Abstract
A novel aliphatic nitrilase, REH16, was found in Ralstonia eutropha H16 and overexpressed in Escherichia coli BL21(DE3), and its enzymatic properties were studied. The temperature and pH optima were 37 °C and 6.6, respectively, and the best thermostability of the nitrilase was observed at 25 °C, which preserved 95% of activity after 120 h of incubation. REH16 has a broad hydrolytic activity toward aliphatic and heterocyclic nitriles and showed high tolerance of 3-cyanopyridine; this enzyme could hydrolyze as high as 100 mM 3-cyanopyridine completely. To improve the 3-cyanopyridine conversion efficiency in an aqueous reaction system, water-miscible organic solvents were tested, and ethanol (10% v/v) was chosen as the optimal co-solvent. Finally, under optimized conditions, using the fed-batch reaction mode, total of 1050 mM 3-cyanopyridine was hydrolyzed completely in 20.8 h with eight substrate feedings, yielding 129.2 g/L production of nicotinic acid and thus showing a potential for industrial application.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nicotinic acid (vitamin B3) has a strong demand among manufacturers of foodstuffs and feed additives and is used as an intermediate for the synthesis of isoniazid, inositol hexanicotinate, and N,N-diethylnicotinamide in the pharmaceutical industry. As a vitamin supplement, nicotinic acid can help to lower the risk of cardiovascular diseases, pellagra, and arthritis [1]. The mainstream process of chemical production of nicotinic acid involves high energy consumption due to the reaction temperature [2]. Based on the advantages of biotransformation in green production, several biotransformation processes such as the amidase pathway and nitrilase pathway have been reported as an alternative to the chemical process of production of nicotinic acid [3,4,5,6,7,8]. According to some reports, the process of 3-cyanopyridine hydrolysis by nitrilases holds great promise for industrial applications [9].
Nitrilases (EC 3.5.5.1) have been recognized as high-commercial-value biocatalysts for hydrolysis of various nitriles for formation of the corresponding carboxylic acids [10]. In recent years, many nitrilases were discovered in bacteria, fungi, and plants and studied in terms of the substrate specificity, enantioselectivity, and amide formation [11], and some of these nitrilases have been successfully applied to commercial production of carboxylic acids, such as 4-cyanopentanoic acid [12] and (R)-(−)-mandelic acid [13]. On the basis of the great synthetic possibilities related to nitrilases and carboxylic acids, screening of nitrilases for nicotinic acid production has a great potential. In the last decades, several nitrilases have been used to produce nicotinic acid [3,4,5,6,7, 14,15,16,17,18,19]. Among them, several nitrilase producing strains have been isolated and utilized as whole-cell catalysts on 3-cyanopyridine hydrolysis, including Bacillus pallidus Dac521 [5], Rhodococcus sp. NDB 1165 [7], Nocardia globerula NHB-2 [3, 4], Fusarium proliferatum ZJB-09150 [14] and Stenotrophomonas maltophilia AC21 [15]. By the use of the strains Rhodococcus sp. NDB 1165, as high as 1.6 M of nicotinic acid was obtained in fed-batch reaction mode. However, the biochemical properties of the corresponding nitrilases remain unclear. On the other hand, several nitrilases with potential on 3-cyanopyridine hydrolysis were cloned and purified, and their biochemical properties were studied in details, including Gibberella intermedia GA3-1 [16], Rhodobacter sphaeroides LHS-305 [17], Pseudomonas putida CGMCC3830 [18], Acidovorax facilis 72W [19] and Alcaligenes faecalis MTCC 126 [6]. Among them, the highest specific activity toward 3-cyanopyridine was obtained as 71.8 U/mg with the purified nitrilase from P. putida CGMCC3830 [18].
