Abstract
Ecto-5′-nucleotidase (eN) is the major extracellular adenosine-producing ecto-enzyme in mouse brain. Via the production of adenosine, eN participates in many physiological and pathological processes, such as wakefulness, inflammation, nociception and neuroprotection. The mechanisms regulating the expression of eN are therefore of considerable neurobiological and clinical interest. Having previously described a modulatory effect of melatonin in the regulation of eN mRNA levels, we decided to analyze the melatonin receptor subtype involved in the regulation of eN mRNA levels by comparing eN mRNA patterns in melatonin-proficient transgenic mice lacking either the melatonin receptor subtype 1 (MT1 KO) or both melatonin receptor subtypes (MT1 and MT2; MT1/2 KO) with the corresponding melatonin-proficient wild-type (WT) controls. By means of radioactive in situ hybridization, eN mRNA levels were found to be diminished in both MT1 and MT1/2 KO mice compared with WT controls suggesting stimulatory impacts of melatonin receptors on eN mRNA levels. Whereas eN mRNA levels increased during the day and peaked at night in WT and MT1 KO mice, eN mRNA levels at night were reduced and the peak was shifted toward day-time in double MT1/2 KO mice. These data suggest that the MT2 receptor subtype may play a role in the temporal regulation of eN mRNA availability. Notably, day-time locomotor activity was significantly higher in MT1/2 KO compared with WT mice. Our results suggest melatoninergic signaling as an interface between the purinergic system and the circadian system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the mammalian brain, the enzyme ecto-5′-nucleotidase (eN) is the predominant enzyme producing extracellular adenosine via hydrolysis of adenosine 5′-monophosphate (AMP) at physiological extracellular pH and purine concentrations (Langer et al. 2008; Kulesskaya et al. 2013). This glycosyl phosphatidylinositol (GPI)-anchored membrane protein (Zimmermann 1992) is located on the surface of a variety of cell types and is involved (via adenosine production) in the regulation of many processes under physiological and pathological conditions, such as hypoxia, ischemia and tumor progression (Allard et al. 2012; Colgan et al. 2006; Haskó and Cronstein 2004). Typically, adenosine activates G-protein-coupled P1 adenosine receptors A2A and A2B or A1 and A3, which can stimulate or inhibit adenylyl cyclase and, thereby, increase or decrease intracellular cyclic AMP (cAMP) levels, respectively (Alam et al. 2015). However, adenosine receptors might also activate other signaling pathways via inwardly rectifying K+ channels, phospholipase C (PLC), Ca2+ channels and mitogen-activated protein kinases (Jacobson and Gao 2006; Boison 2008).
Although many factors have been shown to induce or suppress eN expression (Narravula et al. 2000; Regateiro et al. 2013; Spychala and Kitajewski 2004; Wang et al. 2012), our knowledge of the mechanisms regulating eN expression in the brain is still incomplete. We recently demonstrated time-dependent changes in eN mRNA levels and strain-dependent differences in temporal patterns of eN mRNA availability between melatonin-proficient and melatonin-deficient mice. We have therefore suggested that melatonin is involved in the regulation of eN mRNA levels (Homola et al. 2015). In mammals, a rhythmic and exclusively nocturnal secretion of melatonin from the pineal gland transduces photic information to the entire organism and represents an important hormonal output signal from the circadian master clock located in the suprachiasmatic nucleus (SCN; Bartness et al. 1993; Korf and von Gall 2013).
Melatonin in mammals exerts its effects via two high-affinity G-protein-coupled receptors denoted as melatonin receptor subtype 1 (MT1) and melatonin receptor subtype 2 (MT2; von Gall et al. 2002b). These receptors are highly expressed in the hypophysial pars tuberalis and the SCN but are also present in other brain regions, including regions showing high eN expression, such as the hippocampus and the basal ganglia (Adamah-Biassi et al. 2013; Lacoste et al. 2015). Via MT1 and MT2, melatonin can act upon multiple signal transduction cascades (Chen et al. 2014; von Gall et al. 2002b; Tosini et al. 2014). One of the best investigated effects of MT receptor activation is the inhibition of the cAMP-dependent pathway via protein Gi. However, melatonin might also play a dual role: as shown for the hypophysial pars tuberalis, melatonin not only inhibits but also sensitizes the cAMP-dependent pathway via heterologous sensitization involving the A2B adenosine receptor (von Gall et al. 2002a).
Although several studies have provided evidence that purinergic signals affect the circadian system (Bhattacharya et al. 2013; Perez-Aso et al. 2013; Sigworth and Rea 2003), a reciprocal interaction between the circadian system and the purinergic system has thus far received little attention. To test the hypothesis that the circadian and purinergic systems reciprocally interact, we analyze whether the melatoninergic system affects the regulation of eN mRNA levels in the mouse prosencephalon. To this end, we applied radioactive in situ hybridization (ISH) by using a triplet of specific oligoprobes and compared the location and the time-dependent eN mRNA patterns observed in melatonin-proficient transgenic mice lacking the MT1 (MT1 KO) or both melatonin receptor subtypes (MT1/2 KO) with the corresponding melatonin-proficient wild-type (WT) controls.
