Abstract
Adipose-derived mesenchymal stem cells (ADSCs) are considered to be seed cells in bone tissue engineering and emerging evidence indicates that circular RNAs (circRNAs) function in the osteogenic differentiation of ADSCs. The mechanisms of osteoblastic differentiation of ADSCs from the perspective of circRNA modulation are examined in this study. First, circRNA-23525 was upregulated during osteoblastic differentiation of ADSCs. Second, overexpression of circRNA-23525 increased Runx2, ALP and OCN at both mRNA and protein levels. Alkaline phosphatase (ALP) and Alizarin Red staining indicated a similar tendency. Silencing circRNA-23525 produced the opposite effect. Bioinformatics analysis with luciferase assays confirmed that circRNA-23525 functioned as a sponge for miR-30a-3p. In the osteoblastic differentiation of ADSCs, the dynamic expression of miR-30a-3p and circRNA-23525 resulted in an opposite trend at 3, 7 and 14 days. Overexpression of circRNA-23525 downregulated miR-30a-3p and knockdown of circRNA-23525 promoted the expression of miR-30a-3p. Bioinformatics methods and luciferase assays suggested that miR-30a-3p modulated Runx2 expression by targeting 3′UTR. Knockdown of miR-30a-3p facilitated osteogenesis in ADSCs and enhancing miR-30a-3p interfered with the osteogenic process. Finally, circRNA-23525 overexpression significantly increased Runx2 expression, while co-transfection of miR-30a-3p mimics reversed it. Runx2 expression was decreased in circRNA-23525-knockdown ADSCs but expression was rescued by including the miR-30a-3p inhibitor in the osteoblastic process. ALP activity and mineralized bone matrix confirmed the function of circRNA-23525/miR-30a-3p in osteogenesis. Taken together, the current study demonstrated that circRNA-23525 regulates Runx2 expression via targeting miR-30a-3p and is thus a positive regulator in the osteoblastic differentiation of ADSCs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bone remodeling, the process of resorption and formation of bone, takes place throughout adulthood (Martin et al. 2009). Various bone-related diseases, such as osteoporosis, osteoarthritis and severe trauma, may cause decreased bone strength, reduced bone mass and microarchitectural degeneration of bone tissue (Compston et al. 2019; Vicente et al. 2016). Adipose-derived mesenchymal stem cells (ADSCs), characterized by multi-differentiation and self-renewal, are mesenchymal stem cells (MSCs) isolated from adipose tissue and capable of bone regeneration and reconstruction (Griffin et al. 2017; Bhumiratana et al. 2016). Due to several factors such as the wide range of adipose tissue, convenient material extraction, limited damage to the donor areas and ease of genetic modification (Zuk et al. 2001; Kern et al. 2006), ADSCs have become extensively useful as seed cells in bone tissue engineering, suggesting their far-reaching potential in clinical applications. However, the mechanisms of osteogenic differentiation in ADSCs is a highly complicated and delicate regulatory process (Kim et al. 2015; Wen et al. 2014), involving a large number of signaling pathways and the interaction of related biological molecules, orchestrated with spatiotemporal-specific expression. Hence, further study of ADSCs in osteogenic differentiation may support the advancement of bone tissue engineering.
Unlike linear RNAs, circular RNAs (circRNAs) are a type of closed continuous loop that do not have 5′ caps or 3′ tails and mainly exist in eukaryotic cells (Chen et al. 2015). The expression profiles of circRNAs are tissue-specific and widely detected in different tissues and organs in humans. Moreover, circRNAs are highly stable and evolutionarily conserved in mammalian cells (Rybak-Wolf et al. 2015). New bioinformatics technologies have facilitated the study of circRNA function. Apart from very small amounts of circRNA encoding protein (Legnini et al. 2017; Yang et al. 2017), most circRNAs resemble other non-coding RNAs (ncRNAs), such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) and greatly influence the regulation of gene function. CircRNAs can function as miRNA sponges to indirectly interfere with mRNA translation, regulate gene splicing and transcription and interact with RNA-binding proteins (Hansen et al. 2013; Ashwal-Fluss et al. 2014; Du et al. 2016). CircRNAs also play an important role in biological processes and participate in the occurrence and advancement of diseases (Han et al. 2018).
Researchers have reported that circRNAs participate in the osteogenic differentiation of MSCs. Microarray analysis or RNA sequencing techniques to analyze the expression profiles of circRNAs have been utilized in periodontal ligament stem cells (PDLSCs) (Zheng et al. 2017; Gu et al. 2017), ADSCs (Long et al. 2018) and bone marrow mesenchymal stem cells (BMSCs) (Zhang et al. 2019). Specifically, Li et al. demonstrated that the circRNA CDR1as downregulates miR-7 expression, resulting in the upregulation of GDF5 and facilitates the osteogenesis of PDLSCs through a Smad-dependent and p38 MAPK pathways (Li et al. 2018a, b). However, few studies have examined the association between the molecular function of circRNAs and the osteoblastic differentiation of ADSCs.
Our previous research (Long et al. 2018) using microarray analysis demonstrated that dozens of circRNAs were differentially expressed during the osteoblastic differentiation of ADSCs, indicating the involvement of circRNAs in this process. Among the circRNAs identified with this differential expression, circRNA-23525, a key circRNA, was authenticated. In the current study, how circRNA-23525 modulates osteogenesis in ADSCs is examined. The mechanisms by which circRNAs influence the osteogenic process of ADSCs are investigated to characterize this potential biomarker and therapeutic target for bone regeneration.