In the present study, a novel nitrilase, REH16, with high 3-cyanopyridine hydrolytic activity was found in Ralstonia eutropha H16. Then, REH16 was cloned and overexpressed in Escherichia coli BL21(DE3). The biochemical properties and substrate specificity of REH16 were investigated. Furthermore, for development of a nicotinic acid production process, the bioprocess parameters of 3-cyanopyridine hydrolysis were optimized. Finally, bench scale conversion of 3-cyanopyridine to nicotinic acid using REH16 in substrate fed-batch mode was performed.
Materials and methods
Materials
All the recombinant nitrilases introduced into E. coli BL21(DE3) were obtained from our previous studies [20, 21]. Nicotinic acid and 3-cyanopyridine were purchased from Sigma-Aldrich Co., LLC. (USA). All other nitriles and acids were purchased from Sigma-Aldrich Co., LLC (USA) and Tokyo Chemical Industry Co., Ltd. (Japan) and of analytical grade.
Preparing the resting cells
The E. coli strains harboring nitrilases were cultivated in Luria–Bertani (LB) broth in a flask at 37 °C. Isopropyl-β-d-thiogalactoside (IPTG) was used for the gene expression induction at a final concentration of 0.1 mM. The incubation temperature after the induction was decreased to 20 °C. The induced cultures were harvested after 20 h of incubation and centrifuged at 8000×g, 4 °C, for 10 min. The wet cells were collected and washed twice with physiological saline and then stored at −20 °C. The wet cells were used as resting cells for the hydrolysis of 3-cyanopyridine.
The screening procedure
The resting cells (~20 mg) were resuspended in 1 mL of sodium phosphate buffer (pH 7.0, 100 mM) with 5% (v/v) ethanol, and then the reaction was initiated by addition of 5.2 mg 3-cyanopyridine powder with a final concentration of 50 mM. The reaction proceeded at 30 °C for 12 h, and then the resting cells were removed from the mixture by centrifugation. The formation of ammonia in the mixture was determined by the Berthelot method [22].
Sequence analysis of the REH16 gene
The nucleotide sequence of the nitrilase inserted into the vector pET-28a(+) was determined (BGI, Shenzhen, China) and then used in a BLASTN search (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome). The nitrilase REH16 was most active towards 3-cyanopyridine and it was chosen for further investigation. Multiple sequence alignments of nitrilase REH16 with the reported nitrilases were carried out using the ClustalX 2.0 software and then modified by means of ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). The phylogenetic tree analysis of REH16 was performed in the MEGA 7.0 software by the neighbor-joining method.
Protein purification
The obtained E. coli resting cells overexpressing recombinant REH16 were resuspended in sodium phosphate buffer (pH 7.0, 50 mM). The cell lysis was performed by a sonication method in an ice-water bath. The lysate was centrifuged at 12,000×g for 15 min, and then the collected supernatant was loaded onto the Ni–NTA resin. The nitrilase was purified from the supernatant by Ni–NTA chromatography with separation by imidazole gradient elution. The elution profile of the nitrilase was recorded by a UV detector at 280 nm. The eluted nitrilase was desalted by dialysis using a Biotech Cellulose Ester Dialysis Membrane with a molecular weight cutoff of 1 kDa (Spectrum Chemical, USA) and concentrated by ultrafiltration using a 50 mL Amicon Ultra Centrifugal Filter Device with a molecular weight cutoff of 10 kDa (Millipore, USA). Protein concentration was determined by the Bradford method. All the purification procedures were carried out at 4 °C.
Nitrilase assay
The standard nitrilase assay was conducted in a 1-mL reaction system containing sodium phosphate buffer (100 mM, pH 7.0), 50 mM 3-cyanopyridine, and an appropriate amount of nitrilase at 30 °C for 10 min. Samples (100 μL) were taken at various intervals and quenched by adding 10% (v/v) 2 M HCl. The production was determined by high-performance liquid chromatography (HPLC). One unit of the nitrilase activity was defined as the amount of the nitrilase that produced 1 μmol of nicotinic acid per minute under the standard assay conditions. All the experiments were performed in triplicate.
