Abstract
Extracellular purines (ATP, ADP, AMP and adenosine) are important signaling molecules in the CNS. Levels of extracellular purines are regulated by enzymes located at the cell surface referred to as ectonucleotidases. Time-dependent changes in their expression could profoundly influence the availability of extracellular purines and thereby purinergic signaling. Using radioactive in situ hybridization, we analyzed the mRNA distribution of the enzymes NTPDase1, -2 and -3 and ecto-5′-nucleotidase in the prosencephalon of two mouse strains: melatonin-proficient C3H and melatonin-deficient C57Bl. The mRNAs of these enzymes were localized to specific brain regions, such as hippocampus, striatum, medial habenula and ventromedial hypothalamus. NTPDase3 expression was more widely distributed than previously thought. All ectonucleotidases investigated revealed a prominent time-dependent expression pattern. In C3H, the mRNA expression of all four enzymes gradually increased during the day and peaked during the night. In contrast, in C57Bl, ecto-5′-nucleotidase expression peaked at the beginning of the day and gradually decreased to trough levels at night. Recording of locomotor activity revealed higher daytime activity of C57Bl than of C3H. Our results indicate that the expression of ectonucleotidases varies according to time and genotype and suggest that melatonin exerts modulatory effects associated with different regulations of purinergic signaling in the brain. These findings provide an important basis for further examination of the complexity of the purinergic system in the brain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Extracellular purines [adenosine 5′-triphosphate (ATP), adenosine 5′-diphosphate (ADP), adenosine 5′-monophosphate (AMP), and adenosine] are important signaling molecules involved in many physiological processes of the nervous system, such as neurotransmission and -modulation, neural development and protection, neuron–glia reciprocal signaling and responses of the innate immune system (Burnstock 2006, 2009; Abbracchio et al. 2009; Burnstock and Verkhratsky 2012; Mutafova-Yambolieva and Durnin 2014). In particular, the nucleoside adenosine has received considerable attention during the past decades. Adenosine has been shown to directly affect the regulation of sleep, vasodilation and neuroprotection (Dunwiddie and Masino 2001; Haskó and Cronstein 2004), nociception (Zylka 2011), innate immunity and inflammation (Haskó and Cronstein 2004).
The regulation of extracellular purine levels is primarily accomplished by enzymes located at the cell surface, referred to as ectonucleotidases. Ectonucleotidases belong to several enzyme families, including the nucleoside triphosphate diphosphohydrolase family (E-NTPDases; EC 3.6.1.5), ectonucleotide pyrophosphatase/phosphodiesterase family (E-NPPs; EC 3.6.1.9, EC 3.1.4.1), alkaline phosphatases (EC 3.1.3.1), and ecto-5′-nucleotidase (EC 3.1.3.5) (Zimmermann 2000; Robson et al. 2006; Zimmermann et al. 2012). At physiological extracellular pH, the ectonucleoside triphosphate diphosphohydrolases 1, 2 and 3 (NTPDase1/2/3) that hydrolyze ATP and ADP to AMP, and ecto-5′nucleotidase (eN) that converts AMP to adenosine, are considered to be the dominant ectonucleotidases in the brain. NTPDase8 is absent from brain but highly expressed in liver, kidney, and intestine (Langer et al. 2008; Kukulski et al. 2011).
Although various enzyme histochemical and immunohistochemical data are available to date, our knowledge about the exact localization of these ectonucleotidases in the brain is still incomplete. Enzyme histochemistry has been most commonly used to analyze the distribution of these enzymes. Immunohistochemical data are scarce due to the limited availability of specific antibodies.
To investigate the spatial distribution of cells expressing NTPDase1-3 and eN in the mouse prosencephalon, we applied radioactive in situ hybridization using a triplet of specific oligoprobes. Since time-dependent changes in the expression of the enzymes could have profound effects on the availability of extracellular purines and the temporal regulation of extracellular purine-regulated physiological processes, we examined whether the expression of these enzymes revealed day/night changes. We compared C3H mice, which produce melatonin—an important output signal associated with the circadian system—and C57Bl mice, which are melatonin-deficient. These mice have been widely used to decipher mechanisms of the circadian system and melatonin-dependent processes (von Gall et al. 1998, 2000; Korf et al. 2003; Korf and von Gall 2006; Yasuo et al. 2009; Pfeffer et al. 2012; Goto et al. 2013; Wicht et al. 2014).
Our results provide novel information regarding the mRNA localization of these enzymes and reveal conspicuous time-dependent changes in their expression. Furthermore, we demonstrate significant differences between the two mouse strains, which suggest that melatonin exerts a modulatory effect on the expression of ectonucleotidases and purinergic signaling in the mouse brain.
