Abstract
Adipose-derived mesenchymal stem cells (ADSCs) are an important source of stem cells for tissue repair and regeneration but the regulatory mechanism of stem cell differentiation is still unclear. Runt-related gene 2 (Runx2) is a bone-specific transcription factor that plays an important role in promoting osteogenic differentiation. Protein levels of Runx2 are regulated by non-coding RNA. In order to identify the regulatory mechanism underlying non-coding RNA regulation of Runx2, we employed bioinformatics analysis, quantitative reverse transcription PCR (qRT-PCR), osteoblast differentiation induction, immunohistochemical and bifluorescein reporter experiments. The results showed that expression of long non-coding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and Runx2 was increased in ADSCs induced in osteogenic differentiation media for 21 days, while miR-30 expression was downregulated. qRT-PCR and alkaline phosphatase (ALP) histochemical staining assays demonstrated that knockdown of lncRNA MALAT1 or overexpression of miR-30 suppressed Runx2-mediated osteoblast differentiation by suppressing osteocalcin (OCN), osteopontin (OPN) and osterix (OSX) expression. Overexpressing Runx2 reversed the inhibitory effect of miR-30 on osteogenic differentiation of ADSCs. Bifluorescein report experiments confirmed that miR-30 is a potential target of lncRNA MALAT1 and Runx2 is a potential target of miR-30. Taken together, the results suggested that the expression of lncRNA MALAT1 promoted Runx2-mediated osteogenic differentiation of ADSCs by targeting miR-30.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Adipose-derived mesenchymal stem cells (ADSCs) have the capacity of self-renewal and multilineage differentiation and are therefore useful to regenerate or repair bone tissue (Madhurakkat Perikamana et al. 2018; Zhu et al. 2018). The production of ADSCs is greater and proliferation is faster than bone marrow-derived mesenchymal stem cells, so ADSCs are commonly used in clinical applications (Fernandes et al. 2018; Su et al. 2015; Zhang et al. 2009). However, the precise molecular mechanisms of differentiation remain unclear, although a great deal of research has examined ADSC differentiation into specific lineages.
Non-coding RNAs including long non-coding RNA (lncRNA) and microRNA (miRNA) are thought to play a key role in osteoblast differentiation through various regulatory mechanisms including chromatin modification, transcription factor binding, competent endogenous mechanism and other posttranscriptional mechanisms (Niu et al. 2018; Peng et al. 2018; Yang et al. 2018). Increasing evidence has verified that lncRNA plays an important role in regulating osteogenic differentiation. Gao et al. found that lncRNA KCNQ1OT1 promotes osteogenic differentiation to relieve osteolysis via Wnt/β-catenin activation (Gao et al. 2018a). In human ADSCs, microarray detection found that abnormal expression of lncRNA H19 was involved in osteogenic differentiation (Huang et al. 2017). Silencing of H19 caused a significant increase in expression of osteogenesis-related genes, including ALP and RUNX2, while decreased expression was observed after H19 overexpression. Ectopic expression of lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has also been observed in calcific valves and after osteogenic induction of human aortic valve interstitial cells (Xiao et al. 2017). In vitro experiments revealed that lncRNA MALAT1 acted as a positive regulator of osteogenic differentiation by repressing miR-204 expression and activity and thereby promoting expression of osteoblast-specific markers, including alkaline phosphatase (ALP), osteocalcin (OCN) and mineralized bone matrix formation (Xiao et al. 2017). However, the role of lncRNA MALAT1 in the regulation of ADSC osteogenic differentiation is still unclear. So, the aim of this study is to illustrate the role of lncRNA MALAT1 in the osteogenic differentiation of ADSCs.
In the current study, we found that expression of lncRNA MALAT1 was significantly increased during osteogenic differentiation of ADSCs. We demonstrated that lncRNA MALAT1 promoted osteogenesis and its pro-osteogenic function was at least partially mediated by sponging miR-30. Moreover, our results showed that miR-30 not only downregulated Runx2 by interacting with the 3′-UTR of Runx2 but also suppressed Runx2 expression at the mRNA level. These results increased our understanding of the role of the lncRNA-miRNA-mRNA functional network in osteogenesis and bone development.
