV). The elements of EVs represent the selection probabilities of terminals and functions during solution construction in sub-populations. Each EV consists of two parts: \(E_\mathrm{t}\) and \(E_\mathrm{f}\). \(E_\mathrm{t}\) represents the selection probabilities of terminals, while \(E_\mathrm{f}\) represents the selection probabilities of functions. Accordingly, the lengths of \(E_\mathrm{t}\) and \(E_\mathrm{f}\) are equal to the size of the terminal set and function set, respectively. By assigning different EVs to different sub-populations, the sub-populations can focus on finding solutions in different sub-search spaces. To facilitate sharing information among sub-populations and to improve the search efficiency, three operations are proposed in EMGP: (1) sub-search space initialization, (2) sub-search space adaption, and (3) learning between sub-populations. The general relationship of the three operations and the pseudocode of EMGP are illustrated in Fig. 1 and Algorithm 1, respectively. Firstly, the solution space is divided into different sub-spaces. Each sub-space is defined by an EV and is assigned to a sub-population. After that, each sub-population focuses on evolving solutions in the corresponding sub-search space. During the evolution, EVs of sub-populations, on the one hand, are adjusted adaptively. On the other hand, they learn from each other to find out more suitable EVs. The details of the proposed EMGP are described as follows.

3.2 Sub-search space initialization
In this operation, the EV of each sub-population is initialized. Specifically, the terminal part of EV of the first sub-population (i.e., \(E_{\mathrm{t},0}\)) is set according to the entropy of each feature (Zhong and Cai 2015). Features with smaller entropy are more important. The top-10% most important features are set to 1, while the others are set to 0 in \(E_{t,0}\). Because the functions have no entropy measurement, all the elements in \(E_{\mathrm{f},0}\) are set to 1. To improve global searching ability, the other EVs are scattered evenly in the value ranges. To ensure that the sub-search spaces assigned to sub-populations are scattered evenly in the whole search space, the number of terminals (or functions) assigned to each sub-population, denoted as \(\nu _\mathrm{t}\) (or \(\nu _\mathrm{f}\)), is initialized by :
where \(N_s\) is the number of sub-populations, T represents the number of terminals, and F represents the number of functions. Every sub-population gets \(\nu _\mathrm{t}\) terminals and \(\nu _\mathrm{f}\) functions from the terminal set and function set randomly. These \(\nu _\mathrm{t}\) terminals and \(\nu _\mathrm{f}\) functions are called the “compulsory” primitives of sub-populations. All primitives are assigned to sub-populations properly so that each primitive is the “compulsory” terminal (or function) of at least one sub-population. The elements of \(E_\mathrm{t}\) and \(E_\mathrm{f}\) are initialized by:
where k is the index of the sub-population, \(u_\mathrm{t}\) and \(u_\mathrm{f}\) are the indexes of terminals and functions, respectively.
3.3 Sub-search space adaptation
This operation is used to evolve the sub-population independently and update the EV of each sub-population adaptively during the evolution. Specifically, a EV is updated based on the distributions of terminals and functions in the current sub-population in each generation. Since the surviving individuals have better fitness values, they are more likely to contain important terminals and functions that are used to construct promising solutions. Hence, a EV is updated by:
where l is the generation index, \(\pi \) is the number of generations between two migrations, \(P_{t,k}\) and \(P_{f,k}\) are the proportions of terminals and functions in the current sub-population, respectively. In each generation, the elements \(p_{t,k,u_\mathrm{t}}\) and \(p_{f,k,u_\mathrm{f}}\) of \(P_{t,k}\) and \(P_{f,k}\) are updated according to the scaled frequency of terminals and functions in the current sub-population as shown in (4).
\(n_a\) and \(n_b\) are the scaled frequency of terminals and functions computed by (5); c is a small constant value ensuring that all terminals and functions have a small probability to be selected. In (5), I is the set of indexes of individuals which contain terminal a (or function b); \({\bar{f}}\) and \(\delta \) are the average and the standard variation of fitness value in a certain sub-population, respectively; and \(f_i\) is the fitness value of the ith individual.
After each update, the EV is normalized by:
3.4 Learning between sub-populations
To efficiently utilize the searching information and determine a more suitable EV, two operations, namely the migration and Monte-Carlo-based EV generation, are performed every \(\pi \) generations during the evolution of each sub-population. The migration operation contains two phases: immigration and emigration. In the immigration phase, the best individual of each sub-population is substituted by the best-so-far individual \(J^*\) of the whole population. In the emigration phase, a random individual is selected from each sub-population and compared with another random individual in other sub-populations. The latter will be replaced by the former if the former is better.
To preserve the diversity of sub-populations, the cosine distance (7) is adopted to measure the similarity between \(EV_G\) and \(EV_k\) (denoted as \(\varOmega (G,k)\)) in immigration, while (8) is adopted to measure the similarity between \(EV_i\) and \(EV_j\) (denoted as \(\varOmega (i,j)\)) in emigration. G is the index of the sub-population of the best individual \(J^*\).