Effects of temperature and pH on the activity of purified REH16
To determine the optimal temperature and pH for the nitrilase activity of REH16 toward 3-cyanopyridine, the reaction was conducted with 10 μg of purified REH16 and 50 mM 3-cyanopyridine in a 1-mL reaction system for 10 min at different temperatures or at different pH levels (various buffers). The optimal temperature for REH16 was determined by measuring the nitrilase activity during 3-cyanopyridine hydrolysis carried out at different temperatures (10–70 °C) in 100 mM sodium phosphate buffer (pH 7.0). The optimal pH of REH16 was determined by measuring the nitrilase activity during 3-cyanopyridine hydrolysis carried out at different pH levels (5.0–9.0) and 25 °C. Sodium citrate/citric acid buffer (pH 5.0–6.0, 100 mM), sodium phosphate buffer (pH 6.0–8.0, 100 mM), and Tris-HCl buffer (8.0–9.0, 100 mM) were used in these experiments.
To determine thermostability of the nitrilase activity of REH16, the purified nitrilase was incubated at different temperatures (20, 30, 37, and 50 °C). Samples were withdrawn at various intervals and then the residual activity toward 3-cyanopyridine was determined under standard assay conditions. The purified nitrilase was dissolved in 100 mM sodium phosphate buffer at different pH levels (6.0, 6.6, and 7.0) and incubated at 20 °C. Samples were taken at various intervals for determination of the residual activity.
Effects of metal ions on REH16 activity
The effects of metal ions and ethylenediaminetetraacetic acid (EDTA) on the nitrilase REH16 activity were determined at a final concentration of 1 mM. The nitrilase activity was determined under the standard assay conditions. The control reaction did not contain any additive.
Substrate specificity of REH16
The specific activities of REH16 were studied under standard conditions with a total of 28 nitriles (separately). The conversion was determined by assaying the ammonia amount produced in the reaction by the Berthelot method. All the experiments were carried out in triplicate.
Effects of co-solvents on REH16 activity
To determine the effects of organic solvents on the E. coli cells expressing REH16, various solvents in a final concentration range of 5–25% (v/v) were added (individually) into the reaction mixture containing sodium phosphate buffer (pH 6.6, 100 mM), 100 mM 3-cyanopyridine, and resting cells at 100 mg/mL. The mixture was incubated at 30 °C with shaking at 200 rpm for 60 min and then the conversion was determined by HPLC.
To identify an optimal co-solvent for the 3-cyanopyridine bioconversion, the mixture containing 100 mg/mL resting cells, sodium phosphate buffer (pH 6.6, 100 mM), and 10% (v/v) of various solvents was incubated at 30 °C for different periods, and then the reaction was initiated by adding 100 mM 3-cyanopyridine. The conversion was determined after 60 min of incubation with shaking at 200 rpm.
The effect of 3-cyanopyridine concentration on nitrilase activity
To determine the effect of substrate concentration on nitrilase activity, the reaction was performed at various substrate concentrations (50–200 mM) in a 10-mL reaction system, which contained sodium phosphate buffer (pH 6.6, 100 mM), 100 mg/mL resting cells, and ethanol (10%, v/v). Samples were taken at various intervals and quenched with 2 M HCl (10%, v/v). The formation of nicotinic acid was determined by HPLC.
The effect of concentration of resting cells on the 3-cyanopyridine conversion
To determine the optimal resting cell concentration for hydrolysis of 100 mM 3-cyanopyridine, the conversion was conducted with resting cells at 50, 75, 100, 150, or 200 mg/mL, at 30 °C for 60 min, and then the formation of nicotinic acid was determined.