Materials and methods
Cell culture and identification
Primary human ADSCs (ScienCell, San Diego, CA, USA, http://sciencellonline.com) were cultured in growth medium (GM) consisting of a minimum essential medium supplemented with 10% fetal bovine serum and 1% antibiotics. Osteoblast differentiation of human ADSCs was induced at 70–80% confluence using osteogenic medium (OM; standard GM supplemented with 100 nM dexamethasone, 200 μM L ascorbic acid and 10 mM β-glycerophosphate). The OM was changed every 2 days and cells were harvested at the indicated times after osteoblast differentiation.
For ADSC identification, ADSCs reaching 80–90% confluency were detached with 0.02% ethylenediaminetetraacetic acid/0.25% trypsin (Sigma-Aldrich, St. Louis, MO, USA) for 5 min at room temperature and then replated. For phenotypic analysis, fluorescein isothiocyanate (FITC-F), or phycoerythrin (PE) was used. The expression of the following markers was investigated: CD29, CD90, CD44, CD105 and von Willebrand factor (vWF). An IgG-matched isotype served as the internal control for each antibody. For normoxic cultures, ADSCs were cultured in 95% air (20% O2) and 5% CO2. For hypoxia studies, ADSCs were cultured in a multigas incubator (ASTEC) that was flushed with a humidified gas mixture composed of 2% O2:5% CO2:93% N2.
To identify the capacity for multilineage differentiation of ADSCs, ADSCs were cultured under the following differentiation conditions. For adipocyte differentiation, ADSCs were cultured in adipogenic differentiation medium and Oil Red O staining was performed to assess adipocyte differentiation after 14 days. For osteoblast differentiation, ADSCs were cultured in osteogenic differentiation medium and cells were stained with Alizarin Red after 21 days to monitor osteogenesis.
The 293T cells were obtained from the American Type Culture Collection (Manassas, VA, USA; http://www.atcc.org) and cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum and 1% antibiotics.
Alkaline phosphatase staining and activity
Cells cultured for 21 days under PM or OM were assayed for ALP staining and activity, as described previously (Jin et al. 2016). ALP staining was performed according to the protocol of the NBT/BCIP staining kit (CoWin Biotech, Beijing, China). ALP activity was analyzed using an ALP activity colorimetric assay kit (BioVision, Milpitas, CA, USA).
Cell transfection
Silencing RNA (siRNA) against lncRNA MALAT1, the Runx2 overexpression vector and miR-30 mimic vector were designed and synthesized by GenePharma Company (Shanghai, China). Cell transfection was performed using Lipofectamine 2000 reagent (Life Technologies Corporation, Bethesda, USA) following the manufacturer’s protocol. Quantitative reverse transcription PCR (qRT-PCR) was used to confirm the transfection efficiency after transfection for 48 h.
qRT-PCR
Total RNA in cells was extracted using the TRIzol™ Plus RNA Purification Kit (Invitrogen, CA, USA) in accordance with the manufacturer’s protocol. The SuperScript™ One-Step RT-PCR System with pTRUEscript 1st Strand cDNA Synthesis Kit (Aidlab, Beijing, China) was used for the real-time PCR analysis to detect the expression of relative gene or on-coding RNA. The primer sequences for miR-30, LncRNA MALAT1, siLncRNA MALAT1, osteocalcin (OCN), osteopontin (OPN), osterix (OSX), runt-related transcription factor 2 (Runx2), U6 and D-glyceraldehde-3-phosphate dehydrogenase (GAPDH) are shown in Table 1. U6 was used for internal controls of miR-130; others used GAPDH as internal controls.