If \(\varOmega \) is larger than a random threshold \(\omega =rand(0,1)\), the migration between these two sub-populations will be rejected.
In the Monte-Carlo-based EV generation, a new EV is generated considering the existing EVs by Monte-Carlo methods (MC) and is assigned to a random sub-population. Since the effectiveness of an EV can be estimated according to the performance of the corresponding sub-population, better performance of the sub-population implies the higher probability of containing the valuable primitives the EV has. Following this mind, the importance of primitives is designed as the product of elements of EV and the corresponding rankings. Thus, the newly generated EV (labeled as \(EV'\)) can be calculated by:
where r is the rank of each sub-population in descending order based on the fitness of the best individual in the sub-population. Finally, the \(EV'\) is normalized by (6) and assigned to a random sub-population.
4 Proposed HSL-GEP
In this section, a hierarchical parallel genetic programming named high-performance self-learning gene expression programming (HSL-GEP) is developed based on the proposed EMGP framework mentioned above and the GPU-CUDA computing platform. In the following parts, the basic concepts of GPU are introduced at first. Then, the general architecture of HSL-GEP and its implementations are presented.
4.1 Basic concepts of GPU
A GPU contains three types of hierarchical layers: grid, block, and thread. Generally, a thread is a basic unit of GPU, and a common GPU can contain thousands of threads. A block contains a number of threads, and a grid contains a number of blocks. There are a number of streaming multiprocessors (SMs) on each chip of GPU, and one SM can run several blocks simultaneously. Similar to CPU, each SM has its own cache, which is shared by all the blocks in the SM. Each SM contains a limited number of registers. Thus, the more blocks there are, the less registers a block can occupy on the SM.
There are five common kinds of memory in GPU: cache, register, global memory, shared memory, and constant memory Cook (2012). The cache and register are the fastest memory in GPU, but their capacity is the smallest. The global memory, which can be read/written by both CPU and GPU, is the largest memory, but its bandwidth is the lowest in GPU. The shared memory is a segment of user-programmed cache in GPU. The shared memory can be read by multiple threads simultaneously, but it can only be written sequentially. The shared memory is commonly used for data reutilization and data sharing between threads. The constant memory is a virtual address of the global memory. It has no physical memory block, but it utilizes the cache and a broadcasting mechanism to speed up the data reading ratio of multi-thread.
4.2 The architecture of HSL-GEP
The architecture of HSL-GEP contains three major modules: main evolution module, independent evolution module, and GPU fitness evaluation module, as shown in Fig. 2. The main evolution module is responsible for the initialization of the whole algorithm and the outer evolution procedure, including scheduling CPU threads to perform independent evolution module, synchronization, and learning operation among sub-populations. The independent evolution module is responsible for evolving a sub-population using the five reproduction steps in EMGP (i.e., environment updating, mutation and crossover, chromosome decoding, fitness evaluation, and selection). A multi-core CPU is applied in the independent evolution module to pipeline the independent evolution of all sub-populations to shadow the latency time of CPU and GPU communication. The GPU fitness evaluation module is used to evaluate the fitness values of individuals in sub-populations through GPU. The model utilizes the threads in CPU (denoted as TC) and those in GPU (TG) to work cooperatively during the evolution. The pseudocode of HSL-GEP is illustrated in Algorithm 2, and the related symbols are listed in Table 1. Firstly, the model initializes the computational resources (e.g., CPU threads and GPU memory allocation), EVs, and the population. Then, the model performs EV updating and genetic operations to evolve sub-populations independently. If such independent evolution satisfies the “trigger” \(\pi \), the migration and the Monte-Carlo-based EV generation are performed. Finally, the evolution is terminated when the model meets the termination condition.

4.3 Main evolution module
The initialization of the main evolution module contains three parts: the whole population initialization, threads creation, and memory allocation in GPU. Before the population initialization, an ingredient type vector \(C_{k,j}\) and the EV need to be initialized at first.
The vector \(C_{k,j}=(c_{k,j,0},c_{k,j,1},\ldots ,c_{k,j,L-1})\) of each individual describes the type of all genes in the individual. The \(C_{k,j}\) is introduced because SL-GEP has different type requirements in different parts of the individual. For example, a gene belonging to head part can be either function or terminal, while a gene belonging to tail part can only be terminal. The \(C_{k,j}\) is initialized randomly based on the available gene types defined by the individual representation. The EVs of all sub-populations are also initialized by the method mentioned in Sect. 3.2. Since different sub-populations are controlled by different EVs, an EV-based assignment is developed to initialize individuals, as shown in Algorithm 3.