Materials and methods
Isolation and culture of ADSCs
All experiments were confirmed by the Ethical Committees of the State Key Laboratory of Oral Diseases, West China School of Stomatology, Sichuan University (Chengdu, China). Six-week-old male C57BL/6 mice were purchased from the Sichuan University Animal Experimental Center. Mouse inguinal fat pads (bilateral) were cut into small pieces (1–2 mm3) and digested by 0.2% collagenase type I (Gibco, Waltham, MA, USA). The samples were shaken in a 37 °C water bath for 50 min. After centrifugation at 1000 RPM for 8 min, the cells were resuspended in medium consisting of α-MEM (HyClone, Logan, UT, USA), 10% FBS (Gibco, Waltham, MA, USA) and penicillin/streptomycin (100 U/mL/100 μg/mL) (Sigma-Aldrich, St. Louis, MO, USA). Cells were cultured at 37 °C in a humidified, 5% CO2/95% air environment and the medium was refreshed every 2–3 days. After achieving 80–90% confluence, cells were passaged at a 1:3 ratio and cells in the third passage were used for further study. Our previous study identified the surface markers of cells acquired utilizing flow cytometry. Results demonstrated that the isolated ADSCs had high purity based on positivity for CD29, CD105 and Sca-1 and negativity for CD45 and CD31 (Li et al. 2015a, b).
Induction of osteogenic differentiation in vitro
ADSCs were seeded into 6-well plates for osteoblastic differentiation. Regular growth medium was substituted for osteogenic-specific medium. The induction medium contained α-MEM, 10% FBS, 10 mM β-glycerophosphate, 1 × 10–8 M dexamethasone, 0.01 μM 1,25-dihydroxyvitamin D3, penicillin/streptomycin (100 U/mL/100 μg/mL) and 50 μg/mL L-ascorbic acid (Sigma-Aldrich, St. Louis, MO, USA). Cells in each group were induced with osteoblastic medium (Figs. 2 and 3). Alkaline phosphatase (ALP) staining was performed 7 days after induction and Alizarin Red S (ARS) staining was analyzed 14 days after induction. The expression of osteogenic marker genes (ALP and Runx2) was measured using quantitative real-time polymerase chain reaction (qRT-PCR) at days 0, 3, 7 and 14 after osteogenic induction.
Bioinformatics analysis
Prediction of circRNAs target was based on Arraystar’s miRNA target prediction software based on TargetScan and miRanda (Arraystar, Inc., Rockville, MD, USA). TargetScan (https://www.targetscan.org) and miRDB (https://www.mirdb.org/) databases were used to predict miRNA targets. The common genes in the two algorithms were chosen for the following experiments.
Cell transfection
Lentivirus vectors overexpressed circRNA-23525 (circRNA-23525), small interfering (si) RNAs (the sequences of siRNAs: 5′-GGCTATAAAAGTCTTACAG-3′) targeting the backsplice junction of circRNA-23525 (si-circRNA-23525) and the negative control of each (NC, si-NC) were obtained from BSD Labs (Chengdu, China). Cells were seeded in 6-well plates (5 × 105 cells/well) before transfection. To perform transduction, at 60–80% confluence, murine ADSCs were infected at a multiplicity of infection of 50. Transfection was conducted using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. After 48 h of infection, the efficiency of the lentivirus vector was estimated to be 80–90%. Cells were visualized on a fluorescent microscope (Olympus, Tokyo, Japan) using green fluorescent protein (GFP). MiR-30a-3p mimics, miR-30a-3p inhibitor and a nontargeting control miRNA mimic (miR-NC) were obtained from BSD Labs (Chengdu, China) and utilized for further study.
Dual-luciferase reporter assay
Reporter constructs containing miR-30a-3p-binding sites, wild-type circRNA-23525 (circRNA-23525 WT) and the WT Runx2 3′UTR (Runx2 WT), or a mismatch sequence, as well as their respective mutant forms (circRNA-23525 Mu and Runx2 Mu), were constructed and inserted into psiCHECK2 luciferase vectors (Promega, Madison, WI, USA). HEK-293 cells were transfected with two different groups of constructed vectors. In accordance with the manufacturer’s instructions, we implemented all transfections using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Luciferase activity was assayed using a Double-Luciferase Reporter Gene Assay Kit (TransGen Biotech, Beijing, China) at 48 h after transfection. Each experiment was repeated at least three times.
ALP staining and activity
ADSCs were cultured in osteoblastic induction medium for 7 days, then fixed in 4% paraformaldehyde for 30 min. ALP staining was executed using a BCIP/NBT ALP Color Development Kit (Beyotime, Shanghai, China). After adding BCIP/NBT staining buffer, samples were cultivated in a dark room for 30 min. They were then washed with double-distilled water and observed with the naked eye, as well as with a microscope. The absorbance of each group was detected at 405 nm according to the manufacturer’s instructions. ALP activity was evaluated using absorbance levels.