The expression level of lncRNA MALAT1, miR-30, Runx2, osteopontin (OPN), OCN and osterix (OSX) in cells was measured using a TaqMan MicroRNA Reverse Transcription Kit and TaqMan Universal Master Mix II (Applied Biosystems, Foster City, CA, USA). All experiments were performed three times and the relative RNA expression rates were calculated using the 2-△△Ct method (Zhao et al. 2018).
Bifluorescein report experiments
The 3′-UTR fragment of Runx2 or lncRNA MALAT1 carrying the predicted target site of miR-30 was amplified using PCR and inserted into the PYr-MirTarget luciferase vector (Promega, Madison, WI, USA). The resultant constructs were named Runx2-wild-type (Runx2 3′-UTR-WT) or lncRNA MALAT1-wild-type (lncRNA MALAT1-WT). A mutated 3′-UTR fragment of Runx2 or lncRNA MALAT1 lacking the predicted target site of miR-30 was similarly amplified and inserted into the PYr-MirTarget luciferase vector. These constructs were named Runx2-mutated-type (Runx2 3′-UTR-Mut) or lncRNA MALAT1-mutated-type (lncRNA MALAT1-Mut). The 293T cells were co-transfected with miR-30 negative control (NC)/mimic and Runx2 3′-UTR-WT/Mut or lncRNA MALAT1-WT/Mut using Lipofectamine 2000 following the manufacturer’s protocol. Reporter assays were performed using the dual-luciferase assay system (Promega) following the manufacturer’s instructions.
Western blotting
Protein was extracted from cells using RIPA lysis buffer containing proteinase inhibitor (Sigma-Aldrich). Protein concentration was determined using the BCA Protein Assay Kit (Vigorous Biotechnology Beijing Co. Ltd., Beijing, China). Equal amounts of protein lysates (20 μg per lane) were separated using 10% SDS-PAGE gels and then electroblotted onto nitrocellulose membranes (Millipore, Danvers, MA, USA). Membranes were blocked for 2 h with 5% non-fat dry milk in Tris-buffered saline containing 0.1% Tween 20 and incubated at 4 °C overnight with the following primary antibodies: mouse monoclonal anti-human Runx2 (1:500, Santa Cruz Biotechnology) and mouse monoclonal anti-human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:5000, Santa Cruz Biotechnology). GAPDH was used as an internal control for protein loading. The membrane was then incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:5000, Santa Cruz Biotechnology) for 1 h at room temperature. The immune complexes were detected by enhanced chemiluminescence (ECL, Cell Signaling Technology, Danvers, MA, USA). The integrated density of the band was quantified by Quantity One software (Bio-Rad, Hercules, CA, USA).
Statistical analysis
Continuous variables are presented as means ± standard deviation (SD). One-way analysis of variance was carried out for multiple comparisons using GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). P values ≤ 0.05 indicate a statistically significant difference.
Results
lncRNA MALAT1 was upregulated but its target miR-30 was downregulated during osteoblast differentiation of human ADSCs
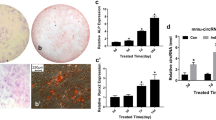
In order to identify if the expression of lncRNA MALAT1 and miR-30 was related to osteogenic differentiation of ADSCs, primary human ADSCs were examined. The results showed that ADSCs were positive for the mesenchymal stem cell markers CD29, CD90, CD44 and CD105, and negative for the endothelial marker vWF (Fig. 1a–f); meanwhile, the phase contrast image showed that the cells had a long filamentous structure (Fig. 1g), indicating that the cells were ADSCs. Oil Red O and ALP staining indicated that the ADSCs acquired adipogenic (Fig. 1h) and osteogenic differentiation (Fig. 1i) features under adipogenic or osteogenic induction conditions.