After the population initialization, threads for parallelization and the GPU memory for fitness evaluation are allocated. Specifically, five arrays for each GPU are allocated for symbolic regression problems. Arrays TI and TO are used to store training inputs and outputs, respectively, while array DEV, FIT, and CV are used to store the decoded individuals, the calculated fitness values, and the temporary data during calculation, respectively. TI is a two-dimension array whose size is \(MI \times MV\), where MI is the maximum number of input samples and MV is the dimension of input vectors. TO and FIT are two one-dimension arrays with MI elements and \(N_B\) elements, respectively. \(N_B\) is the number of GPU blocks which will be mentioned later. DEV is a two-dimension array storing the decoded individual, and its dimension is \(N_B \times L\), where L is the length of the individual. DEV is essentially the buffer between GPU and CPU which is introduced above. CV is a two-dimension array with a size of \(N_B \times MI\). CV mainly stores the temporary value during computation. All these five arrays are allocated to the global memory of GPU to apply to solving problems with large-scale data.
Following the initialization, the outer evolution procedure performs the following four steps iteratively: evoking independent evolution of sub-populations, synchronization barriers, learning operations between sub-populations mentioned in Sect. 3.4, and termination judgment, until the termination conditions are met.
4.4 Independent evolution module
The independent evolution module is performed by TCs to evolve sub-populations. One TC is responsible for one or multiple sub-populations. Specifically, each TC is responsible for at most \(\lceil \frac{N_s}{N_{TC}}\rceil \) sub-populations, where \(N_s\) is the number of sub-populations and \(N_{TC}\) is the number of TC. During the independent evolution of sub-populations, the EVs are updated according to (3) and (6), and the genetic operations like mutation and crossover are performed to generate new individuals.
The mutation and crossover in HSL-GEP are the same as SL-GEP. Specifically, the extended “DE/current-to-best/1” mutation proposed in SL-GEP is adopted to generate a mutant vector for each target vector (i.e., parent individual). The “DE/current-to-best/1” mutation can be expressed by:
where \(\xi \) is a scaling factor which is randomly set by:
Each gene in the target vector will mutate to a new value with certain probability. If a gene is to be mutated, its value is also set by the EV-based assignment (Algorithm 3). In the EV-based assignment, the route-wheel selection is utilized to determine \(C_{k,j}\) based on \(PT_k\). \(PT_k\) is the probability vector of ingredient types and is updated by
where \(m_a\) is the frequency of three ingredient types (i.e., terminals, functions, and ADFs). c is a small constant.
The crossover operation generates a trial vector \(U_{k,j}\) for each \(X_{k,j}\) by
where CR is a random number ranging from 0 and 1, and \(\kappa \) is a random integer ranging from 0 to \(M_s-1\).
After the genetic operations, the GPU fitness evaluation module is invoked to evaluate the fitness of individuals. The fitness evaluation model invokes the GPU asynchronously to reduce the waiting time of both CPU and GPU. Once the fitness of a newly generated trial vector is calculated, the selection operation is performed to choose the better one between each pair of trial vector and target vector by (14) to form the new population.
where \(f(\cdot )\) is the objective function. Without loss of generality, we consider the problem at hand as a minimization problem (e.g., to minimize the fitting error). Thus, the individual with smaller objective function value is regarded as the better one.
4.5 Fitness evaluation module
This module is used to evaluate the fitness values of individuals in sub-populations using GPU. The fitness of each newly generated individual is calculated in four steps, as shown in Fig. 3. Firstly, one row of DEV (a decoded individual) is read into a block cache. Then, TGs in this block calculate the outputs of different rows of TI based on DEV and store the outputs into CV. After that, TGs calculate the error between TO and different columns of CV, which are then stored back to CV. Finally, the errors of a row in CV are summed up and stored to FIT. This arrangement guarantees when one decoded individual is read into the cache of one block, TGs in this block can always hit the memory of the decoded individual. When TGs need to use the same decoded individual to calculate the output repetitively, the frequent memory hit cuts down the times of swapping between cache and global memory and reduces the data transportation time in GPU. After the outputs of an individual on all inputs are calculated, another unfinished individual will be loaded into the block to perform fitness evaluation until all individuals are calculated. Once all submitted individuals are evaluated, the FIT is copied back to CPU to set the fitness values of the submitted individuals. By this mean, multi-thread CPU can pipeline the evocation of fitness evaluation in GPU and make full use of both computing resources.
To reduce the data transmission times between CPU and GPU, a buffer is introduced in this module to store sub-populations ready for fitness evaluation. Once the buffer is full, or all sub-populations are checked and there is at least one sub-population in the buffer, the GPU will be evoked immediately to evaluate the fitness of individuals stored in the buffer.
5 Experimental studies
In this section, we test the proposed HSL-GEP on benchmark problems and real-world problems and analyze its results. First, the experimental settings, including benchmark problems, compared algorithms, and performance metrics, are described. Then, the results and discussions are presented.