The fed-batch reaction for nicotinic acid production
A 500 mL three-necked round-bottom flask was used for a 200 mL fed-batch reaction. The reaction mixture contained sodium phosphate buffer (100 mM, pH 6.6), 10% (v/v) ethanol, 20 g of the resting cells, and 100 mM 3-cyanopyridine. The initial parameters were set up as follows: 30 °C, pH 6.60–6.65, and agitation speed of 300 rpm. The temperature was maintained by means of a water bath. The pH was maintained by the ammonia addition controlled by a pH auto controller; 100 mM 3-cyanopyridine per batch was mixed into the reaction after the current batch converted completely. Samples were taken at various intervals and quenched by addition of 10% (v/v) 2 M HCl. The product was analyzed by HPLC.
Analytical methods
Nicotinic acid and 3-cyanopyridine were quantified by HPLC using a Zorbax SB-Aq column (4.6 mm × 250 mm, 5 μm; Agilent Technologies, Ltd., USA) eluted with 0.1% phosphate solution versus acetonitrile (75:25 v/v) at a flow rate of 0.8 mL/min. The elution was monitored at 210 nm.
Results and discussion
Screening of recombinant nitrilases for 3-cyanopyridine hydrolysis
For identification of novel nitrilases, a data-mining approach based on NCBI BLAST was adopted in our previous works [20, 21]. Thirteen nitrilases were cloned and overexpressed in E. coli BL21(DE3) in soluble form. Each E. coli strain harboring a nitrilase was cultivated in LB broth and harvested as resting cells to be screened for activity toward 3-cyanopyridine hydrolysis. However, using whole-cell as catalyst, the catalysis efficiency was determined not only by the specific activity of the enzyme but also the expression level of the nitrilase. Therefore, we search the optimal biocatalyst based on the specific activity of the cells irrespective of the nitrilase expression level during this screening process. Among these enzymes, a new nitrilase, REH16 from R. eutropha H16 (GenBank accession number: AM260479.1), showed the highest specific activity (6.9 U/mg dcw) toward 3-cyanopyridine (Table 1). After that, REH16 was used for further experiments.
Phylogenetic relations of REH16
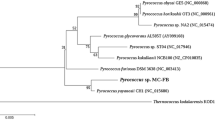
For the phylogenetic analysis of nitrilase REH16, 19 nitrilases with different substrate specificity from bacteria, fungi, and plants (that have been reported before) were selected. As shown in Fig. 1a, the nitrilases could be grouped into three clusters. The arylacetonitrilases that use arylacetonitriles as optimal substrates could be grouped exactly into cluster I in the phylogenetic tree and all these arylacetonitrilases are from bacteria. The aliphatic nitrilases could be grouped into cluster II of the phylogenetic tree, and most of them are from bacteria except for F. proliferatum and F. oxysporum f. sp. cubense (fungal nitrilases). Most nitrilases in cluster III are from fungi and plants. The approximate position of REH16 in the phylogenetic tree indicated that REH16 may be an aliphatic nitrilase and has more sequence similarity with eukaryotic nitrilases. Furthermore, multiple sequence alignments of conserved regions to REH16 with closely related nitrilases were performed, and the results are shown in Fig. 1b. The results revealed that REH16 contains the typical nitrilase conserved catalytic triad E47-K129-C163.
Bioinformatic analysis of REH16. a Phylogenetic analysis of REH16 with closely related nitrilases. The scale bar represents 0.1 changes per amino acid. b Multiple sequence alignment of conserved regions of nitrilases. The conserved sequences are indicated with boxes. The nitrilase catalytic triads are indicated as triangles
Expression and purification of REH16
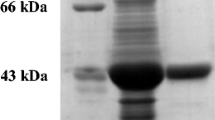
Nitrilase REH16 with a hydrolytic activity toward 3-cyanopyridine was overexpressed in soluble form in E. coli and purified. In SDS-PAGE analysis, the purified nitrilase yielded a single band with a size of approximately 40 kDa (Fig. 2), which was in agreement with the predicted size.