Materials and methods
Animals
All animal experiments were conducted in accordance with accepted standards of human animal care and were consistent with Federal guidelines and Directive 2010/63/EU of the European Union. Mice with a targeted deletion of the MT1 gene (MT1 KO; Liu et al. 1997), mice deficient in both melatonin receptor subtypes (MT1/2 KO; obtained by crossing MT1 KO with MT2 KO mice) and the corresponding WT mice were bred onto a melatonin-proficient C3H/HeN genetic background for at least 10 generations (Pfeffer et al. 2012). Their genotypes were confirmed by polymerase chain reaction (PCR). Because of fertility issues, MT2 KO mice were not used in these experiments. Experiments were performed with adult male mice (11-14 weeks old). Animals were adapted to a 12 L/12D photoperiod (light phase: 230 μW/cm2; dark phase: dim red light <5 μW/cm2) with access to food and water ad libitum for at least two weeks prior to experiments. To avoid masking artifacts, animals were transferred to constant darkness (DD) for one complete day/night cycle one day before tissue sampling. Circadian time (CT) 00 was defined as “lights on” in the previous light–dark cycle. Mice were deeply anesthetized with isoflurane and then decapitated under dim red light (<5 μW/cm2) at four different time points (CT00, CT06, CT12, CT18; four animals per genotype and time point, with the exception of MT1 KO at CT18, for which data were obtained from three animals). The eyes were first removed under dim red light and then the brains were harvested and immediately frozen in isopentane on dry ice. Serial frozen sections (20 μm thick) were cut in the coronal plane by using a cryostat (Leica, Wetzlar, Germany).
Genotyping by PCR analysis
Genotypes were confirmed by PCR analysis of genomic DNA isolated from tail biopsies. Mixtures of three primers were used for each reaction allowing the detection of both WT and the targeted alleles in a single reaction. For DNA amplification, the following primers were used: MT1: WT forward: 5′- GAG TCC AAG TTG CTG GGC AGT GGA -3′, KO forward: 5′- CCA GCT CAT TCC TCC ACT CAT GAT CTA -3′, common reverse for WT and KO: 5′- GAA GTT TTC TCA GTG TCC CGC AAT GG -3′; MT2: common forward for WT and KO: 5′- CCA GGC CCC CTG TGA CTG CCC GGG -3′, WT reverse: 5′- CCT GCC ACT GAG GAC AGA ACA GGG -3′, KO reverse: 5′- TGC CCC AAA GGC CTA CCC GCT TCC-3′. Separated reactions were performed for each receptor subtype. Amplification of the MT1 targeted allele was performed by using a program of 94 °C (45 s), 60 °C (45 s) and 72 °C (120 s) × 38 cycles. The program used for the amplification of the MT2 targeted allele was 94 °C (30 s), 60 °C (30 s) and 72 °C (60 s) × 40 cycles. The amplified DNA products were separated on 2 % agarose gels.
Radioactive ISH
The mRNA encoding eN was detected in the mouse prosencephalon by means of radioactive ISH as previously described (Homola et al. 2015). Briefly, three antisense 45-mer oligonucleotide probes or their corresponding sense probes were labeled with [33P] dATP (PerkinElmer, Waltham, Mass., USA) by using terminal deoxynucleotidyl transferase (Invitrogen, Karlsruhe, Germany) according to the manufacturer’s instructions and pooled. The following antisense probes were used: antisense (AS) 1: 5′-GAG TGG AGC CAT CCA GGT AGA CGA TCG TTC TCC CGA GTT CCT GGG-3′, AS2: 5′-GTC CCT CCA AAG GGC AGC ACA GCA GCC AGG TTC TCC CAG GTG ATG-3′, AS3: 5′-AGA AGC AGC CAC TTG GGT ACC TTA GCG GCC GCG GGA CGC ATG GCT-3′. For hybridization, 65 μl hybridization buffer (50 % deionized formamide, 0.03 M TRIS HCl pH 8, 0.2 mg/ml yeast tRNA, 1× Denhardt’s solution, 0.6 M NaCl, 0.25 % sodium dodecyl sulfate, 0.25 M ethylenediaminetetraacetic acid pH 8 and 1× dextran sulfate) containing the pooled probes (activity: 6 × 105 cpm per 100 μl) were applied to sections fixed onto slides with 4 % paraformaldehyde (PFA) dissolved in phosphate-buffered saline (PBS). The slides were incubated in a humid chamber at 42 °C overnight. On the following day, high-stringency washes in 2× SSC (standard sodium citrate)/0.1 % sarkosyl at room temperature and in 0.1× SSC/0.1 % sarkosyl at 55 °C were performed. The sections were rinsed in 70 % and 100 % ethanol, air-dried and exposed to a Kodak BioMax MR X-ray film (Sigma, Eching, Germany) for 40 days.
By use of Adobe Photoshop software (Adobe Systems, San Jose, Calif., USA), the relative optical densities (OD) of the autoradiographic signals were measured to determine mRNA levels. The signals from the region of interest in two to three sections were analyzed and averaged to yield a single value for each mouse. The autoradiographic signals were corrected for non-specific background signal calculated from the background of the entire film. The signals from the striatum were measured in the most caudal region, whereas the signals from the dentate gyrus were measured in its most rostral region. The data are expressed as means ± SD. To obtain an average value over 24 h (24-h value) for each mouse strain, the values from all four time points investigated were pooled. The control sense probes did not generate a signal in basal ganglia or meninges. A weak signal was detected in the hippocampus but this signal was much less intense than that generated by the antisense probes. The size, shape and location of the various brain regions were confirmed by using sections counterstained with 0.125 % cresyl fast violet (Chroma/Waldeck, Muenster, Germany). The mouse brain atlas of Franklin and Paxinos (2008) served as a reference.