Characteristics of ADSCs and multipotent differentiation potential. (a–f) Detection of cell surface markers by immunofluorescence staining with antibodies against CD29 (a), CD90 (b), CD44 (c), CD105 (d) and vWF (e) labeled with FITC (green color) or PE (red color). A negative FITC- and PE-labeled mouse IgG isotype control (f) was included (magnification × 200). Scale bars 50 μm. (g) ADSCs showed a typical cobblestone-like morphology. (h and i) Differentiation potential of ADSCs assessed by Oil Red O (h) and Alizarin Red staining (i). Scale bars 100 μm
To investigate the functions of lncRNA MALAT1 and miR-30, we examined their dynamic expression profiles in ADSCs after induction to the osteogenic lineage. The results showed that compared with the control group, the expression of lncRNA MALAT1 was increased over time (Fig. 2a), while the expression of miR-30 was downregulated (Fig. 2b). The qRT-PCR experiments showed that expression of osteoblastic markers Runx2, OPN, OCN and OSX was increased during osteogenesis (Fig. 2c–f).
Expression patterns of lncRNA MALAT1 and miR-30 during osteoblast differentiation of ADSCs. (a) qRT-PCR analysis was used to detect the expression of lncRNA MALAT1 during osteoblast differentiation of ADSCs at days 0, 3, 7, 14 and 21. RNA expression at the indicated time points was normalized to day 0. GAPDH was used as an internal control. (b) Relative miR-30 transcript levels at the indicated time points. U6 was used as an internal control. (c–f) qRT-PCR detection shows the expression of osteoblastic markers Runx2 (c), OPN (d), OCN (e) and OSX (f) on selected days. GAPDH was used as internal control. Data are presented as means ± SD. Asterisk indicates P < 0.05, triple asterisks indicate P < 0.001 vs. control. Runx2, runt-related transcription factor 2; OPN, osteopontin; OCN, osteocalcin; OSX, osterix
Expression of lncRNA MALAT1 promoted osteogenic differentiation of ADSCs
In order to determine if lncRNA MALAT1 expression played a role in promoting osteogenic differentiation, siRNA against lncRNA MALAT1 was constructed and transfected into ADSCs. Expression of lncRNA MALAT1 was significantly downregulated (fold change 1, 0.25) compared with non-transfected control cells or the NC group (Fig. 3a). ADSCs with or without lncRNA MALAT1 knockdown were then cultured in GM or OM for 21 days. The results showed that ALP activity was increased in OM compared with GM. In contrast, knockdown of lncRNA MALAT1 suppressed OM-induced osteogenic differentiation (Fig. 3b–b″′ and c). qRT-PCR assays also showed that knockdown of lncRNA MALAT1 reversed OM-induced osteogenic differentiation by inhibiting expression of osteoblastic markers Runx2, OPN, OCN and OSX (Fig. 3d–g).
The expression of lncRNA MALAT1 promoted osteogenic differentiation of ADSCs. (a) qRT-PCR detection shows expression of lncRNA MALAT1 in ADSCs after transfection with siRNA against lncRNA MALAT1 (siMALAT1) or the negative control (NC) vector for 48 h. GAPDH was used as an internal control. Data are presented as means ± SD. Triple asterisks indicate P < 0.001 vs. control. (b–b″′) Images of ALP staining in ADSCs after culture in GM or OM for 14 days. Scale bars 100 μm. (c) Histograms show ALP activity and quantification of Alizarin Red staining by spectrophotometry. Data are presented as means ± SD. Triple asterisks indicate P < 0.001 vs. control. (d–g) Relative mRNA expression of Runx2 (d), OPN (e), OCN (f), and OSX (g) was measured by qRT-PCR at day 14 of OM induction. GAPDH was used for normalization. Data are presented as means ± SD. Double asterisks indicate P < 0.01, triple asterisks indicate P < 0.001. ALP, alkaline phosphatase
Expression of miR-30 suppressed osteogenic differentiation of ADSCs
To examine if miR-30 expression played a role in promoting osteogenic differentiation, a miR-30 overexpression vector (mimic) was constructed and transfected into ADSCs. The results showed that the expression of miR-30 was significantly increased (fold change 1, 7) in mimic-transfected cells compared with the control or NC group (Fig. 4a). We then cultured ADSCs with or without miR-30 overexpression in GM or OM for 21 days. The results showed that ALP activity was increased in OM compared with GM induction conditions, while overexpression of miR-30 suppressed OM-induced osteogenic differentiation (Fig. 4b–b″′ and c). qRT-PCR assays also demonstrated that overexpressing miR-30 reversed OM-induced osteogenic differentiation by inhibiting expression of osteoblastic markers Runx2, OPN, OCN and OSX expression (Fig. 4d–g).