Effects of temperature and pH on nitrilase activity
The optimal pH and temperature of the purified REH16 were determined. The optimal temperature was found to be 37 °C (as shown in Fig. 3a), and more than 80% of maximal activity was retained between 30 and 50 °C. When the temperature was above 50 °C, the activity of REH16 decreased sharply. REH16 exerted a higher activity (>80% of maximal activity, as shown in Fig. 3b) between pH 6 and 7, and the highest activity was observed at pH 6.6. This behavior is different from that of other nitrilases reported before. To the best of our knowledge, nitrilases have the highest activity in a weakly basic environment generally.
Effects of temperature and pH on enzymatic activity and stability. a Optimal temperature: enzymatic activity was measured at various temperatures (20–70 °C) in 100 mM sodium phosphate buffer (pH 7.0). b Optimal pH: enzymatic activity was measured in different buffers: sodium citrate/citric acid buffer (pH 5.0–6.0, 100 mM, triangles), sodium phosphate buffer (pH 6.0–8.0, 100 mM, squares), Tris/HCl buffer (pH 8.0–9.0, 100 mM, circles). c Thermal stability: the purified enzyme was incubated at various temperatures for a given period, and the activity was measured at 25 °C. Squares 25 °C, triangles 30 °C, circles 37 °C. d pH stability: the purified enzyme was incubated in sodium phosphate buffers (different pH levels) at 4 °C for a given period, and the activity was measured at 25 °C. Circles pH 6.0, squares pH 6.6, triangles pH 7.0
The thermostability of REH16 was determined by analyzing the residual activity of the purified nitrilase after incubation at different temperatures for various periods. REH16 showed the best stability during storage at 25 °C: only 5% of activity was lost after 120-h incubation (Fig. 3c). Although the optimal temperature of REH16 is 37 °C, the half-life of activity was only 5 h at this temperature. Compared to the results at 37 °C, the half-life of the activity at 30 °C was more than 120 h. Additionally, the half-life of the activity at 50 °C was less than 20 min (data not shown). Therefore, 30 °C was chosen as the optimal temperature for nitrile hydrolysis by REH16. The results on pH stability of REH16 are shown in Fig. 3d. REH16 has excellent stability in the pH range of 6.0–7.0, an activity loss was barely detectable after 120 h of incubation. According to the results above, nitrilase REH16 is suitable for biotransformation at 30 °C and pH 6.6.
Effects of metal ions and EDTA
The effects of various metal ions and ethylenediaminetetraacetic acid (EDTA) on nitrilase REH16 were assessed (Table 2). The metal-chelating agent EDTA had little or no effect on REH16 activity, suggesting that metal-binding sites do not exist in the structure of REH16 (or are not important). The nitrilase activity was inhibited by thiol-binding metal ions (Ag+, Cu2+, and Zn2+), indicating that the thiol group is indispensable for the nitrilase activity. Most of the metal ions caused obvious inhibition of the REH16 activity, thus showing that the nitrilase activity of REH16 does not require the presence of metal ions.
Substrate specificity
The results on the specific activities of REH16 toward 28 nitriles are presented in Table 3. REH16 showed a broad hydrolytic activity toward aliphatic and heterocyclic nitriles. The highest activity among these aliphatic nitriles (entries 16–28) was observed toward fumaronitrile, which was 36-fold higher than that toward 3-cyanopyridine. No hydrolytic activities toward malononitrile and dodecanenitrile were detected. These results indicated that the carbon chain length of the substrate has a strong influence on the nitrilase activity of REH16. For most of the aromatic nitriles (entries 5–15), the hydrolytic activities of nitrilase REH16 were not detected except for 3-phenylpropionitrile, 2-phenylbutyronitrile, and cinnamonitrile. Compared to these three aromatic nitriles, REH16 exerted strong action on cinnamonitrile. These results probably suggest that the hydrolytic activity toward these aromatic nitriles may be due to the fact that the cyano group is far enough from the benzene ring.