Locomotor activity rhythms
Animals were entrained to a 12L/12D photoperiod for at least 1 week before experiments. An infrared recording system (Mouse-E-Motion, Hamburg, Germany) was used continuously to record spontaneous locomotor activity at 10-min intervals for at least ten consecutive days under LD conditions (Pfeffer et al. 2012). Actograms and activity profiles with acrophases were generated by using Clocklab software (Actimetrics, Wilmette, Ill., USA) as previously described (Pfeffer et al. 2012).
Statistical analysis
Statistical analyses were performed by using the BIAS. program package (Ackermann 1991). Time-dependent variances were tested by using the non-parametric Kruskal-Wallis test followed by the Conover-Iman (Bonferroni-Holm correction) multiple comparison test (comparing the time points examined). When n = 3 with a strong effect size (MT1 KO, CT18), the following test was used: Headrick's quasi-exacter p-Wert for a low number of cases. For analysis of locomotor activity rhythms, an unpaired t-test was employed to compare the WT with either of the transgenic mouse genotypes after confirmation was obtained that variances were not significantly different. Differences were considered significant at P < 0.05.
Results
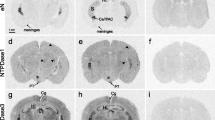
To examine whether the melatoninergic system might influence eN mRNA levels, we investigated and tested transgenic mice deficient in the MT1 melatonin receptor subtype (MT1 KO) or both MT1 and MT2 melatonin receptor subtypes (MT1/2 KO) and the corresponding WT controls by using ISH with specific eN oligoprobes. In all three strains investigated, eN mRNA was found in the striatum, meninges, hippocampus including the dentate gyrus, fasciola cinerea and indusium griseum, the lateral and capsular areas of the central amygdala and the interstitial nucleus of the posterior limb of the anterior commissure (Fig. 1).
Distribution of ecto-5′-nucleotidase (eN) mRNA in coronal sections of the prosencephalon of melatonin receptor-deficient mice and the corresponding wild-type (WT) controls a-l Representative autoradiographs depicting signals from radioactive in situ hybridization (ISH) with antisense probes for eN in the prosencephalon of animals killed at CT12 (a-f), namely WT (a, d), MT1 KO (b, e) and MT1/2 KO (c, f) and at CT18 (g-l), namely WT (g, j), MT1 KO (h, k) and MT1/2 KO (i, l). In all mouse strains, eN mRNAs were found in the striatum (S), hippocampus (HI) including the fasciola cinerea (FC) and indusium griseum (IG), the lateral and capsular areas of the central amygdala, the interstitial nucleus of the posterior limb of the anterior commissure (Ce/IPAC) and the meninges. At CT12, eN mRNA levels were lowest in MT1 KO mice and intermediate in MT1/2 KO compared with WT. At CT18, mRNA levels were lowest in MT1/2 KO and intermediate in MT1 KO. m–o No signal was generated with sense probes with the exception of a weak signal in the hippocampus. Bar 1 mm
The eN mRNA levels in the striatum and the dentate gyrus were then analyzed at four different time points during the day (CT00, CT06, CT12 and CT18). mRNA levels pooled from all four time points in order to obtain an average value over 24 h (24 h-value) were significantly weaker in MT1 KO and MT1/2 KO than in WT mice. Notably, no difference was seen between MT1 KO and MT1/2 KO mice (Fig. 2a, b). However, differences between MT1 KO and MT1/2 KO mice became apparent when mRNA levels were analyzed separately for each of the four different time points to demonstrate the time-dependent mRNA patterns (Figs. 2c, d, 3).
Levels of eN mRNA in the striatum and hippocampal dentate gyrus of melatonin receptor-deficient mice and the corresponding WT controls. a, b Mean values of eN mRNA levels pooled from all four time points investigated to obtain an average value over 24 h (24-h value) in the dentate gyrus (DG; a) and in the striatum (b). The 24-h values were significantly lower in MT1 KO and MT1/2 KO than in WT mice but no differences were found between MT1 KO and MT1/2 KO mice. c, d Levels of eN mRNA in the DG (c) and the striatum (d) of the different genotypes at the four different time points investigated. The differences in mRNA levels between the genotypes at the different time points did not reveal a consistent pattern. Values are expressed as means ± SD (n ≥ 3 per time point and genotype). ***P < 0.001, ****P < 0.0001; Kruskal-Wallis test
Time-dependent patterns of eN mRNA in the prosencephalon of melatonin receptor-deficient mice and the corresponding WT controls. a Temporal patterns of eN mRNA in the hippocampal DG. b Temporal patterns of eN mRNA in the striatum. a, b In both the DG and the striatum, a graded time-dependent pattern was observed in WT. The mRNA levels were lowest at the beginning of the subjective day (CT00), increased throughout the day and peaked in the middle of the subjective night (CT18). In MT1 KO mice, mRNA levels peaked at CT18; however, no clear increase during the day was observed. In MT1/2 KO mice, mRNA levels decreased between CT12 and CT18 and the peak in mRNA levels shifted to the day-time. The relative optic density (rel. OD) of the autoradiographic signal was used to determine mRNA levels at the four different time points of the day. Values are expressed as means ± SD (n ≥ 3 per time point and genotype). *P < 0.05, **P < 0.01, ***P < 0.001; Kruskal-Wallis test
WT mice (bearing a C3H background) displayed clear time-dependent changes in eN mRNA levels (Fig. 3), similar to those previously shown for the C3H mouse strain (Homola et al. 2015). mRNA levels were lowest at the beginning of the subjective day (CT00), increased during the day and were highest in the middle of the subjective night (CT18). This temporal pattern was changed in the melatonin receptor-deficient mice (Fig. 3). The MT1 KO mice revealed peak levels at CT18 similar to WT mice but no significant rise in mRNA levels was observed from CT00 to CT12. In the MT1/2 KO mice, mRNA levels rose during the subjective day; however, instead of a peak at CT18 (middle of the subjective night), a drop was observed and the peak in mRNA levels was shifted toward day-time. These time-dependent alterations in mRNA levels were detected in both the hippocampal dentate gyrus (Fig. 3a) and the striatum (Fig. 3b).