Expression of miR-30 suppressed osteogenic differentiation of ADSCs. (a) qRT-PCR detection shows expression of miR-30 in ADSCs after transfection with miR-30 mimics or NC for 48 h. U6 was used as an internal control. Data are presented as means ± SD. Triple asterisks indicate P < 0.001 vs. control. (b–b″′) Images of ALP staining in ADSCs after culture in OM for 14 days. Scale bars 100 μm. (c) Histograms show ALP activity and quantification of Alizarin Red staining by spectrophotometry. Data are presented as means ± SD. Triple asterisks indicate P < 0.001 vs. control. (d–f) Relative mRNA expression of Runx2, OPN, OCN and OSX was measured by qRT-PCR at day 14 of OM induction. GAPDH was used for normalization. Double asterisks indicate P < 0.01, triple asterisks indicate P < 0.001
miR-30 is a potential target of lncRNA MALAT1
To determine the possible interaction between miR-30 and Runx2, we first performed a bioinformatics screen for possible miR-30 target genes, using an online 3′-UTR binding site prediction database (http://starbase.sysu.edu.cn/). Overlap analyses showed that miR-30 has a broadly conserved binding site with lncRNA MALAT1. A mutated version of lncRNA MALAT1 was constructed in which 15 complementary nucleotides in the binding site were altered (Fig. 5a). This mutated construct was fused to the luciferase coding region and co-transfected into 293T cells along with miR-30 mimics. The relative luciferase activity showed that when the wild-type lncRNA MALAT1 was co-transfected with miR-30 mimics, luciferase activity was significantly decreased compared with co-transfection with the control miRNA (Fig. 5b). However, this effect was not observed after mutation of lncRNA MALAT1, indicating that miR-30 specifically targets lncRNA MALAT1. The qRT-PCR analyses further confirmed that downregulation of lncRNA MALAT1 expression significantly enhanced miR-30 expression in vitro (Fig. 5c).
miR-30 is a potential target of lncRNA MALAT1. (a) Complementary bases between the sequences are labeled with red font. The sequence of the mutant lncRNA MALAT1 construct is also shown as underlined. (b) Dual-luciferase reporter assay of 293T cells co-transfected with lncRNA MALAT1-WT, or lncRNA MALAT1-Mut and with miR-30 mimic or miR-NC. (c) miR-30 expression in ADSCs was measured by qRT-PCR following transfection with siRNA against lncRNA MALAT1 for 48 h. Data are presented as means ± SD. Asterisk indicates P < 0.05, double asterisks indicate P < 0.01, triple asterisks indicate P < 0.001 vs. control
Runx2 overexpression reversed the inhibitory effect of miR-30 on osteogenic differentiation of ADSCs
We sought to test the role of Runx2 in miR-30-mediated inhibition of osteogenic differentiation by transfecting a Runx2 overexpression vector into ADSCs. Compared to non-transfected control or NC cells, transfection of the overexpression vector significantly increased Runx2 expression (Fig. 6a). ADSCs overexpressing Runx2 or miR-30 were then cultured in OM for 21 days, resulting in decreased levels of ALP activity when miR-30 was overexpressed under OM conditions. However, overexpressing Runx2 reversed the inhibitory effect of miR-30 on osteogenic differentiation of ADSCs under OM induction (Fig. 6b–b″ and c). qRT-PCR analysis further demonstrated that overexpression of Runx2 reversed the inhibitory effect of miR-30 on osteogenic differentiation by promoting expression of osteoblastic markers OPN, OCN and OSX (Fig. 6d–f).