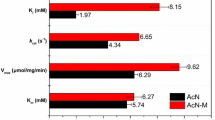
Kinetic parameters of the purified REH16 were determined using 3-cyanopyridine as a substrate. The K m and V max values were 0.40 mM and 36.0 μmol min−1 mg−1, respectively. Compared the K m values with other purified nitrilases from G. intermedia CA3-1 (12.11 mM) [16], R. sphaeroides LHS-305 (45 mM) [17], P. putida CGMCC3830 (27.9 mM) [18], and A. facilis 72W (28 mM) [19]. REH16 presented the lowest K m value and demonstrated the highest affinity toward 3-cyanopyridine among these nitrilases.
Effects of co-solvent on REH16 activity
Organic solvents are generally used as a co-solvent to increase catalytic efficiency by enhancing the solubility of a nitrile substrate in an aqueous reaction system. Therefore, six organic solvents were tested to study the effects of organic solvents on the bioconversion of 3-cyanopyridine mediated by the recombinant REH16 overexpressed in E. coli (Fig. 4). Dimethyl sulfoxide (DMSO), acetone, and tetrahydrofuran (THF) caused significant inhibition of the 3-cyanopyridine hydrolysis, signifying denaturation of the nitrilase in these organic solvents. An enhancement of the hydrolysis was observed after the addition of methanol, ethanol, or isopropanol as a co-solvent. Especially in the presence of 5–10% (v/v) of methanol or ethanol, 100% conversion of 3-cyanopyridine was achieved.
The nitrilase stability was another consideration in the co-solvent-mediated biocatalysis. On the basis of the results above, methanol, ethanol, and isopropanol were chosen for the nitrilase stability experiments. The resting E. coli cells were incubated with 10% (v/v) of one of the co-solvents for various periods and then the conversion of 3-cyanopyridine was performed to assay the residual nitrilase activity. The results are shown in Fig. 5. The resting cells showed high tolerance toward methanol and ethanol. Because there was not much difference between methanol and ethanol in the effects on the nitrilase activity, ethanol was chosen as the optimal co-solvent for the 3-cyanopyridine hydrolysis after we compared environmental effects and toxicity.
The effect of 3-cyanopyridine concentration on REH16 activity
3-Cyanopyridine is detrimental to a nitrilase, necessitating a low substrate concentration during the biotransformation. To alleviate the activity decrease, an optimal substrate concentration in the reaction needs to be determined exactly. To study the effect of 3-cyanopyridine concentration on the activity of recombinant REH16 overexpressed in E. coli, the hydrolysis at various 3-cyanopyridine concentrations (25–150 mM) was performed in a 10-mL reaction mixture.
Nitrilase REH16 could tolerate as much as 100 mM 3-cyanopyridine (Fig. 6a). At 150 mM substrate, obvious inhibition of the conversion was observed: the conversion was not completed after 90 min. Therefore, 100 mM 3-cyanopyridine in the reaction mixture may be the optimal concentration for the bioconversion.
The effect of resting-cell concentration on conversion of 3-cyanopyridine
Hydrolysis of 100 mM 3-cyanopyridine was catalyzed by 50, 75, 100, 150, and 200 mg/mL resting cells (Fig. 6b). The conversion rate of 3-cyanopyridine was increased by the increase in the resting-cell concentration. At 100 mg/mL resting cells in the reaction mixture, 3-cyanopyridine was hydrolyzed completely in 60 min. Therefore, 100 mg/mL resting cells was chosen as the optimal biocatalyst concentration for conversion of 100 mM 3-cyanopyridine.