5.1 Experimental settings
Table 2 lists fifteen symbolic regression problems (SRPs) for testing, where \(F_0\) to \(F_{10}\) are benchmark problems and the other four problems are real-world problems. The selected benchmark problems have been widely used for symbolic regression in the literature (McDermott et al. 2012; Vladislavleva et al. 2009; Keijzer 2003; Yao et al. 1999). They contain various function primitives (e.g., \(+\), −, \(\sin \), \(\ln \), \(\exp \), and so on) and have different difficulty levels for investigation. In addition, the selected real-world problems are also adopted in the papers of the compared methods (i.e., the SL-GEP (Zhong et al. 2016) and GPPI (Chen et al. 2017)) and obtained from various applications (i.e., \(F_{11}\) to \(F_{14}\) are obtained from the gas chromatography measurements of the composition of a distillation tower,Footnote 1 the Communities and Crime unnormalized dataset (CCUN),Footnote 2 and the Diffuse Large-B-Cell Lymphoma (DLBCL) dataset provided by Rosenwald et al. (2002), respectively). Especially, the data in \(F_{12}\), the Communities and Crime normalized dataset (CCN), is obtained by standardizing the data of \(F_{13}\) from the website by z-score. Besides, the non-predictive features in CCN and CCUN are removed and the number of murders in 1995 (the 130th feature) is used as the goal attribute. All of the benchmark problems and the real-world ones are useful to investigate the effectiveness of the proposed method. The data associated with each problem are divided into training data and testing data, and the 10-fold cross-validation method is adopted, except for the \(F_{14}\) whose size of training data and testing data has been specified in Rosenwald et al. (2002). Each fold of training and testing data is run 5 times independently (i.e., totally 50 independent runs for \(F_0\) to \(F_{13}\) and 20 independent runs for \(F_{14}\)). All the missing values in the data are substituted by the average value of the attribute.
We compare the proposed HSL-GEP with five other methods. The first one is the recently published SL-GEP (Zhong et al. 2016), which has been shown quite effective in solving SRPs. The second one is “SL-GEP+GPPI,” which integrates SL-GEP with a feature selection technique (named GPPI). GPPI is the state-of-the-art feature selection method proposed in Chen et al. (2017), to improve GPs on SRPs. In SL-GEP+GPPI, GPPI is firstly performed to identify the importance of different features (i.e., terminal primitives). After that, SL-GEP evolves solutions based on these feature importance. The third one is EM-SLGEP which directly uses the SL-GEP as the GP solver for each sub-population in EMGP framework. The main difference between SL-GEP and EM-SLGEP lies in the application of the proposed multi-population framework. The fourth competitor is “SL-GEP+2DG” which accelerates SL-GEP using a recently proposed GPU implementation (i.e., design a two-dimension stack for GPU computation and it is denoted as 2DG) (Chitty 2016a). The main difference between 2DG and the proposed hierarchical parallel framework is the utilization of computing resources. 2DG only focuses the utilization of GPU, but HSL-GEP considers CPU, GPU, and their cooperation. The fifth competitor is “EMSLGEP+2DG,” which adopts the GPU implementation in Chitty (2016a) to implement EM-SLGEP. The sixth method is the proposed HSL-GEP, which combines both the multi-population framework and the hierarchical parallel computing mechanism and adopts the SL-GEP as the basic GP solver for each sub-population.
The important parameter settings of all methods are listed in Table 3. The other parameters of SL-GEP and GPPI are set the same as those in the original paper (Zhong et al. 2016; Chen et al. 2017). To fully utilize the computing resources of GPU, all the methods implemented on GPU have a relative large S, which is 512. The time interval between two migrations (\(\pi \)) is 100 generations. The number of threads in CPU (\(N_{TC}\)) is 1 for all methods but 4 for HSL-GEP. The number of blocks (\(N_B\)) in GPU is 256, and the number of threads in each block (\(N_T\)) is 128. The experiment platform is one chip of GeForce GTX 1070 GPU and Intel Core i7-7700 4-core CPU with 3.60GHz. All algorithms terminate when the optimal fitness value is reached or the limited running time is reached. In this study, the optimal fitness value is reached when the Rmse is less than \(1e^{-4}\). And the limited running time is 60 s and 180 s for benchmark problems and real-world problems, respectively.
For all problems and all compared algorithms, the function set is set to { \(+,-,\times ,\div ,\max ,\min ,pow,\mod , \sin , \cos ,\exp ,\ln , \sqrt{ },sgn,\lceil \rceil ,\lfloor \rfloor ,|\cdot |,opp,+5,\times 5\)}, where the sgn returns the sign of the input and the opp returns the opposite number of the input. The \(+5\) and \(\times 5\) serve as the redundant functions in the function set. The terminal set is determined according to the specific problem. For example, the number of terminals in \(F_0\) is only one, while that of \(F_6\) is twenty-five (i.e., \(x_0\) to \(x_{24}\)). Three performance metrics are utilized for algorithm comparison. The first metric is the root-mean-square error (RMSE) on the testing data, which is calculated by:
where \(x_{ti}\) and \(y_{ti}\) are the input vector and output of testing data, respectively, and \(N_\mathrm{t}\) is the number of testing data. When the RMSE of an independent run is less than \(1e^{-4}\), the algorithm is regarded as achieving the perfect hit (i.e., achieving the optimal fitness value). The second one is the success rate on the training data (Suc), which describes the percentage of the perfect hits. The Suc is calculated by (16).