ARS staining and quantification
ARS staining was performed 14 days after the osteoblastic induction of ADSCs. ADSCs were fixated with 4% formalin solution for 20 min, then subjected to 0.1% ARS (Sigma-Aldrich, St. Louis, MO, USA) at room temperature for 10 min). Samples were then rinsed with distilled water to determine the mineralized nodules. In order to quantify matrix mineralization, the stain was incubated in 10% cetylpyridinium chloride (Sigma-Aldrich, St. Louis, MO, USA) for 30 min and the absorbance was detected at 570 nm using a spectrophotometer. The concentration of total protein was measured to normalize ARS quantification.
qRT-PCR analysis
In accordance with the manufacturer’s protocol, total cellular RNA was obtained using TRIzol (Life Technologies, Carlsbad, CA, USA). RNAs of ADSCs were then reverse transcribed into complementary DNA (cDNA) using a Geneseed® II First Strand cDNA Synthesis Kit (Geneseed, Guangzhou, China). Subsequently, qRT-PCR was conducted with Geneseed® qPCR SYBR® Green Master Mix (Geneseed, Guangzhou, China). The housekeeping gene β-actin served as the control for circRNAs and mRNAs and U6 served as a reference gene for miRNAs. qRT-PCR was performed as follows: incubation at 95 °C for 5 min, followed by 40 cycles at 95 °C for 10 s and 60 °C for 34 s. BSD Labs (Chengdu, China) designed all primer sequences (Table 1). The 2−ΔΔCt method was used to compute the relative expression levels of RNAs. All assays were repeated at least three times.
Western blotting analysis
Protein samples were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a PVDF membrane (Membrane Solutions, Shanghai, China). The membranes were blocked and then incubated at 4 °C overnight with primary antibodies (anti-β-actin, anti-ALP, anti-Runx2, anti-OCN, or anti-GAPDH). The samples were then incubated with horseradish peroxidase-conjugated secondary antibody (Sungene Biotech, Tianjin, China). The bands were detected using an enhanced chemiluminescence reagent (PerkinElmer Inc., Waltham, MA, USA). Protein levels in each sample were normalized to β-actin or GAPDH levels in the different groups.
Statistical analyses
Experiments were performed in triplicate and results are presented as the mean ± standard deviation (SD). Comparisons between the two groups were calculated using Student’s t test. For differences among multiple groups, a one-way ANOVA was employed. Statistical significance was defined as a P value of < 0.05. All statistics were analyzed using SPSS Statistics for Windows, Version 20.0 (IBM Corp., Armonk, NY, USA) and illustrated using GraphPad Prism (version 7.0; GraphPad Software Inc., La Jolla, CA, USA).
Results
CircRNA-23525 expression during osteogenic differentiation of ADSCs
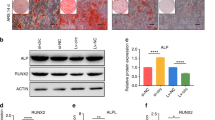
In our previous microarray analysis (Long et al. 2018), circRNA-23525 was significantly upregulated on days 3, 7 and 14 during osteogenic differentiation of ADSCs. To confirm the expression level of circRNA-23525 during osteogenesis, the osteogenic differentiation of ADSCs was confirmed. ALP staining and ARS staining demonstrated that ADSCs did indeed show osteogenic differentiation (Fig. 1a–a’, b–b’). The expression of two osteogenesis-specific markers (Runx2 and ALP) was measured. Compared with day 0, ALP expression was increased at days 3, 7 and 14 (Fig. 1c) and Runx2 expression was significantly increased at days 7 and 14 (Fig. 1c’). Furthermore, the dynamic expression of circRNA-23525 was increased relative to the control group at three time points (3, 7 and 14 days). The control group did not demonstrate osteogenic induction. CircRNA-23525 expression gradually increased until day 14 with induction (Fig. 1d).
CircRNA-23525 expression during osteogenic differentiation of adipose-derived mesenchymal stem cells (ADSCs). (a–a’) Osteogenic differentiation was determined using alkaline phosphatase (ALP) staining at day 7. (b–b’) Osteogenic differentiation was investigated by Alizarin Red S (ARS) staining after osteoblastic induction for 14 days. (c–c’) qRT-PCR analysis indicated increased expression of osteogenic-related markers (ALP and Runx2) during osteoblastic process in ADSCs. Expression was normalized to β-actin and expressed as means ± standard deviations (SD) (n = 3; *p < 0.05 compared with day 0). (d) After induction (Ind) for 3, 7, or 14 days, the dynamic expression of circRNA-23525, as measured using qRT-PCR, was significantly increased compared with the control group (Con); *p < 0.05
Overexpression of circRNA-23525 promotes osteoblastic differentiation of ADSCs
To determine the biological functions of circRNA-23525 in terms of osteoblastic differentiation of ADSCs, a lentiviral vector was constructed to overexpress circRNA-23525 in ADSCs. The expression of GFP by fluorescent microscopy was measured to assess transfection efficiency. Data indicated that transfection was successful in 80–90% of cells (Fig. 2a–a’). circRNA-23525 expression was increased greater than 10-fold compared with the control group and the vector group (Fig. 2b). After 3 days of osteogenic differentiation, qRT-PCR and Western blot analysis revealed that circRNA-23525 overexpression significantly increased the expression of Runx2, OCN and ALP at both the mRNA and protein levels (Fig. 2f, g).