Fed-batch production of nicotinic acid
To achieve a high nicotinic acid concentration (g product/L), a fed-batch mode was designed for 3-cyanopyridine bioconversion (Fig. 7). At the first stage, 100 mM 3-cyanopyridine was fed into the reaction system when the substrate was hydrolyzed completely in each batch. The first stage was run for eight cycles, and a total of 800 mM 3-cyanopyridine was hydrolyzed completely in 10.3 h. During the first four feedings, no nitrilase activity decrease was observed (hydrolysis rate did not decrease). Then the rate of hydrolysis of 3-cyanopyridine decreased slightly between the 4th and 7th feeding. The hydrolysis rate of 3-cyanopyridine decreased sharply after the 8th feeding: 2.4 h was needed for complete conversion of 100 mM substrate. Thus, on the second stage, the substrate feeding rate was adjusted to 50 mM per batch after the 8th feeding. And the reaction lasted for another five cycles in 10.5 h. Considering the bioconversion efficiency, the substrate feeding was terminated after the 13th feeding. Finally, a total of 1050 mM 3-cyanopyridine was hydrolyzed completely in 20.8 h, leading to 129.2 g/L production of nicotinic acid.
Several potential nitrilases from N. globerula NHB-2 [3, 4] and Rhodococcus sp. NDB 1165 [7] have been reported as biocatalysts in the form of whole cells for 3-cyanopyridine hydrolysis in fed-batch reaction mode. Using N. globerula NHB-2, a total concentration of nicotinic acid of 800 mM was formed in 18 h after 25 substrate feedings (40 mM at 20-min intervals) in a fed-batch reaction. The highest production of nicotinic acid was 1.6 M in a fed-batch reaction with 32 substrate feedings (50 mM at 20-min intervals) in 11 h using Rhodococcus sp. NDB 1165. The lower tolerance to the substrate concentration necessitated the high frequency of substrate feeding during the hydrolysis process. Compared to the feeding rate of 40 and 50 mM with the corresponding feeding intervals, 100 mM feedings were performed in the present work, suggesting that REH16 has better substrate tolerance.
Conclusions
A new aliphatic nitrilase, REH16 from R. eutropha H16, was identified. REH16 has high activity toward aliphatic and heterocyclic nitriles. By means of resting cells of E. coli overexpressing nitrilase REH16, a total of 1050 mM 3-cyanopyridine was hydrolyzed completely in 20.8 h in periodical fed-batch reaction mode, yielding 129.2 g/L production of nicotinic acid and pointing to a good potential for industrial application.
References
Carlson LA (2005) Nicotinic acid: the broad-spectrum lipid drug. A 50th anniversary review. J Intern Med 258:94–114
Hatanaka M, Tanaka N (1993) Process for producing pyridinecarboxylic acid. WO patent 93/05022
Sharma NN, Sharma M, Kumar H, Bhalla TC (2006) Nocardia globerula NHB-2: bench scale production of nicotinic acid. Process Biochem 41:2078–2081
Sharma NN, Sharma M, Bhalla TC (2011) An improved nitrilase-mediated bioprocess for synthesis of nicotinic acid from 3-cyanopyridine with hyperinduced Nocardia globerula NHB-2. J Ind Microbiol Biotechnol 38:1235–1243
Almatawah QA, Cowan DA (1999) Thermostable nitrilase catalyzed production of nicotinic acid from 3-cyanopyridine. Enzym Microb Technol 25:718–724
Pai O, Banoth L, Ghosh S, Ghisti Y, Banerjee UC (2014) Biotransformation of 3-cyanopyridine to nicotinic acid by free and immobilized cells of recombinant Escherichia coli. Process Biochem 49:655–659
Prasad S, Misra A, Jangir VP, Awasthi A, Raj J, Bhalla TC (2007) A propionitrile-induced nitrilase of Rhodococcus sp. NDB 1165 and its application in nicotinic acid synthesis. World J Microbiol Biotechnol 23:345–353