In (16), \(D_s\) is the number of perfect hits and D is the total number of independent runs. The third metric is the average running time (\(T_{r}\)), which measures the computational time efficiency of the algorithms.
5.2 Comparison with state-of-the-art algorithms
Table 4 shows the comparison results of these six algorithms. The bold data are the best results. The Wilcoxon rank-sum test results between other methods and HSL-GEP are labeled out. Symbols \(\approx ,+,\) and − represent the corresponding method is similar to, significantly better, and worse than HSL-GEP (EM-SLGEP) according to the Wilcoxon rank-sum test at \(\alpha =10\%\). Symbols in brackets are the Wilcoxon rank-sum test results between other methods and EM-SLGEP. The symbol b, s(b, s) demonstrates the number of cases in which HSL-GEP (EM-SLGEP) performs better than, or similar to another method.
It can observed in Table 4 that EM-SLGEP outperforms SL-GEP and SL-GEP+GPPI in most problems. In some cases, such as \(F_{1}, F_2\) and \(F_9\), EM-SLGEP has much better Suc values compared with SL-GEP and SL-GEP+GPPI. Although in those tough problems such as \(F_3\) and \(F_6\), all methods, including the basic SL-GEP, have the same Suc and \(T_r\), EM-SLGEP still has a better Rmse than SL-GEP and SL-GEP+GPPI, which means a better solution quality. The above results validate the effectiveness of the proposed multi-population framework. As for GPU-based methods, HSL-GEP performs significantly better than other two methods (i.e., SL-GEP+2DG, EMSLGEP+2DG) in most of these fifteen problems, especially the real-world ones. HSL-GEP obtains the best RMSE and Suc results among all GPU-based methods in all testing problems. Since SL-GEP+2DG and EMSLGEP+2DG cannot fully utilize the GPU and multi-core CPU (i.e., CPU and GPU must wait to each other alternatively), they are not efficient enough to accelerate the search. The above results demonstrate the effectiveness of our two main novelties. Both the proposed EV-based multi-population GP framework and the developed hierarchical parallel computing mechanism can improve the searching efficiency.
To investigate the convergence speed of all compared algorithms, we plot the convergence curves of the best-of-run RMSE obtained by the algorithms in all the problems. Figure 4 illustrates the RMSE versus time convergence curves of the compared algorithms. It is observed that the results of EM-SLGEP (i.e., the green curve) can converge faster and deeper than that of SL-GEP (i.e., the black curve) and SL-GEP+GPPI (i.e., the purple curve) in most problems such as \(F_1, F_2, F_6, F_7, F_8, F_9\), and \(F_{10}\). These also validate the effectiveness of the proposed multi-population mechanism. Meanwhile, the convergence curves of the proposed HSL-GEP (i.e., the red curve) converge faster than all the other curves in most cases. Besides, the curve of the proposed method can always converge to a smaller RMSE, which means that the proposed method can find better quality solutions. For the simple problems, although the feature selection ability in HSL-GEP cannot bring too much benefit to the evolution process, HSL-GEP also achieves better (or at least similar) results. As for the tough real-world problems, HSL-GEP converges faster and deeper than others, including the two GPU-based methods. These results demonstrate the efficacy of HSL-GEP.
5.3 Primitive selection analysis
In this subsection, we investigate the effectiveness of primitive selection in HSL-GEP. The primitive selection results of six problems and the target primitives (functions and terminals) of selected benchmark problems are listed in Table 5. These six benchmark problems are selected as examples because they cover nearly the whole function set and cover different difficulty levels (i.e., from the simplest \(F_0\) to the hardest \(F_{10}\)). The components of \(E_{t,G}\) and \(E_{f,G}\) are regarded as the importance of each primitive. Five most important functions and terminals (i.e., those with larger weights) are shown in Table 5. Since \(F_0,F_1\), and \(F_3\) have less than five terminals, the selection results of terminals are omitted in Table 5. It is observed that all methods, including the basic GP solver, SL-GEP, have a certain ability of primitive selection. But EM-SLGEP and HSL-GEP can find out more target primitives (i.e., those marked in bold font). For example, in \(F_{10}\), EM-SLGEP and HSL-GEP find out two to four target functions, while others can only find out one function. Besides, the weights of the target primitives found by EM-SLGEP and HSL-GEP are also higher than those of others. For example, in the most complicate task \(F_5\), the sum of weights of target terminals found by EM-SLGEP and HSL-GEP are 0.24 and 0.34, respectively, which are much higher than those found by others.