In an attempt to relate the time-dependent differences in mRNA levels to physiological output, we compared the spontaneous locomotor activity rhythms of the transgenic mice (MT1 KO and MT1/2 KO) with their WT controls. Notably, MT1/2 KO mice were significantly more active during day-time (the inactive phase) than WT mice (Fig. 4). This was evident from both the actograms (Fig. 4a, a’, a’’) and from the plots of the averages of day-time activity (Fig. 4b).
Locomotor activity rhythms of melatonin receptor-deficient mice and their corresponding WT controls in a standard photoperiod. a Double-plotted actograms of spontaneous locomotor activity under 12 h light:12 h dark conditions of representative WT (a), MT1 KO (a’) and MT1/2 KO (a’’) mice. Gray areas indicate periods of darkness. b Bar plots depicting the relative day-time activity of mice of the various genotypes during the inactive phase. MT1/2 KO mice were significantly more active during the day-time than were WT mice. Values are expressed as means ± SEM (n = 6 per genotype). *P < 0.05; unpaired t-test
Discussion
In a recent study, we demonstrated that the temporal patterns of ectonucleotidase mRNA availability in the prosencephalon are distinctly different between melatonin-proficient and melatonin-deficient mice, suggesting that melatonin influences eN and NTPDase-family ectonucleotidase mRNA availability (Homola et al. 2015). To gain insight into the melatonin receptor subtype mediating this effect, we have now compared eN mRNA levels in melatonin-proficient WT mice and in melatonin-proficient mice deficient in the melatonin receptor subtype MT1 (MT1 KO) or in both melatonin receptor subtypes (MT1/2 KO) at four different time points.
In general, eN mRNA distribution in the brain did not differ between WT, MT1 KO and MT1/2 KO mice. The eN-positive brain regions included the meninges, striatum, hippocampus with dentate gyrus, fasciola cinerea and indusium griseum, lateral and capsular areas of the central amygdala and interstitial nucleus of the posterior limb of the anterior commissure. However, eN mRNA levels revealed marked strain- and time-dependent differences as determined in the striatum and dentate gyrus. The averaged 24-h values were significantly reduced in both the MT1 KO and MT1/2 KO mice indicating that the melatoninergic system increased eN mRNA levels. Apparently, the MT1 receptor subtype primarily mediated this response, since the 24-h values of the eN mRNA levels were reduced in MT1 and MT1/2 KO mice to a similar extent. However, interaction between the two receptor subtypes should also be considered, as such interaction might lead to a similar effect (a reduction within both types of transgenic mice). Therefore, further investigations of this mechanism are necessary in order to determine the relevant receptor subtype/subtypes involved.
On the other hand, the MT2 receptor subtype seems to play a role in setting the maximum of the eN mRNA levels to the night-time. In the absence of this receptor subtype, the elevated night-time eN mRNA levels observed in WT and MT1 KO are abrogated and the peak in eN mRNA levels is shifted toward day-time in MT1/2 KO mice. The latter observation is in agreement with the suggested role of melatonin and MT2 in sleep regulation (Comai and Gobbi 2014; Fisher and Sugden 2009; Ochoa-Sanchez et al. 2011). Adenosine is known to accumulate during wakefulness and to increase sleep pressure. In the absence of MT2 and the elevated night-time eN mRNA levels, this accumulation would be impaired and sleep pressure would be reduced. This hypothesis is also supported by our behavioral experiments: the day-time locomotor activity (during the resting phase) is increased in the MT1/2 KO mice as compared with WT mice but not in the MT1 KO mice.
Our study focused on the effects of the melatoninergic system on eN mRNA levels and future experiments are needed to demonstrate whether the melatoninergic system elicits similar changes in protein levels and adenosine concentration. Nevertheless, previous studies have shown a direct relationship between increased eN mRNA levels and adenosine production (Narravula et al. 2000). Furthermore, increased night-time adenosine levels as compared with day-time levels have been demonstrated in the rat basal forebrain (Murillo-Rodriguez et al. 2004). Therefore, it may be assume that the elevated night-time eN mRNA levels shown here lead to higher extracellular adenosine levels.
The molecular mechanisms by which melatonin promotes and gates the elevation of eN mRNA levels remain to be elucidated. Melatonin can act upon many signal transduction pathways depending on the receptor subtype, on the G-protein subunits involved and on the tissue and cell type. MT1 can couple to both pertussis-toxin-sensitive Gi and -insensitive Gq/11 proteins and thereby inhibit adenylyl cyclase or stimulate PLC (isoforms β or η) respectively. MT1 activation can also stimulate mitogen-activated protein kinase/extracellular signal-regulated kinase (MEK1/2-ERK1/2) pathways in non-neuronal cells and modulate ion fluxes via the activation or inhibition of specific ion channels, such as the inward rectifier potassium channels Kir3 or the potassium channel BKCa, in a tissue-dependent manner. MT2 can inhibit cAMP and cGMP formation, activate protein kinase C and interact with PLC to regulate the diacylglycerol signaling pathway. Both MT1 and MT2 can stimulate c-Jun N-terminal kinase (JNK) activity (for reviews, see Dubocovich et al. 2010; Hardeland et al. 2011; Zawilska et al. 2009). Interestingly, a novel concept of the MT1-dependent activation of the cAMP pathway via coupling to Gs proteins has emerged (Chen et al. 2014; Shiu et al. 2010). Furthermore, the propensity of MT1 and MT2 to form homo- and heteromers (Tosini et al. 2014) and their ability to bind melatonin in an uncoupled state (Legros et al. 2014) should be kept in mind when considering possible melatonin-dependent mechanisms. In addition to the various and convergent signaling pathways attributed to MT1 or MT2, melatonin-receptor-independent responses have been reported (Jan et al. 2011; Reiter et al. 2014) and may also play a role in melatonin-dependent mechanisms.