The function of upregulated circRNA-23525 in osteogenic differentiation of ADSCs. (a–a’) Transfection efficiency of circular RNA (circRNA) was 80–90% based on the expression of GFP in ADSCs observed by fluorescence microscope. (b) The relative circRNA-23525 expression after transfection with circRNA-23525 overexpression-vector in ADSCs. (c) Images of ALP staining in the control (Con), circRNA-23525-overexpression negative control (NC) and circRNA-23525-overexpression (circRNA-23525) groups (top row). Cells were induced with osteoblastic medium for 7 days. Images of ARS staining show calcium nodes in Con, NC and circRNA-23525 groups (bottom row). ADSCs were induced for 14 days. (d, e) Histograms illustrate ALP activity and intensity of ARS by spectrophotometry (compared with the Con and NC groups). (f) The relative mRNA expression of Runx2, OCN and ALP was analyzed using qRT-PCR after transfection with circRNA-23525 overexpression vector at day 3 of osteoblastic differentiation. β-Actin was used for normalization. (g) The protein expression of Runx2, OCN, ALP and the control β-actin was measured using Western blot analysis at day 3 of osteoblastic differentiation. ADSCs in the three groups were all induced to osteogenic differentiation. The results are presented as mean ± SD, n = 3, *p < 0.05 (vs. negative and blank control)
Staining and activity assays of ALP revealed that overexpression of circRNA-23525 increased ALP activity 7 days after induction (Fig. 2c, d). After 14 days of osteogenic induction, the quantification of ARS staining was increased in the circRNA-23525 overexpression group, demonstrating elevated mineralized bone matrix formation (Fig. 2c, e).
Knockdown of circRNA-23525 decreases osteoblastic differentiation of ADSCs
The effect of downregulated circRNA-23525 on osteoblastic differentiation of ADSCs was also investigated. The expression of circRNA-23525 was downregulated after transfection and qRT-PCR demonstrated that circRNA-23525 expression was decreased by approximately 40% in the circRNA-23525 knockdown group (Fig. 3a). In contrast to circRNA-23525 upregulation, circRNA-23525 knockdown decreased the levels of osteoblastic markers Runx2, OCN and ALP on day 3 compared with the control group and the vector group (Fig. 3e, f).
The function of downregulated circRNA-23525 in osteogenic differentiation of ADSCs. a Relative circRNA-23525 expression after transfection with circRNA-23525 knockdown vector in ADSCs. b Images of ALP staining in the control (Con), small interfering RNA negative (si-NC) and small interfering RNAs targeting circRNA-23525 (si-circRNA-23525) groups (top row). Images of ARS staining for calcium deposition in the Con, si-NC and si-circRNA-23525 groups (bottom row). c, d Comparison of ALP activity and intensity of ARS in different groups by spectrophotometry (compared with the Con and negative control (si-NC) groups). e The relative expression of osteogenic markers at the mRNA level was measured after transfection with si-circRNA-23525 vector at day 3 of osteoblastic induction. f Western blot analysis of osteogenic markers at day 3 of osteogenic induction. β-Actin was used for normalization. Cells in all groups were induced with osteoblastic medium. Expressions are presented as mean ± SD (n = 3, *p < 0.05 vs. negative and blank control)
ALP staining, ALP activity assay and ARS staining revealed that ALP activity and calcium deposition were downregulated by si-circRNA-23525 (Fig. 3b–d). The above data imply that circRNA-23525 is involved in the osteoblastic differentiation of ADSCs.
CircRNA-23525 functions as a sponge of miR-30a-3p
To further explore the underlying mechanisms by which circRNA-23525 modulates the osteoblastic differentiation of ADSCs, the miRNA response elements (MREs) of circRNA-23525 were predicted using Arraystar’s miRNA target prediction software. miR-30a-3p had an excellent predictive score and was selected for further analysis (Fig. 4a). qRT-PCR assays illustrated that altered expression of miR-30a-3p was significantly decreased compared with the control group at three time points (3, 7 and 14 days); the control group showed no osteogenic induction. miR-30a-3p expression was downregulated more than 1.7-fold until day 7 and remained low with induction until day 14 despite a modest rise (Fig. 4b). qRT-PCR results then indicated that miR-30a-3p was decreased in ADSCs with circRNA-23525-overexpression(Fig. 4c), while knockdown of circRNA-23525 enhanced miR-30a-3p expression (Fig. 4c’). To determine whether circRNA-23525 directly modulates miR-30a-3p, luciferase vectors of circRNA-23525 (circRNA-23525-WT) and a mutated form (circRNA-23525-Mut) were constructed. The results showed that luciferase expression decreased significantly after co-transfection of circRNA-23525-WT and miR-30a-3p mimics in HEK-293T cells; however, there was not a significant change after co-transfection of circRNA-23525-Mut and miR-30a-3p mimics (Fig. 4d). These results indicated a direct reciprocity of circRNA-23525 with miR-30a-3p at the putative binding site.