Further, to show the efficiency of the feature selection ability of the proposed method, the importance versus time changing process of terminals in \(F_{10}\) is shown in Fig. 5. The \(F_{10}\) is selected because it is one of the most complex benchmark problems in our experiments (i.e., it has more than one target primitives, different value range, and a large number of noisy features.). The target terminals (i.e., \(x_0,x_1\), and \(x_2\)) are labeled as black, red, and green curves, respectively, while the curves with other colors belong to noisy features. As shown in Fig. 5, the \(x_1\) is missed by SL-GEP and the importance of \(x_0\) and \(x_2\) is not high enough at the end of the evolution. It can be observed that SL-GEP discovers \(x_0\) and \(x_2\) during the evolution. However, the primitive selection results of SL-GEP are not accurate and efficient enough. The importance of primitives in SL-GEP changes slowly. This not only shows a limited ability of SL-GEP to select primitives, but also makes SL-GEP get trapped into local optima easily. Similar results are obtained by SL-GEP+GPU. Besides, SL-GEP+GPPI has a better terminal selection ability than SL-GEP. It can discover \(x_0\) and \(x_2\) faster than SL-GEP, but the accuracy of SL-GEP+GPPI is still not high enough. EM-SLGEP, EMSLGEP+2DG, HSL-GEP have much better primitive selection results. For all these three algorithms, all three target terminals are successfully identified, and the importances of these three terminals become relatively high quickly during the evolution. Among them, the importances of \(x_0\) and \(x_2\) in HSL-GEP and EMSLGEP+2DG are more balanced than EM-SLGEP, and the convergent speed of importance of HSL-GEP is faster than that of EMSLGEP+2DG. Therefore, the proposed HSL-GEP performs significantly better than EM-SLGEP and EMSLGEP+2DG. To sum up, the proposed HSL-GEP can effectively select important primitives (both functions and terminals) in the early evolution stage, which significantly improves the search efficiency.
5.4 Scalability analysis
In this subsection, we investigate the impact of the computing resources on HSL-GEP by varying the number of CPU threads (\(N_{TC}\)), the number of threads in each block (\(N_T\)), and the number of GPU blocks (\(N_B\)). The basic settings of \(N_{TC}\), \(N_T\) and \(N_B\) are 4, 128, and 256, respectively. By controlling the variates, \(N_{TC}\) varies from 1 to 4, \(N_T\) varies from 64 to 512, and \(N_B\) varies from 128 to 1024. When one of these three parameters is modified, the others are set as the basic settings. We choose \(F_9\) and \(F_{10}\) for case study since they are the most challenging benchmark problems. Figure 6 shows the experimental results. In the scalability experiment of \(N_{TC}\), the curves of \(N_{TC}=1\) (i.e., the black curve) have the slowest convergence speed in both \(F_4\) and \(F_5\). On the contrary, the curves of \(N_{TC}=4\) (i.e., the blue curve) have the fastest convergence speed. This is because the larger \(N_{TC}\) means HSL-GEP can utilized more CPU computation resources.
Meanwhile, the parameters of GPU computation resources have a more robust performance. As shown in Fig. 6, the curves of \(N_T\) and \(N_B\) are very close to each other, which means these GPU parameter settings have similar performance. However, to better utilize the GPU, \(N_\mathrm{t}\) and \(N_b\) should not be set too small. For example, the curves of the smallest setting (i.e., the black curves) in both \(N_\mathrm{t}\) and \(N_b\) have the slowest convergent speed compared to other parameter settings. This is because when the parameters are set too small, a part of GPU resources will not be scheduled during running. This phenomenon is also mentioned in Cook (2012). It is also worth mentioning that the red curves in \(N_\mathrm{t}\) and \(N_b\) are always very close to the fastest convergent speed or reach the deepest RMSE in both two problems. The above results suggest that \(N_\mathrm{t}=128\) and \(N_b=256\) are promising parameter settings in this application.
6 Conclusion
This paper proposed a fast parallel genetic programming framework by using the environment-vector-based multi-population mechanism and the hierarchical parallel computing mechanism. A recently published GP variant (named SL-GEP) has been integrated with the proposed fast GP framework to derive an efficient implementation named HSL-GEP. The developed HSL-GEP is capable of discovering the valuable primitives during the evolution process. Besides, the hierarchical parallel computing mechanism in HSL-GEP can fully utilize the heterogeneous computation resources (i.e., GPU and multi-core CPU) to improve the search efficiency. To investigate the efficiency of the proposed HSL-GEP, eleven benchmark problems and four real-world problems have been used for testing. Five other GP variants, including the state-of-the-art GPs with feature selection techniques and GPU-based parallel GPs, are used for comparison. The empirical results have demonstrated that the proposed HSL-GEP has a significantly better (or at least competitive) performance than the other methods. As for future work, we plan to apply the proposed HSL-GEP to large-scale symbolic regression problems with big data and validate its generality on various problems, such as classification and planning problems. Besides, extending the proposed method by using other parallel computing platforms such as Spark is another promising research direction.
Notes
The dataset of \(F_{11}\) can be downloaded from http://symbolicregression.com/sites/default/files/DataSets/towerData.txt.