Based on these observations, melatonin possibly affects eN mRNA levels via various mechanisms. Three possible scenarios will be discussed in the following. (1) The melatonin-dependent stimulation of eN mRNA levels might be accomplished via a signal transduction cascade involving MT1, Gs proteins, increases in cAMP levels and activation of a cAMP response element (CRE), which has previously been demonstrated in the eN promotor (Hansen et al. 1995). Such a mechanism would imply that MT1 receptors are located on eN-expressing cells. (2) Both the induction and the gating of elevation of eN mRNA levels to the night-time might be mediated by melatonin via a sensitizing effect, whereby melatonin would potentiate the inducing effect of an additional factor on eN transcription. Such a role for melatonin has been shown in the hypophysial pars tuberalis where melatonin sensitizes the day-time inducing effect of adenosine on mPer1 expression (von Gall et al. 2002a). Notably, Narravula et al. (2000) obtained evidence that adenosine can induce eN transcription and, consequently, an increase in surface protein. These authors demonstrated the induction of eN mRNA, eN surface expression and eN function by adenosine and adenosine analogues in the vascular endothelium, mediated predominantly via the A2B receptor and suggested a feedback mechanism of direct regulation of eN expression via adenosine. A mechanism whereby melatonin sensitizes the inducing effect of adenosine on eN expression would require the presence of both melatonin and adenosine receptors on eN-expressing cells. (3) Melatonin may elicit an inhibitory action upon inhibitory neurons that are synaptically connected to eN-expressing neurons, thus causing the disinhibition of eN-expressing cells. Such a mechanism would imply that the MT receptor subtypes are located on neurons that are different from the eN-expressing nerve cells. Limited information concerning eN cellular distribution is available; however, Augusto et al. (2013) reported the colocalization of eN and the adenosine receptor A2A in the basal ganglia. At the cellular and subsynaptic level, they colocalized eN and A2A in astrocytes and postsynaptic sites in the striatum. Ena et al. (2013) found eN in striatopallidal but not in striatonigral neurons. Expression of melatonin receptors in the basal ganglia has been reported, although some controversy regarding the exact localization of the different receptor subtypes exists (Adamah-Biassi et al. 2013; Lacoste et al. 2015; Ochoa-Sanchez et al. 2011; Uz et al. 2003). Notably, the occurrence of various complementary or even opposing mechanisms in the same region is also possible as previously reported (Doolen et al. 1998; Farez et al. 2015; Wan et al. 1999). Clearly, additional examination of the molecular basis of the melatonin-mediated modulation of eN expression is required in order to fully understand the mechanisms by which melatonin stimulates and gates the elevation of eN mRNA levels.
The impact of the MT2 receptor on the elevation of eN mRNA levels at night-time and the locomotor activity during day-time demonstrated in this study might be of relevance to the suggested role of this receptor in the regulation of sleep (Comai and Gobbi 2014; Fisher and Sugden 2009; Ochoa-Sanchez et al. 2011). As is well established, adenosine levels, accumulated throughout activity in the basal forebrain (Murillo-Rodriguez et al. 2004), induce the sensation of tiredness and promote sleep (Brown et al. 2012). Since eN functions as the major ectonucleotidase hydrolyzing AMP to extracellular adenosine at physiological pH (Kulesskaya et al. 2013; Lacoste et al. 2015) and since increased eN mRNA levels can be correlated with elevated adenosine production (Narravula et al. 2000), eN mRNA levels might mirror extracellular adenosine concentrations in the rodent brain. This notion is reinforced by the finding that both eN expression levels and adenosine concentrations are elevated at night-time (Murillo-Rodriguez et al. 2004). Extremely high levels of eN mRNA are present in the basal ganglia, specifically in the striatum, interstitial nucleus of the posterior limb of the anterior commissure (IPAC) and central amygdala (Homola et al. 2015). The proximity of these regions to the basal forebrain, together with reports of projections connecting these regions to areas in the basal forebrain (Shammah-Lagnado et al. 2001), makes it reasonable to assume that eN in the striatum, IPAC and central amygdala are also involved in the regulation of adenosine levels in the basal forebrain. Expression of both melatonin receptors has been reported for the striatum (Uz et al. 2003). However, a recent study (Adamah-Biassi et al. 2013) excluded MT1 expression in this region, implying that MT2 is the only melatonin receptor subtype present. Albeit, Lacoste et al. (2015) detected neither MT1 nor MT2 immunoreactivity in the striatum but rather MT2 immunostaining in the globus pallidus, consistent with a previous report showing neuronal labeling of MT2 in the ventral pallidum and globus pallidus (Ochoa-Sanchez et al. 2011). The presence of melatonin receptors in the basal ganglia (Adamah-Biassi et al. 2013; Lacoste et al. 2015; Ochoa-Sanchez et al. 2011; Uz et al. 2003) and the impact of melatonin on eN mRNA levels presented here imply a functional interplay between melatonin and adenosine in the mechanism of sleep regulation.