The relationship between the expression of circRNA-23525 and miR-30a-3p in ADSCs. (a) The predicted binding sites between circRNA-23525 and miR-30a-3p. (b) After induction (Ind) for 3, 7, or 14 days, the altered expression of miR-30a-3p, as performed using qRT-PCR, was decreased compared with the no osteogenic induction group (Con). (c–c’) Expression levels of miR-30a-3p in osteogenically differentiated ADSCs after transfection with circRNA-23525 overexpression and small interfering- (si-) circRNA-23525 vector. U6 was used for normalization; *p < 0.05 vs. negative control and blank control. d The luciferase activity of wild-type circRNA-23525 (circRNA-23525-WT) and mutant circRNA-23525 (circRNA-23525-Mut) in HEK-293T cells was analyzed after co-transfection with miR-30a-3p mimics or NC. The data represent Firefly luciferase normalized to Renilla luciferase. *p < 0.05
MiR-30a-3p binds with Runx2 and is involved in osteogenic differentiation of ADSCs
It has been suggested that miRNAs perform their function primarily through their target genes. Hence, bioinformatics methodology was used to analyze the predictive binding sites of miR-30a-3p. Runx2 is a well-accepted osteogenic master gene (Long 2011). The 3′UTR of mouse Runx2 mRNA has a putative binding element for miR-30a-3p (Fig. 5a). Luciferase reporter constructs of the 3′UTR of Runx2 (Runx2-WT) and a mutated form (Runx2-Mut) were generated to investigate whether miR-30a-3p directly regulates the 3′UTR of Runx2. Luciferase activity was reduced by transfection of the miR-30a-3p mimics and the wild-type 3′UTR of Runx2 in comparison with the controls, which showed no significant differences after transfection of miR-30a-3p and the mutant 3′UTR of Runx2 in 293T cells (Fig. 5b).
MiR-30a-3p decreased Runx2 expression and was involved in regulating the osteogenic differentiation of ADSCs. a Schematic of miR-30a-3p putative target sites in the 3′-UTR of Runx2. b Confirmation of target using luciferase reporter assay in HEK-293T cells. The cells were co-transfected with miR-30a-3p or nontargeting control miRNA mimic (miR-NC) and wild-type Runx2 3′-UTR or mutant Runx2 3′-UTR luciferase constructs. Values are reported as the ratio of Renilla luciferase to Firefly luciferase. c–e The expression level of Runx2 and other osteogenic markers (ALP and OCN) was measured using qRT-PCR and WB analysis in ADSCs after transfection with miR-30a-3p mimics or inhibitor. *p < 0.05 vs. blank control
qRT-PCR analysis validated the observation that Runx2 expression in ADSCs was downregulated after transfection with the miR-30a-3p mimics, but became upregulated after transfection with the miR-30-3p inhibitor (Fig. 5c). After cells were transfected with the miR-30-3p mimics or inhibitor, respectively, qRT-PCR results of other osteoblastic markers (OCN and ALP) showed a similar trend (Fig. 5d, d’). Western blot assays of Runx2, OCN, and ALP were in line with qRT-PCR analysis (Fig. 5e). These results suggest that miR-30a-3p impedes Runx2 expression via binding to the 3′UTR of Runx2 and suppresses osteoblastic differentiation in ADSCs.
CircRNA-23525/miR-30a-3p regulates expression of Runx2 and osteogenic differentiation of ADSCs
Since circRNA-23525 decreases the expression of miR-30a-3p in ADSCs and the expression of Runx2 is regulated by miR-30a-3p, whether circRNA-23525 could upregulate Runx2 expression by acting as a sponge for miR-30a-3p was further explored. circRNA-23525 overexpression promoted Runx2 mRNA expression, and co-transfection of miR-30a-3p inhibitor further enhanced Runx2 expression, whereas co-transfection of miR-30a-3p mimics reversed Runx2 expression (Fig. 6a). The expression level of other osteogenic markers (OCN and ALP) showed a similar trend (Fig. 6b, b’). Specifically, circRNA-23525 knockdown decreased OCN and ALP mRNA expression, and co-transfection of miR-30a-3p mimics further lowered expression, whereas co-transfection of miR-30a-3p inhibitor rescued their expression. A Western blot assay at the protein level confirmed these results (Fig. 6f).
CircRNA-23525 modulates Runx2 expression and osteogenesis in ADSCs by inhibiting miR-30a-3p. (a, b, b’, f) Quantitative real-time PCR and western blotting analyses of the expression level of Runx2 and other osteogenic markers (OCN and ALP) in osteogenically differentiated ADSCs after transfection with circRNA-23525 overexpression or circRNA-23525 knockdown plus miR-30a-3p mimics or a miR-30a-3p inhibitor. (c–e) After co-transfection withcircRNA-23525 or si-circRNA-23525 plus miR-30a-3p mimics or a miR-30a-3p inhibitor, osteogenic differentiation of ADSCs was analyzed using ARS staining, ALP staining and ALP activity assay at 7 or 14 days after osteogenic induction, respectively. *p < 0.05 vs. miR-NC group
ALP staining, ALP activity assay, and ARS staining showed that ALP activity and mineralized bone matrix were reversed by miR-30a-3p mimics in the circRNA-23525 overexpression group and were rescued by miR-30a-3p inhibitor in the circRNA-23525 knockdown group (Fig. 6c–e). These data implied that circRNA-23525 serves upstream of miR-30a-3p and inhibits the effects of miR-30a-3p in targeting Runx2 in osteogenically differentiated ADSCs.
Discussion
The plasticity and multipotency of ADSCs have prompted the development of additional relevant research. Many reports have indicated that ADSC-based cell therapy has ideal clinical efficacy and efficiency for both autologous and allogeneic purposes (Mesimäki et al. 2009; Pak et al. 2016; Kastrup et al. 2017). ADSCs possess adipogenic, osteogenic, chondrogenic, cardiogenic, myogenic and neurogenic potential in vitro (Vermette et al. 2007; Choi et al. 2010). Moreover, our previous research demonstrated that ADSCs transplantation could be applied in bone regeneration (Sun et al. 2014). In comparison with BMSCs, the most promising aspect of ADSCs is that the differentiation potential is maintained with aging (Shi et al. 2005). Other advantages include the wide range of adipose tissue, less-invasive material extraction, lower incidence rates of complications, and comparatively low cost. In the present study, a new axis, which may regulate the osteogenic process of ADSCs in vitro, was examined.