The dataset of \(F_{12,13}\) can be downloaded from http://archive.ics.uci.edu/ml/datasets/Communities+and+Crime+Unnormalized.
References
Ahmad F, Isa NAM, Hussain Z, Osman MK, Sulaiman SN (2015) A ga-based feature selection and parameter optimization of an ann in diagnosing breast cancer. Pattern Anal Appl 18(4):861–870
Ahmed S, Zhang M, Peng L (2013) Enhanced feature selection for biomarker discovery in LC-MS data using GP. In: IEEE congress on evolutionary computation (CEC), pp 584–591
Antonio LM, Coello CCA (2018) Coevolutionary multiobjective evolutionary algorithms: survey of the state-of-the-art. IEEE Trans Evol Comput 22(6):851–865
Banzhaf W, Harding S, Langdon WB, Wilson G (2008) Accelerating genetic programming through graphics processing units. In: Genetic programming theory and practice VI, pp 1–19
Brameier MF, Banzhaf W (2007) Linear genetic programming. Springer, Berlin
Cano A, Ventura S (2014) Gpu-parallel subtree interpreter for genetic programming. In: Conference on genetic and evolutionary computation, pp 887–894
Chen B, Chen B, Liu H, Zhang X (2015) A fast parallel genetic algorithm for graph coloring problem based on CUDA. In: International conference on cyber-enabled distributed computing and knowledge discovery, pp 145–148
Chen Q, Xue B, Niu B, Zhang M (2016) Improving generalisation of genetic programming for high-dimensional symbolic regression with feature selection. In Congress on evolutionary computation (CEC), pp 3793–3800
Chen Q, Zhang M, Xue B (2017) Feature selection to improve generalization of genetic programming for high-dimensional symbolic regression. IEEE Trans Evol Comput 21(5):792–806
Chitty DM (2016a) Faster GPU based genetic programming using A two dimensional stack. In: CoRR. arXiv:1601.00221
Chitty DM (2016b) Improving the performance of gpu-based genetic programming through exploitation of on-chip memory. Soft Comput 20(2):661–680
Cook S (2012) CUDA programming: a developer’s guide to parallel computing with GPUs. Elsevier, London
Deng W, Zhao H, Zou L, Li G, Yang X, Wu D (2017) A novel collaborative optimization algorithm in solving complex optimization problems. Soft Comput 21(15):4387–4398
Deng W, Yao R, Zhao H, Yang X, Li G (2019a) A novel intelligent diagnosis method using optimal LS-SVM with improved pso algorithm. Soft Comput 23(7):2445–2462
Deng W, Xu J, Zhao H (2019b) An improved ant colony optimization algorithm based on hybrid strategies for scheduling problem. IEEE Access 7:20281–20292
Dick G, Rimoni AP, Whigham PA (2015) A re-examination of the use of genetic programming on the oral bioavailability problem. In: Proceedings of the genetic and evolutionary computation conference (GECCO)
Espejo PG, Ventura S, Herrera F (2010) A survey on the application of genetic programming to classification. IEEE Trans Syst Man Cybern C Appl Rev 40(2):121–144
Ferreira C (2006) Gene expression programming. Springer, Berlin
Ffrancon R, Schoenauer M (2015) Memetic semantic genetic programming. In: Proceedings of the 2015 annual conference on genetic and evolutionary computation. ACM, pp 1023–1030
Gandomi AH, Sajedi S, Kiani B, Huang Q (2016) Genetic programming for experimental big data mining: a case study on concrete creep formulation. Autom Constr 70:89–97
Gu S, Cheng R, Jin Y (2018) Feature selection for high-dimensional classification using a competitive swarm optimizer. Soft Comput 22(3):811–822
Hancer E, Xue B, Zhang M, Karaboga D, Akay B (2018) Pareto front feature selection based on artificial bee colony optimization. Inf Sci 422:462–479
Harding S, Banzhaf W (2007) Fast genetic programming and artificial developmental systems on GPUs. In: 21st International symposium on high performance computing systems and applications, 2007. HPCS 2007, p 2
Harding S, Banzhaf W (2007) Fast genetic programming on GPUs. In: European conference on genetic programming, pp 90–101
Harvey DY, Todd MD (2015) Automated feature design for numeric sequence classification by genetic programming. IEEE Trans Evol Comput 19(4):474–489
Keijzer M (2003) Improving symbolic regression with interval arithmetic and linear scaling. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics), vol 2610. Essex, UK, pp 70–82
Koza JR, Poli R (2005) Genetic programming
Langdon WB (2010) A many threaded CUDA interpreter for genetic programming. Springer, Berlin
Langdon WB (2011) Graphics processing units and genetic programming: an overview. Soft Comput 15(8):1657–1669
McDermott J, White DR., Luke S, Manzoni L, Castelli M, Vanneschi L, Jaskowski W, Krawiec K, Harper R, De Jong K, O’Reilly U-M (2012) Genetic programming needs better benchmarks. In: Proceedings of the 14th international conference on genetic and evolutionary computation, GECCO’12, pp 791–798