For nocturnal animals such as mice, the rest phase during which the animal sleeps is the day-time. The increased day-time locomotor activity observed in MT1/2 KO mice thus corresponds to reduced sleeping time. The lack of a difference between WT and MT1 KO mice implies that sleep homeostasis is affected in the absence of the MT2 receptor. This notion further corresponds to our previous observation of higher day-time activity in melatonin-deficient (C57Bl) mice as compared with melatonin-proficient (C3H) mice (Homola et al. 2015) and to observations with polysomnographic recordings (electroencephalogram/electromyogram) of elevated day-time wakefulness in MT2 KO mice as compared with WT mice (Ochoa-Sanchez et al. 2011).
Both melatonin and adenosine play a major role in the regulation of important physiological processes. Interestingly, the melatonin- and the adenosine-mediated (patho)physiological functions strongly converge. Many studies describe the involvement of melatonin, adenosine, or both in wakefulness and sleep regulation (Brown et al. 2012; Comai and Gobbi 2014; Huang et al. 2014), nociception (Srinivasan et al. 2012; Zylka 2011), inflammation and immunomodulation (Alam et al. 2015; Esposito and Cuzzocrea 2010; Mauriz et al. 2012) and neuroprotection (Burnstock 2015; Wang 2009). Furthermore, both compounds share similarities in their mode of action. Both melatonin and adenosine receptors are G-protein-coupled receptors mediating adenylyl cyclase inhibition or stimulation and thus modulation of the cAMP signal transduction pathway.
This overlap between the melatonin and adenosine signaling pathways raises the question as to whether the purinergic and the circadian systems interact with each other and, if yes, in what way. Several reports provide strong evidence for an impact of adenosine on the circadian system. Adenosine receptors are present in the SCN (Sigworth and Rea 2003). Adenosine increases melatonin production and augments norepinephrine stimulation in rat pineal explants (Vacas et al. 1989), it regulates the response of the circadian clock to the phase-adjusting effects of light (Elliott et al. 2001), it modulates retinohypothalamic neurotransmission in the SCN (Hallworth et al. 2002) and it regulates the intrinsic circadian clock in immune cells (Perez-Aso et al. 2013).
The data presented in this study further suggest a reciprocal interaction between the two systems rather than a unidirectional relationship. Understanding the precise molecular basis of this interaction could greatly benefit therapeutic approaches for the treatment of the multiple adenosine/melatonin-responsive diseases.
In summary, we present novel data demonstrating the impact of the melatoninergic system on the mRNA levels of the predominantly extracellular adenosine-producing enzyme eN in mouse brain. In the absence of MT1 and of both MT1 and MT2 receptors, eN mRNA levels are reduced implying a MT1-mediated effect. The MT2 receptor is involved in the increase of eN mRNA levels during night-time. Our results suggest the existence of a reciprocal interaction between the circadian and the purinergic systems involving MT1 and MT2 receptors.
References
Ackermann H (1991) BIAS. A program package for biometrical analysis of samples. Comput Stat Data Anal 11:223–224
Adamah-Biassi E, Zhang Y, Jung H, Vissapragada S, Miller R, Dubocovich M (2013) Distribution of MT1 melatonin receptor promoter-driven RFP expression in the brains of BAC C3H/HeN transgenic mice. J Histochem Cytochem 62:70–84
Alam M, Costales M, Cavanaugh C, Williams K (2015) Extracellular adenosine generation in the regulation of pro-inflammatory responses and pathogen colonization. Biomolecules 5:775–792
Allard B, Turcotte M, Stagg J (2012) CD73-generated adenosine: orchestrating the tumor-stroma interplay to promote cancer growth. J Biomed Biotechnol 15:1–8
Augusto E, Matos M, Sevigny J, El-Tayeb A, Bynoe MS, Muller CE, Cunha RA, Chen J (2013) Ecto-5′-nucleotidase (CD73)-mediated formation of adenosine is critical for the striatal adenosine A2A receptor functions. J Neurosci 33:11390–11399
Bartness TJ, Powers JB, Hastings MH, Bittman EL, Goldman BD (1993) The timed infusion paradigm for melatonin delivery: what has it taught us about the melatonin signal, its reception, and the photoperiodic control of seasonal responses? J Pineal Res 15:161–190
Bhattacharya A, Vavra V, Svobodova I, Bendova Z, Vereb G, Zemkova H (2013) Potentiation of inhibitory synaptic transmission by extracellular ATP in rat suprachiasmatic nuclei. J Neurosci 33:8035–44
Boison D (2008) Adenosine as a neuromodulator in neurological diseases. Curr Opin Pharmacol 8:2–7
Brown RE, Basheer R, McKenna JT, Strecker RE, McCarley RW (2012) Control of sleep and wakefulness. Physiol Rev 92:1087–1187
Burnstock G (2015) An introduction to the roles of purinergic signalling in neurodegeneration, neuroprotection and neuroregeneration. Neuropharmacology. doi:10.1016/j.neuropharm.2015.05.031