Runx2 overexpression reversed the inhibitory effect of miR-30 on osteogenic differentiation of ADSCs. (a) qRT-PCR detection shows expression of Runx2 in ADSCs transfected with miR-30 mimics, Runx2 overexpression vector alone, or both vectors combined for 48 h. GAPDH was used as an internal control. Data are presented as means ± SD. Asterisk indicates P < 0.05, double asterisks indicate P < 0.01, triple asterisks indicate P < 0.001 vs. control. (b–b″) Images of ALP staining in ADSCs transfected with miR-30 mimic or Runx2 overexpression vector after culture in OM for 21 days. Scale bars 100 μm. (c) Histograms show ALP activity and quantification of Alizarin Red staining by spectrophotometry. Data are presented as means ± SD. Triple asterisks indicate P < 0.001 vs. control. Triple number signs indicate P < 0.001 vs. the mimic group. (d–f) Relative mRNA expression of OPN (d), OCN (e) and OSX (f) was measured by qRT-PCR at day 14 of OM induction. GAPDH was used for normalization. Data are presented as means ± SD. Triple asterisks indicate P < 0.001 vs. control. Triple number signs indicate P < 0.001 vs. mimic group
To assess the possible interaction between miR-30 and Runx2, we first performed a bioinformatics screen for miR-30 target genes using the previously employed 3′-UTR binding site prediction database. Overlap analyses showed that miR-30 has a broadly conserved binding site in the 3′-UTR of Runx2. A mutated version of the Runx2 3′-UTR was then constructed in which 12 complementary nucleotides in the binding site were altered (Fig. 7a). This mutated construct was fused to the luciferase coding region and co-transfected into 293T cells along with miR-30 mimics. The relative luciferase activity showed that when the wild-type 3′-UTR of Runx2 was co-transfected with miR-30 mimics, luciferase activity was significantly decreased compared with co-transfection with the control miRNA (Fig. 7b). However, this effect was not observed using the mutated 3′-UTR of Runx2, indicating that miR-30 can specifically bind to the 3′-UTR of Runx2. Western blot and qRT-PCR analyses further confirmed that overexpression of miR-30 significantly suppressed Runx2 expression in vitro (Fig. 7c, d).
Runx2 is a potential target of miR-30. (a) Sequence alignment between miR-30 and Runx2. (b) Complementary bases between the sequences are underlined. The sequence of the mutant Runx2 construct is also shown. (c) Dual-luciferase reporter assay of 293T cells co-transfected with Runx2 3′-UTR-WT or Runx2 3′-UTR-Mut and with miR-30 mimic or miR-NC. (d) qRT-PCR and western blot detection show the expression of Runx2 in ADSCs following transfection with miR-30 mimic or miR-NC for 48 h. Data are presented as means ± SD. Triple asterisks indicate P < 0.001 vs. control
Discussion
This study demonstrated that lncRNA MALAT1 promotes the osteogenic differentiation of ADSCs, in agreement with previous studies (Peng et al. 2018; Xiao et al. 2017). In addition, our study also showed that the expression of lncRNA MALAT1 promoted the osteogenic differentiation of ADSCs by sponging miR-30. Knockdown of lncRNA MALAT1 significantly suppressed OM-induced osteogenic differentiation in in vitro experiments. Our study also demonstrated that miR-30 was downregulated during osteogenic differentiation of ADSCs, while overexpression of miR-30 significantly suppressed osteogenic differentiation of ADSCs. Luciferase activity assay experiments further confirmed that lncRNA MALAT1 interacted with miR-30 and suppressed its regulatory function by sponging miR-30 to limit its availability.