CircRNAs are an intriguing type of RNA due to their covalently closed structure, high stability, and implications for further functions in gene regulation (Li et al. 2018a, b). Notably, emerging evidence has revealed that the osteoblastic differentiation of MSCs is modulated by circRNAs, thus making them a possible promising player in controlling the fate of MSCs. CircRNA_33287, for example, plays a positive role in the osteoblastic differentiation of maxillary sinus membrane stem cells (Peng et al. 2019). Similarly, circRNA CDR1as can also promote osteogenesis in PDLSCs (Li et al. 2018a, b). Microarray analysis has previously reported that circRNAs were involved in the osteogenic differentiation of ADSCs (Long et al. 2018). Through analyzing the raw data of microarray results, 10 circRNAs, which were closely bound up with osteogenesis, were selected for further examination. The qRT-PCR assays of three random circRNAs showed that circRNA-23525 was consistently upregulated in the process of osteogenic differentiation. However, a recent study demonstrated that circRNA_28313 was involved in RANKL+CSF1-induced osteoclast differentiation and promotedovariectomy-induced bone resorption (Chen et al. 2019). In addition, knockdown of circIGSF11 enhanced osteogenesis in BMSCs (Zhang et al. 2019). These results suggest that circRNAs play a crucial part in the osteoblastic differentiation of MSCs, although their functions may depend on cell specificity and pathological condition; this point needs to be further clarified.
In the current study, the function of circRNA-23525 in osteogenic differentiation of ADSCs was confirmed using a gain- and loss-of-function strategy. ADSCs were transfected with circRNA-23525 constructs (overexpression and knockdown) and then induced to differentiate into osteoblasts. The expression of osteogenic markers (Runx2, ALP and OCN) was significantly upregulated in the circRNA-23525 overexpression group. ALP and ARS staining demonstrated that overexpression of circRNA-23525 enhanced osteoblastic differentiation of ADSCs in vitro. In contrast, ALP and ARS were decreased in the circRNA-23525 knockdown group, which suggests that circRNA-23525 knockdown intervenes in the osteoblastic process of ADSCs. Moreover, the si-circRNA-23525 group has indicated that ADSCs depleted in regard to circRNA-23525 underwent slightly osteogenic differentiation in the presence of induction medium. Thus, circRNA-23525 can be considered to be an osteogenesis-related circRNA in ADSCs. However, due to lack of control indicating that ADSCs in circRNA-23525 overexpression group differentiated without osteogenic induction medium, the function of circRNA-23525 demonstrated by the existing data is only during osteogenic induction.
Emerging evidence suggests that circRNAs modulate gene expression at both the transcriptional and the posttranscriptional level (Li et al. 2018a, b). Of note, circRNAs act as competitively endogenous RNAs to regulate miRNAs and their target genes (Hansen et al. 2013). For example, CDR1as impacts biological functions by serving as a miR-7 sponge, leading to the upregulation of GDF5, thus promoting osteogenesis in PDLSCs (Li et al. 2018a, b). In the current study, a bioinformatics analysis predicted that miR-30a-3p could bind to circRNA-23525. In ADSCs osteogenesis, the dynamic expression of miR-30a-3p and circRNA-23525 showed an opposite trend at 3, 7 and 14 days. Results further confirmed that miR-30a-3p expression varied with the upregulation or downregulation of circRNA-23525. Moreover, putative binding was demonstrated using a dual-luciferase assay in which overexpression of miR-30a-3p decreased luciferase reporter activity in the circRNA-23525-WT group compared with the mutant group.
miRNAs have been demonstrated to mediate osteogenesis in MSCs (Hoseinzadeh et al. 2016; Li et al. 2015a, b) and it has been demonstrated that the osteogenic key regulator Runx2 can be directly targeted by miRNAs (Zhang et al. 2011). Runx2 is an essential transcription factor in the regulation of osteoblastic differentiation and skeletal development (Long 2011) and plays a role in osteogenesis-related transduction pathways (Bruderer et al. 2014). The current results demonstrated that miR-30a-3p has predictive binding sites with Runx2. Luciferase assays further elucidated that miR-30a-3p can directly bind to the 3′UTR of Runx2. It should be noted that miR-30 family members, including miR-30a-3p, are negative regulators of osteogenic differentiation in MSCs (Wu et al. 2012). Previous studies have reported that miR-30a-3p directly regulates HIF2α, which functions as an oncoprotein in renal carcinomas (Mathew et al. 2014) and miR-30a-3p regulates the expression of its target gene EPAS1, which is involved in tumor progression in lung cancer (Yuan et al. 2019). However, the specific mechanisms by which miR-30a-3p affects the osteogenic differentiation of ADSCs remain unclear. The present study demonstrated that miR-30a-3p overexpression reduced the expression of osteogenic markers (Runx2, ALP, and OCN) at both the mRNA and protein levels, while silencing miR-30a-3p enhanced the expression of these markers. These findings suggest that miR-30a-3p acts as a negative regulator in the osteogenic differentiation of ADSCs.