Mehta PK, Bhatia SK, Bhatia RK, Bhalla TC (2014) Bench scale production of nicotinic acid using a versatile amide-hydrolysing Geobacillus subterraneus RL-2a isolated from thermal spring of Manikaran, India. J Mol Catal B Enzym 105:58–65
Gong JS, Lu ZM, Li H, Shi JS, Zhou ZM, Xu ZH (2012) Nitrilase in nitrile biocatalysis: recent progress and forthcoming research. Microb Cell Fact 11:142
Polaina J, MacCabe AP (2007) Industrial enzymes. Springer, Netherlands, pp 531–547
Martinkova L, Rucka L, Nesvera J, Patek M (2017) Recent advances and challenges in the heterologous production of microbial nitrilases for biocatalytic applications. World J Microbiol Biotechnol 33:8
Hann EC, Sigmund AE, Hennessey SM, Gavagan JE, Short DR, Bassat AB, Chauhan S, Fallon RD, Rayne MS, DiCosimo R (2002) Optimization of an immobilized-cell biocatalyst for production of 4-cyanopentanoic acid. Org Process Res Dev 6:492–496
Wang H, Fan H, Sun H, Zhao L, Wei D (2015) Process development for the production of (R)-(−)-mandelic acid by recombinant Escherichia coli cells harboring nitrilase from Burkholderia cenocepacia J2315. Org Process Res Dev 19:2012–2016
Jin LQ, Liu ZQ, Xu JM, Zheng YG (2013) Biosynthesis of nicotinic acid from 3-cyanopyridine by a newly isolated Fusarium proliferatum ZJB-09150. World J Microbiol Biotechnol 29:431–440
Badoei-Dalfard A, Karami Z, Ramezani-pour N (2016) Bench scale production of nicotinic acid using a newly isolated Stenotrophomonas maltophilia AC21 producing highly-inducible and versatile nitrilase. J Mol Catal B Enzym. doi:10.1016/j.molcatb.2016.11.019
Gong JS, Li H, Zhu XY, Lu ZM, Wu Y, Shi JS, Xu ZH (2012) Fungal His-tagged nitrilase from Gibberella intermedia: gene cloning, heterologous expression and biochemical properties. PLoS One 7(11):e50622
Yang C, Wang X, Wei D (2011) A new nitrilase-producing strain named Rhodobacter sphaeroides LHS-305: biocatalytic characterization and substrate specificity. Appl Biochem Biotechnol 165:1556–1567
Zhu XY, Gong JS, Li H, Lu ZM, Zhou ZM, Shi JS, Xu ZH (2013) Characterization and functional cloning of an aromatic nitrilase from Pseudomonas putida CGMCC3830 with high conversion efficiency toward cyanopyridine. J Mol Catal B Enzym 97:175–183
Li H, Dong W, Zhang Y, Liu K, Zhang W, Zhang M, Ma J, Jiang M (2017) Enhanced catalytic efficiency of nitrilase from Acidovorax facilis 72W and application in bioconversion of 3-cyanopyridine to nicotinic acid. J Mol Catal B Enzym. doi:10.1016/j.molcatb.2017.03.010
Wang H, Sun H, Wei D (2013) Discovery and characterization of a highly efficient enantioselective mandelonitrile hydrolase from Burkholderia cenocepacia J2315 by phylogeny-based enzymatic substrate specificity prediction. BMC Biotechnol 13:14
Sun H, Gao W, Fan H, Wang H, Wei D (2015) Cloning, purification and evaluation of the enzymatic properties of a novel arylacetonitrilase from Luminiphilus syltensis NOR5-1B: a potential biocatalyst for the synthesis of mandelic acid and its derivatives. Biotechnol Lett 37:1655–1661
Weatherburn MW (1967) Phenol-hypochlorite reaction for determination of ammonia. Anal Chem 39:971–974
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 21406068/B060804), the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fan, H., Chen, L., Sun, H. et al. A novel nitrilase from Ralstonia eutropha H16 and its application to nicotinic acid production. Bioprocess Biosyst Eng 40, 1271–1281 (2017). https://doi.org/10.1007/s00449-017-1787-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-017-1787-x