Chen L, He X, Zhang Y, Chen X, Lai X, Shao J, Shi Y, Zhou N (2014) Melatonin receptor type 1 signals to extracellular signal-regulated kinase 1 and 2 via Gi and Gs dually coupled pathways in HEK-293 cells. Biochemistry 53:2827–2839
Colgan SP, Eltzschig HK, Eckle T, Thompson LF (2006) Physiological roles for ecto-5′-nucleotidase (CD73). Purinergic Signal 2:351–360
Comai S, Gobbi G (2014) Unveiling the role of melatonin MT2 receptors in sleep, anxiety and other neuropsychiatric diseases: a novel target in psychopharmacology. J Psychiatry Neurosci 39:6–21
Doolen S, Krause DN, Dubocovich ML, Duckles SP (1998) Melatonin mediates two distinct responses in vascular smooth muscle. Eur J Pharmacol 345:67–69
Dubocovich ML, Delagrange P, Krause DN, Sugden D, Cardinali DP, Olcese J (2010) International Union of Basic and Clinical Pharmacology. LXXV. Nomenclature, classification, and pharmacology of G protein-coupled melatonin receptors. Pharmacol Rev 62:343–380
Elliott KJ, Todd Weber E, Rea MA (2001) Adenosine A1 receptors regulate the response of the hamster circadian clock to light. Eur J Pharmacol 414:45–53
Ena SL, De Backer J, Schiffmann SN, Kerchove d'Exaerde A de (2013) FACS array profiling identifies ecto-5′ nucleotidase as a striatopallidal neuron-specific gene involved in striatal-dependent learning. J Neurosci 33:8794–8809
Esposito E, Cuzzocrea S (2010) Antiinflammatory activity of melatonin in central nervous system. Curr Neuropharmacol 8:228–242
Farez MF, Mascanfroni ID, Méndez-Huergo SP, Yeste A, Murugaiyan G, Garo LP, Balbuena Aguirre ME, Patel B, Ysrraelit MC, Zhu C, Kuchroo VK, Rabinovich GA, Quintana FJ, Correale J (2015) Melatonin contributes to the seasonality of multiple sclerosis relapses. Cell 162:1338–1352
Fisher SP, Sugden D (2009) Sleep-promoting action of IIK7, a selective MT2 melatonin receptor agonist in the rat. Neurosci Lett 457:93–96
Franklin KBJ, Paxinos G (2008) The mouse brain in stereotaxic coordinates. Academic Press, New York
Gall C von, Garabette ML, Kell CA, Frenzel S, Dehghani F, Schumm-Draeger P, Weaver DR, Korf HW, Hastings MH, Stehle JH (2002a) Rhythmic gene expression in pituitary depends on heterologous sensitization by the neurohormone melatonin. Nat Neurosci 5:234–238
Gall C von, Stehle JH, Weaver DR (2002b) Mammalian melatonin receptors: molecular biology and signal transduction. Cell Tissue Res 309:151–162
Hallworth R, Cato M, Colbert C, Rea MA (2002) Presynaptic adenosine A1 receptors regulate retinohypothalamic neurotransmission in the hamster suprachiasmatic nucleus. J Neurobiol 52:230–240
Hansen KR, Resta R, Webb CF, Thompson LF (1995) Isolation and characterization of the promoter of the human 5′-nucleotidase (CD73)-encoding gene. Gene 167:307–312
Hardeland R, Cardinali DP, Srinivasan V, Spence DW, Brown GM, Pandi-Perumal SR (2011) Melatonin-a pleiotropic, orchestrating regulator molecule. Prog Neurobiol 93:350–384
Haskó G, Cronstein BN (2004) Adenosine: an endogenous regulator of innate immunity. Trends Immunol 25:33–39
Homola M, Pfeffer M, Fischer C, Zimmermann H, Robson SC, Korf HW (2015) Expression of ectonucleotidases in the prosencephalon of melatonin-proficient C3H and melatonin-deficient C57Bl mice: spatial distribution and time-dependent changes. Cell Tissue Res 362:163–176
Huang Z, Zhang Z, Qu W (2014) Roles of adenosine and its receptors in sleep–wake regulation. In: Mori A (ed) Adenosine receptors in neurology and psychiatry, vol 119. Elsevier, Amsterdam, pp 349–371
Jacobson KA, Gao Z (2006) Adenosine receptors as therapeutic targets. Nat Rev Drug Discov 5:247–264
Jan JE, Reiter RJ, Wong PKH, Bax MCO, Ribary U, Wasdell MB (2011) Melatonin has membrane receptor-independent hypnotic action on neurons: an hypothesis. J Pineal Res 50:233–240
Korf HW, Gall C von (2013) Circadian physiology. In: Pfaff DW (ed) Neuroscience in the 21st century. From basic to clinical. Circadian physiology. Springer, Berlin Heidelberg New York, pp 1813–1845
Kulesskaya N, Võikar V, Peltola M, Yegutkin GG, Salmi M, Jalkanen S, Rauvala H (2013) CD73 is a major regulator of adenosinergic signalling in mouse brain. PLoS One 8:e66896
Lacoste B, Angeloni D, Dominguez-Lopez S, Calderoni S, Mauro A, Fraschini F, Descarries L, Gobbi G (2015) Anatomical and cellular localization of melatonin MT 1 and MT 2 receptors in the adult rat brain. J Pineal Res 58:397–417
Langer D, Hammer K, Koszalka P, Schrader J, Robson SC, Zimmermann H (2008) Distribution of ectonucleotidases in the rodent brain revisited. Cell Tissue Res 334:199–217
Legros C, Devavry S, Caignard S, Tessier C, Delagrange P, Ouvry C, Boutin JA, Nosjean O (2014) Melatonin MT 1 and MT 2 receptors display different molecular pharmacologies only in the G-protein coupled state. Br J Pharmacol 171:186–201
Liu C, Weaver DR, Jin X, Shearman LP, Pieschl RL, Gribkoff VK, Reppert SM (1997) Molecular dissection of two distinct actions of melatonin on the suprachiasmatic circadian clock. Neuron 19:91–102