Mei Y, Omidvar MN, Li X, Yao X (2016a) A competitive divide-and-conquer algorithm for unconstrained large-scale black-box optimization. ACM Trans Math Softw 42(2):13
Mei Y, Zhang M, Nguyen S (2016b) Feature selection in evolving job shop dispatching rules with genetic programming. In: Proceedings of the genetic and evolutionary computation conference (GECCO). ACM, pp 365–372
Mei Y, Su N, Xue B, Zhang M (2017) An efficient feature selection algorithm for evolving job shop scheduling rules with genetic programming. IEEE Trans Emerg Top Comput Intell 1(5):339–353
Miller JF, Thomson P (2000) Cartesian genetic programming. In: Genetic programming. Springer, Berlin, pp 121–132
Moore JH, Hill DP, Saykin A, Shen L (2013) Exploring interestingness in a computational evolution system for the genome-wide genetic analysis of Alzheimer’s Disease. Springer, New York
Moraglio A, Krawiec K, Johnson CG (2012) Geometric semantic genetic programming. In: International conference on parallel problem solving from nature. Springer, Berlin, pp 21–31
Neshatian K, Zhang M (2009) Pareto front feature selection: using genetic programming to explore feature space. In: Proceedings of the genetic and evolutionary computation conference, GECCO 2009. Montreal, Québec, Canada, pp 1027–1034
O’Neill M, Ryan C (2001) Grammatical evolution. IEEE Trans Evol Comput 5(4):349–358
Riley M, Mei Y, Zhang M (2016) Improving job shop dispatchingrules via terminal weighting and adaptive mutation in genetic programming. Vancouver, BC, Canada, pp 3362 – 3369
Rojas F, Meza F (2015) A parallel distributed genetic algorithm for the prize collecting steiner tree problem. In: International conference on computational science and computational intelligence (CSCI), pp. 643–646
Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM (2002) The use of molecular profiling to predict survival after chemotherapy for diffuse large-b-cell lymphoma. New Engl J Med 346(25):1937–1947
Sandin I, Andrade G, Viegas F, Madeira D (2012) Aggressive and effective feature selection using genetic programming. In: IEEE congress on evolutionary computation (CEC), pp 1–8
Schmidt M, Lipson H (2009) Distilling free-form natural laws from experimental data. Science 324(5923):81–85
Shao S, Liu X, Zhou M, Zhan J, Liu X, Chu Y, Chen H (2012) A gpu-based implementation of an enhanced GEP algorithm. In: Conference on genetic and evolutionary computation, pp 999–1006
Vladislavleva E, Smits G, Den Hertog D (2009) Order of nonlinearity as a complexity measure for models generated by symbolic regression via pareto genetic programming. IEEE Trans Evol Comput 13(2):333–349
Xue B, Zhang M, Browne WN (2013) Particle swarm optimization for feature selection in classification: a multi-objective approach. IEEE Trans Syst Man Cybern 43(6):1656–1671
Xue B, Zhang M, Browne WN, Yao X (2016) A survey on evolutionary computation approaches to feature selection. IEEE Trans Evol Comput 20(4):606–626
Yang Z, Tang K, Yao X (2008) Large scale evolutionary optimization using cooperative coevolution. Inf Sci 178(15):2985–2999
Yao X, Liu Y, Lin GM (1999) Evolutionary programming made faster. IEEE Trans Evol Comput 3(2):82–102
Zhai Y, Ong YS, Tsang IW (2014) The emerging “big dimensionality”. IEEE Comput Intell Mag 9(3):14–26
Zhong J, Cai W (2015) Differential evolution with sensitivity analysis and the powell’s method for crowd model calibration. J Comput Sci 9:26–32
Zhong J, Ong YS, Cai W (2016) Self-learning gene expression programming. IEEE Trans Evol Comput 20(1):65–80
Zhong J, Cai W, Lees M, Luo L (2017a) Automatic model construction for the behavior of human crowds. Appl Soft Comput 56:368–378
Zhong J, Feng L, Ong Y-S (2017b) Gene expression programming: a survey. IEEE Comput Intell Mag 12(3):54–72
Zhou C, Xiao W, Tirpak TM, Nelson PC (2003) Evolving accurate and compact classification rules with gene expression programming. IEEE Trans Evol Comput 7(6):519–531
Acknowledgements
This work is supported by the National Natural Science Foundation of China (Grant No. 61602181), Program for Guangdong Introducing Innovative and Entrepreneurial Teams (Grant No. 2017ZT07X183), Guangdong Natural Science Foundation Research Team (Grant No. 2018B030312003), and the Fundamental Research Funds for the Central Universities (Grant No. D2191200).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by V. Loia.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Huang, Z., Zhong, J., Feng, L. et al. A fast parallel genetic programming framework with adaptively weighted primitives for symbolic regression. Soft Comput 24, 7523–7539 (2020). https://doi.org/10.1007/s00500-019-04379-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-019-04379-4