Previous studies have found that miR-30 family members negatively regulate osteoblast differentiation (Eguchi et al. 2013; Tian et al. 2015; Wu et al. 2012). BMP9-induced osteogenic differentiation is partially inhibited by miR-30a in the mesenchymal stem cell line C3H10T1/2 (Zhang et al. 2015). In the current study, we found that expression of miR-30 can significantly suppress Runx2 expression by interacting with the 3′-UTR of Runx2 at the mRNA level. Dual-luciferase reporter assays confirmed that Runx2 is a direct target of miR-30. Runx2 is an important osteoblast lineage-determining transcription factor involved in directing osteoblastic differentiation and Runx2 appears to be the master gene in osteogenesis as it induces the expression of OCN, OSX and OPN, which are all required for terminal osteoblast differentiation (Ducy et al. 1997; Komori 2002; Liu et al. 2018; Sun et al. 2018). Related to these findings, our study found that the expression of Runx2 was increased during osteogenic differentiation of ADSCs. Overexpression of Runx2 significantly reversed the inhibitory effect of miR-30 on ADSC osteogenic differentiation.
Besides miR-30, Gao et al. recently found that lncRNA MALAT1 promotes osterix expression to regulate osteogenic differentiation by targeting miR-143 in human bone marrow-derived mesenchymal stem cells (hBMSCs) (Gao et al. 2018b). In this study, we found that lncRNA MALAT1 promotes osteogenic differentiation by targeting miRNA-30 and by promotion of Runx2 expression in ADSCs. But miR-143 involved in the lncRNA MALAT1-mediated osteogenic differentiation in ADSCs is still unclear.
In conclusion, our in vitro study using transgenic ADSCs was the first demonstration to the best of our knowledge that lncRNA MALAT1 can induce osteogenesis by sponging miR-30. Our findings were also consistent with the ability of lncRNA MALAT1-modified ADSCs to promote osteoblast differentiation by targeting miR-30/Runx2. Although our results were very promising, further experimentation will still be needed to determine if lncRNA MALAT1-modified ADSCs can also promote osteogenesis in vivo. Despite this limitation, our study supported the therapeutic promise of lncRNA MALAT1 in promoting osteogenic differentiation.
References
Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747–754
Eguchi T, Watanabe K, Hara ES, Ono M, Kuboki T, Calderwood SK (2013) OstemiR: a novel panel of microRNA biomarkers in osteoblastic and osteocytic differentiation from mesencymal stem cells. PLoS One 8:e58796
Fernandes M, Valente SG, Sabongi RG, Gomes Dos Santos JB, Leite VM, Ulrich H, Nery AA, da Silva Fernandes MJ (2018) Bone marrow-derived mesenchymal stem cells versus adipose-derived mesenchymal stem cells for peripheral nerve regeneration. Neural Regen Res 13:100–104
Gao X, Ge J, Li W, Zhou W, Xu L (2018a) LncRNA KCNQ1OT1 promotes osteogenic differentiation to relieve osteolysis via Wnt/beta-catenin activation. Cell Biosci 8:19
Gao Y, Xiao F, Wang C, Cui P, Zhang X, Chen X (2018b) Long noncoding RNA MALAT1 promotes osterix expression to regulate osteogenic differentiation by targeting miRNA-143 in human bone marrow-derived mesenchymal stem cells. J Cell Biochem 119:6986–6996
Huang G, Kang Y, Huang Z, Zhang Z, Meng F, Chen W, Fu M, Liao W (2017) Identification and characterization of long non-coding RNAs in osteogenic differentiation of human adipose-derived stem cells. Cell Physiol Biochem 42:1037–1050
Jin C, Jia L, Huang Y, Zheng Y, Du N, Liu Y, Zhou Y (2016) Inhibition of lncRNA MIR31HG promotes osteogenic differentiation of human adipose-derived stem cells. Stem Cells 34:2707–2720