Mauriz JL, Collado PS, Veneroso C, Reiter RJ, González-Gallego J (2012) A review of the molecular aspects of melatonin’s anti-inflammatory actions: recent insights and new perspectives. J Pineal Res 54:1–14
Murillo-Rodriguez E, Blanco-Centurion C, Gerashchenko D, Salin-Pascual R, Shiromani P (2004) The diurnal rhythm of adenosine levels in the basal forebrain of young and old rats. Neuroscience 123:361–370
Narravula S, Lennon PF, Mueller BU, Colgan SP (2000) Regulation of endothelial CD73 by adenosine: paracrine pathway for enhanced endothelial barrier function. J Immunol 165:5262–5268
Ochoa-Sanchez R, Comai S, Lacoste B, Bambico FR, Dominguez-Lopez S, Spadoni G, Rivara S, Bedini A, Angeloni D, Fraschini F, Mor M, Tarzia G, Descarries L, Gobbi G (2011) Promotion of non-rapid eye movement sleep and activation of reticular thalamic neurons by a novel MT2 melatonin receptor ligand. J Neurosci 31:18439–18452
Perez-Aso M, Feig JL, Aránzazu M, Cronstein BN (2013) Adenosine A2A receptor and TNF-α regulate the circadian machinery of the human monocytic THP-1 cells. Inflammation 36:152–162
Pfeffer M, Rauch A, Korf HW, Gall C von (2012) The endogenous melatonin (MT) signal facilitates reentrainment of the circadian system to light-induced phase advances by acting upon MT2 receptors. Chronobiol Int 29:415–429
Regateiro FS, Cobbold SP, Waldmann H (2013) CD73 and adenosine generation in the creation of regulatory microenvironments. Clin Exp Immunol 171:1–7
Reiter RJ, Tan DX, Galano A (2014) Melatonin: exceeding expectations. Physiology (Bethesda) 29:325–333
Shammah-Lagnado SJ, Alheid GF, Heimer L (2001) Striatal and central extended amygdala parts of the interstitial nucleus of the posterior limb of the anterior commissure: evidence from tract-tracing techniques in the rat. J Comp Neurol 439:104–126
Shiu SYW, Pang B, Tam CW, Yao K (2010) Signal transduction of receptor-mediated antiproliferative action of melatonin on human prostate epithelial cells involves dual activation of Gαs and Gαq proteins. J Pineal Res 49:301–311
Sigworth L, Rea M (2003) Adenosine A1 receptors regulate the response of the mouse circadian clock to light. Brain Res 960:246–251
Spychala J, Kitajewski J (2004) Wnt and β-catenin signaling target the expression of ecto-5′-nucleotidase and increase extracellular adenosine generation. Exp Cell Res 296:99–108
Srinivasan V, Lauterbach EC, Ho KY, Acuña-Castroviejo D, Zakaria R, Brzezinski A (2012) Melatonin in antinociception: its therapeutic applications. Curr Neuropharmacol 10:167–178
Tosini G, Owino S, Guillaume J, Jockers R (2014) Understanding melatonin receptor pharmacology: latest insights from mouse models, and their relevance to human disease. Bioessays 36:778–787
Uz T, Akhisaroglu M, Ahmed R, Manev H (2003) The pineal gland is critical for circadian period1 expression in the striatum and for circadian cocaine sensitization in mice. Neuropsychopharmacology 28:2117–2123
Vacas MI, Sarmiento MI, Pereyra EN, Cardinali DP (1989) Effect of adenosine on melatonin and norepinephrine release in rat pineal explants. Acta Physiol Pharmacol Latinoam 39:189–195
Wan Q, Man H, Liu F, Braunton J, Niznik HB, Pang SF, Brown GM, Wang YT (1999) Differential modulation of GABAA receptor function by Mel1a and Mel1b receptors. Nat Neurosci 2:401–403
Wang X (2009) The antiapoptotic activity of melatonin in neurodegenerative diseases. CNS Neurosci Ther 15:345–357
Wang H, Lee S, Lo Nigro C, Lattanzio L, Merlano M, Monteverde M, Matin R, Purdie K, Mladkova N, Bergamaschi D, Harwood C, Syed N, Szlosarek P, Briasoulis E, McHugh A, Thompson A, Evans A, Leigh I, Fleming C, Inman GJ, Hatzimichael E, Proby C, Crook T (2012) NT5E (CD73) is epigenetically regulated in malignant melanoma and associated with metastatic site specificity. Br J Cancer 106:1446–1452
Zawilska JB, Skene DJ, Arendt J (2009) Physiology and pharmacology of melatonin in relation to biological rhythms. Pharmacol Rep 61:383–410
Zimmermann H (1992) 5′-Nucleotidase molecular structure and functional aspects. Biochem J 285:345–365
Zylka MJ (2011) Pain-relieving prospects for adenosine receptors and ectonucleotidases. Trends Mol Med 17:188–196
Acknowledgments
The authors thank Elke Laedtke and Iris Habazettl for their excellent work and Dr. Hanns Ackermann (Institute for Biostatistics and Mathematic Modelling, Goethe University, Frankfurt am Main, Germany) for his kind help with the statistical analyses.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work was supported by the Dr. Senckenbergische Stiftung, Frankfurt am Main.
Rights and permissions
About this article
Cite this article
Homola, M., Pfeffer, M., Robson, S.C. et al. Melatonin receptor deficiency decreases and temporally shifts ecto-5′-nucleotidase mRNA levels in mouse prosencephalon. Cell Tissue Res 365, 147–156 (2016). https://doi.org/10.1007/s00441-016-2378-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-016-2378-x