Considering circRNA-23525 acts as a miR-30a-3p sponge and Runx2 is suppressed by miR-30a-3p expression, we next examined whether circRNA-23525 modulates osteoblastic differentiation by regulating Runx2 expression. Results demonstrated that circRNA-23525 overexpression plus miR-30a-3p inhibition enhanced Runx2 expression, whereas miR-30a-3p overexpression reversed Runx2 expression. Moreover, OCN and ALP separately showed similar trends in expression. ALP and ARS staining revealed that circRNA-23525 knockdown plus miR-30a-3p mimics suppressed ALP activity and calcium nodule formation. Adding the knockdown of miR-30a-3p could rescue this process. Taken together, these data demonstrate that the circRNA-23525/miR-30a-3p pathway enhances the osteogenesis of ADSCs by modulating Runx2. On the other hand, due to the lack of controls such as ADSCs expressing circRNA-23525 upregulation plus miR-30a-3p mimics or an inhibitor without the osteogenic induction medium, the role of the circRNA-23525/miR-30a-3p pathway only confirms under osteogenic induction in this study. Therefore, ADSCs expressing high levels of circRNA-23525 differentiated without osteogenic induction medium should be further executed to elucidate the mechanism in the circRNA-23525/miR-30a-3p/Runx2 functional axis. In addition, questions remain regarding circRNA-23525 as an osteogenesis-related circRNA in vivo. Further research will propose additional evidence for the role of this functional axis in the osteogenic differentiation of ADSCs.
In summary, the current study was the first to identify an association between circRNA-23525 with osteogenesis in ADSCs. It clarifies the regulatory role of the circRNA-23525/miR-30a-3p/Runx2 functional axis in ADSCs osteogenic differentiation. Under osteoblastic induction, circRNA-23525 suppresses miR-30a-3p expression, thus relieving the expression of Runx2 targeted by miR-30a-3p, which enhances the osteoblastic differentiation of ADSCs (Fig. 7). The current results revealed a novel mechanism in stimulating the osteogenic induction of ADSCs and suggest a promising avenue of application in tissue engineering and treatment for bone diseases and trauma.
A proposed model illustrating the functional axis of circRNA-23525/miR-30a-3p/Runx2 during osteogenic differentiation of ADSCs. CircRNA-23525 acts to regulate Runx2 expression via targeting miR-30a-3p and is thus considered as a positive regulator in the osteogenic process of ADSCs. ↑, promote or activate; ⊥, inhibit
Conclusions
In conclusion, the current study revealed that circRNA-23525 inhibits miR-30a-3p expression, thereby rescuing the expression of Runx2 targeted by miR-30a-3p and promoting the osteogenic differentiation of ADSCs.
References
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, MemczakS RN, Kadener S (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66
Bhumiratana S, Bernhard JC, Alfi DM, Yeager K, Eton RE, Bova J, Shah F, Gimble JM, Lopez MJ, Eisig SB, Vunjak-Novakovic G (2016) Tissue-engineered autologous grafts for facial bone reconstruction. Sci Transl Med 8:343ra83
Bruderer M, Richards RG, Alini M, Stoddart MJ (2014) Role and regulation of RUNX2 in osteogenesis. Eur Cell Mater 28:269–86
Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12:381–8
Chen X, Ouyang Z, Shen Y, Liu B, Zhang Q, Wan L, Yin Z, Zhu W, Li S, Peng D (2019) CircRNA_28313/miR-195a/CSF1 axis modulates osteoclast differentiation to affect OVX-induced bone absorption in mice. RNA Biol 19:1–14
Choi YS, Dusting GJ, Stubbs S, Arunothayaraj S, Han XL, Collas P, Morrison WA, Dilley RJ (2010) Differentiation of human adipose-derived stem cells into beating cardiomyocytes. J Cell Mol Med 14:878–89
Compston JE, McClung MR, Leslie WD (2019) Osteoporosis Lancet 393:364–76
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 44:2846–58
Griffin MF, Ibrahim A, Seifalian AM, Butler PEM, Kalaskar DM, Ferretti P (2017) Chemical group-dependent plasma polymerisation preferentially directs adipose stem cell differentiation towards osteogenic or chondrogenic lineages. Acta Biomater 50:450–61
Gu XG, Li MY, Jin Y, Liu D, Wei F (2017) Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet 18:100
Han B, Chao J, Yao HH (2018) Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther 187:31–44
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–8
Hoseinzadeh S, Atashi A, Soleimani M, Alizadeh E, Zarghami N (2016) MiR-221-inhibited adipose tissue-derived mesenchymal stem cells bioengineered in a nano-hydroxy apatite scaffold. Vitro Cell Dev Biol Anim 52:479–87
Kastrup J, Haack-Sørensen M, Juhl M, HararySøndergaard R, Follin B, Drozd Lund L (2017) Cryopreserved off-the-shelf allogeneic adipose-derived stromal cells for therapy in patients with ischemic heart disease and heart failure-a safety study. Stem Cells Transl Med 6:1963–71
Kern S, Eichler H, Stoeve J, Klüter H, Bieback K (2006) Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem cells 24:1294–301
Kim TH, Shah S, Yang L, Yin PT, Hossain MK, Conley B, Choi JW, Lee KB (2015) Controlling differentiation of adipose-derived stem cells using combinatorial graphene hybrid-pattern arrays. ACS Nano 9:3780–90
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I (2017) Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66:22–37