Komori T (2002) Runx2, a multifunctional transcription factor in skeletal development. J Cell Biochem 87:1–8
Liu L, Wang J, Song X, Zhu Q, Shen S, Zhang W (2018) Semaphorin 3A promotes osteogenic differentiation in human alveolar bone marrow mesenchymal stem cells. Exp Ther Med 15:3489–3494
Madhurakkat Perikamana SK, Lee J, Ahmad T, Kim EM, Byun H, Lee S, Shin H (2018) Harnessing biochemical and structural cues for tenogenic differentiation of adipose derived stem cells (ADSCs) and development of an in vitro tissue interface mimicking tendon-bone insertion graft. Biomaterials 165:79–93
Niu G, Zhuang H, Li B, Cao G (2018) Long noncoding RNA linc-UBC1 promotes tumor invasion and metastasis by regulating EZH2 and repressing E-cadherin in esophageal squamous cell carcinoma. J BUON 23:157–162
Peng S, Cao L, He S, Zhong Y, Ma H, Zhang Y, Shuai C (2018) An overview of long noncoding RNAs involved in bone regeneration from mesenchymal stem cells. Stem Cells Int 2018:8273648
Su X, Liao L, Shuai Y, Jing H, Liu S, Zhou H, Liu Y, Jin Y (2015) MiR-26a functions oppositely in osteogenic differentiation of BMSCs and ADSCs depending on distinct activation and roles of Wnt and BMP signaling pathway. Cell Death Dis 6:e1851
Sun M, Chi G, Li P, Lv S, Xu J, Xu Z, Xia Y, Tan Y, Li L, Li Y (2018) Effects of matrix stiffness on the morphology, adhesion, proliferation and osteogenic differentiation of mesenchymal stem cells. Int J Med Sci 15:257–268
Tian Y, Liu J, Bai X, Tan X, Cao Y, Qin K, Zhao Z, Zhang Y (2015) MicroRNA expression profile of surgical removed mandibular bone tissues from patients with mandibular prognathism. J Surg Res 198:127–134
Wu T, Zhou H, Hong Y, Li J, Jiang X, Huang H (2012) miR-30 family members negatively regulate osteoblast differentiation. J Biol Chem 287:7503–7511
Xiao X, Zhou T, Guo S, Guo C, Zhang Q, Dong N, Wang Y (2017) LncRNA MALAT1 sponges miR-204 to promote osteoblast differentiation of human aortic valve interstitial cells through up-regulating Smad4. Int J Cardiol 243:404–412
Yang Q, Jia L, Li X, Guo R, Huang Y, Zheng Y, Li W (2018) Long noncoding RNAs: new players in the osteogenic differentiation of bone marrow- and adipose-derived mesenchymal stem cells. Stem Cell Rev 14:297–308
Zhang P, Luo X, Wang H (2009) Clinical transplantation of a tissue-engineered airway. Lancet 373:718 author reply 718-719
Zhang R, Weng Y, Li B, Jiang Y, Yan S, He F, Chen X, Deng F, Wang J, Shi Q (2015) BMP9-induced osteogenic differentiation is partially inhibited by miR-30a in the mesenchymal stem cell line C3H10T1/2. J Mol Histol 46:399–407
Zhao J, Zhang C, Gao Z, Wu H, Gu R, Jiang R (2018) Long non-coding RNA ASBEL promotes osteosarcoma cell proliferation, migration, and invasion by regulating microRNA-21. J Cell Biochem 119:6461–6469
Zhu Y, Wu Y, Cheng J, Wang Q, Li Z, Wang Y, Wang D, Wang H, Zhang W, Ye J, Jiang H, Wang L (2018) Pharmacological activation of TAZ enhances osteogenic differentiation and bone formation of adipose-derived stem cells. Stem Cell Res Ther 9:53
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
1. The expression of lncRNA MALAT1 promoted osteogenic differentiation of ADSCs.
2. The expression of miR-30 suppressed osteogenic differentiation of ADSCs.
3. The miR-30 is a potential target of lncRNA MALAT1.
Rights and permissions
About this article
Cite this article
Yi, J., Liu, D. & Xiao, J. LncRNA MALAT1 sponges miR-30 to promote osteoblast differentiation of adipose-derived mesenchymal stem cells by promotion of Runx2 expression. Cell Tissue Res 376, 113–121 (2019). https://doi.org/10.1007/s00441-018-2963-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-018-2963-2