Li H, Li T, Fan J, Li T, Fan L, Wang S, Weng X, Han Q, Zhao RC (2015) miR-216a rescues dexamethasone suppression of osteogenesis, promotes osteoblast differentiation and enhances bone formation, by regulating c-Cbl-mediated PI3K/AKT pathway. Cell Death Differ 22:1935–45
Li J, Hu C, Han L, Liu L, Jing W, Tang W, Tian W, Long J (2015) MiR-154-5p regulates osteogenic differentiation of adipose derived mesenchymal stem cells under tensile stress through the Wnt/PCP pathway by targeting Wnt11. Bone 78:130–41
Li X, Yang L, Chen LL (2018) The biogenesis, functions and challenges of circular RNAs. Mol Cell 71:428–42
Li XB, Zheng YF, Zheng Y, Huang Y, Zhang Y, Jia L, Li W (2018) Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther 9:232
Long F (2011) Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol 13:27–38
Long T, Guo ZY, Han L, Yuan XY, Liu L, Jing W, Tian WD, Zheng XH, Tang W, Long J (2018) Differential expression profiles of circular RNAs during osteogenic differentiation of mouse adipose-derived stromal cells. Calcif Tissue Int 103:338–52
Martin TJ, Gooi JH, Sims NA (2009) Molecular mechanisms in coupling of bone formation to resorption. Crit Rev Eukaryot Gene Expr 19:73–88
Mathew LK, Lee SS, Skuli N, Rao S, Keith B, Nathanson KL, Lal P, Simon MC (2014) Restricted expression of miR-30c-2-3p and miR-30a-3p in clear cell renal cell carcinomas enhances HIF2α activity. Cancer Discov 4:53–60
Mesimäki K, Lindroos B, Törnwall J, Mauno J, Lindqvist C, Kontio R, Miettinen S, Suuronen R (2009) Novel maxillary reconstruction with ectopic bone formation by GMP adipose stem cells. Int J Oral Maxillofac Surg 38:201–9
Pak J, Lee JH, Park KS, Jeong BC, Lee SH (2016) Regeneration of cartilage in human knee osteoarthritis with autologous adipose tissue-derived stem cells and autologous extracellular matrix. Biores Open Access 5:192–200
Peng W, Zhu S, Chen J, Wang J, Rong Q, Chen S (2019) Hsa_circRNA_33287 promotes the osteogenic differentiation of maxillary sinus membrane stem cells via miR-214-3p/Runx3. Biomed Pharmacother 109:1709–17
Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N (2015) Circular RNAs in the mammalian brain are highly abundant, conserved and dynamically expressed. Mol Cell 58:870–85
Shi YY, Nacamuli RP, Salim A, Longaker MT (2005) The osteogenic potential of adipose-derived mesenchymal cells is maintained with aging. PlastReconstr Surg 116:1686–96
Sun JJ, Zheng XH, Wang LY, Liu L, Jing W, Lin YF, Tian WD, Tang W, Long J (2014) New bone formation enhanced by ADSCs overexpressing hRunx2 during mandibular distraction osteogenesis in osteoporotic rabbits. J Orthop Res 32:709–20
Vermette M, Trottier V, Ménard V, Saint-Pierre L, Roy A, Fradette J (2007) Production of a new tissue-engineered adipose substitute from human adipose-derived stromal cells. Biomaterials 28:2850–60
Vicente R, Noël D, Pers YM, Apparailly F, Jorgensen C (2016) Deregulation and therapeutic potential of microRNAs in arthritic diseases. Nat Rev Rheumatol 12:211–20
Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ (2014) Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater 13:979–87
Wu T, Zhou H, Hong Y, Li J, Jiang X, Huang H (2012) MiR-30 family members negatively regulate osteoblast differentiation. J Biol Chem 287:7503–11
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z (2017) Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 27:626–64
Yuan S, Xiang Y, Wang G, Zhou M, Meng G, Liu Q, Hu Z, Li C, Xie W, Wu N, Wu L, Cai T, Ma X, Zhang Y, Yu Z, Bai L, Li Y (2019) Hypoxia-sensitive LINC01436 is regulated by E2F6 and acts as an oncogene by targeting miR-30a-3p in non-small cell lung cancer. Mol Oncol 13:840–56
Zhang MJ, Jia LF, Zheng YF (2019) circRNA expression profiles in human bone marrow stem cells undergoing osteoblast differentiation. Stem Cell Rev 15:126–38
Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, Stein GS (2011) A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A 108:9863–8
Zheng YF, Li XB, Huang YP, Jia L, Li W (2017) The Circular RNA Landscape of Periodontal Ligament Stem Cells During Osteogenesis. J Periodontol 88:906–14
Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH (2001) Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng 7:211–28
Funding
This study was supported by grants from the National Nature Science Foundation of China (Nos. 31570950, 10502037, and 31070833), the Science and Technology Foundation of Sichuan Province (Nos. 2017SZ0032, 2010GZ0225, 2011GZ0335, and 2009SZ0139) and National Key Research and Development Program of China (No. 2016YFC1101404).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All applicable international, national and institutional guidelines for the care and use of animals were followed. All procedures in our studies involving animals were executed in accordance with a protocol approved by the Animal Research Committee of Sichuan University (Chengdu, China).
Rights and permissions
About this article
Cite this article
Guo, Z., Zhao, L., Ji, S. et al. CircRNA-23525 regulates osteogenic differentiation of adipose-derived mesenchymal stem cells via miR-30a-3p. Cell Tissue Res 383, 795–807 (2021). https://doi.org/10.1007/s00441-020-03305-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-020-03305-7