Materials and methods
Animals
All animal experiments were conducted in accordance with accepted standards of human animal care and were consistent with federal guidelines and Directive 2010/63/EU of the European Union. Adult C3H/HeN and C57BL/6N mice were obtained from Charles River (Sulzfeld, Germany). NTPDase2 knockout mice and the corresponding wild-type (WT) have been described previously (Vandenbeuch et al. 2013; Gampe et al. 2014). Their genotypes were confirmed by PCR. Experiments were performed with 11- to 17-week-old male mice. Animals experienced a photoperiod of 12 h light (L) and 12 h dark (D) (light phase: 230 μW/cm2; dark phase: dim red light <5 μW/cm2). Food and water were available ad libitum. For in situ hybridization (ISH), 1 day before tissue sampling animals were transferred to constant darkness (DD) for one complete day/night cycle. Circadian time (CT) 00 was defined as lights on in the previous light–dark schedule. Mice were anesthetized with isofluran and then decapitated at four different time points in the darkness (CT00, CT06, CT12 and CT18; four animals per strain and time point). Eyes were removed before light was switched on and the brains were dissected and immediately frozen in isopentane on dry ice. Tissue was kept at −80 °C until serial coronal 20-μm-thick cryostat sections were prepared using a cryotome (Leica, Wetzlar, Germany).
Genotyping
Genomic DNA was isolated from tail biopsies using the PEQLAB Direct PCR® Lysis Reagent Tail (Erlangen, Germany) according to the manufacturer’s instructions. A PCR reaction was performed using the following primer pairs: WT: forward: 5′-GTT AGG GGG TGT CTG TCT GG-3′, reverse: 5′-AAA TGC TGC CTG GAA ATGG-3′; NTPDase2 KO: forward: 5′-CCG TAA AGC ACG AGG AAGC-3′, reverse: 5′-CTG GAC GAA GAG CAT CAGG-3′. The PCR conditions used were: 95 °C (30 s), 60 °C (30 s) and 72 °C (90 s) × 35 cycles.
Radioactive in situ hybridization
For each enzyme, three antisense 45-mer oligonucleotide probes or their corresponding sense probes were pooled. The following antisense probes were used: eN antisense1: (AS1) 5′-GAG TGG AGC CAT CCA GGT AGA CGA TCG TTC TCC CGA GTT CCT GGG-3′, AS2: 5′-GTC CCT CCA AAG GGC AGC ACA GCA GCC AGG TTC TCC CAG GTG ATG-3′, AS3: 5′-AGA AGC AGC CAC TTG GGT ACC TTA GCG GCC GCG GGA CGC ATG GCT-3′; NTPDase1 AS1: 5′-GGC TTG TTG ACA CTG CAG CCA GGA CCT CGT CTG CCG ATT GTT CGC-3′, AS2: 5′- GCC TGC TGT GGC TCC CAG GTA GAC AGG AGT CTG GTG ATG CTT GGA-3′, AS3: 5′- GCT GGT CTA GAC TGT CCC GGG TAA GCA GGA AAG GGT GAC TCC GGC-3′; NTPDase2 AS1: 5′- CCA ACT TTC CAG CCA TGG GAG CAC GCG GGA GAA CAC AGC AGG GAC-3′, AS2: 5′- TCT GTG TCA CTG CCT CCA GCA CCT TGG CTG TGG CCT CTG GGC TGG-3′, AS3: 5′- GAC AGA GGG CAG CGT TGC TGG TCC CTG ACA GGC TGA CAG TGG CAC-3′; NTPDase3 AS1: 5′-CAC AGA AGC CAC CTT CTC CCT GCA CAG TCG TGG GTC ACC AGT TCC-3′, AS2: 5′-GCT GAG CAT GTA GCC AAG GGA CCA GGC GAT GCT GCT GTT CCC CAC-3′, AS3: 5′-AGT CCC CAC TGG GTG GAT GAG GGA CTC CCA GGA GGT GCT CTG GCA-3′. Each probe was labeled with [33P] dATP (PerkinElmer, Waltham, MA, USA) using terminal deoxynucleotidyl transferase (Invitrogen, Karlsruhe, Germany) according to the manufacturer’s instructions. The sections were fixed on the slides with 4 % PFA/PBS for 10 min and washed with PBS for 5 min and with 50 mM glycine/PBS for 20 min. Blocking of non-specific binding was achieved by incubation with 0.1 M triethanolamine and 0.25 % acetic anhydride for 10 min. Slides were then washed with 2× sodium chloride sodium citrate (SSC) solution containing 0.1 % sarkosyl for 10 min, followed by a 5-min wash with 0.1× SSC/0.1 % sarkosyl and subsequently immersed in 70 and 100 % ethanol for 5 min each. For hybridization, 65 μl hybridization buffer [50 % deionized formamide, 0.03 M Tris HCL (pH 8), 0.2 mg/ml yeast tRNA, 1× Denhardt’s solution, 0.6 M NaCl, 0.25 % sodium dodecyl sulfate, 0.25 M ethylenediaminetetraacetic acid (pH 8) and 1× dextran sulfate] containing the designated pooled probes (concentration: 6 × 105 cpm per 100 μl) were applied to each slide. The slides were incubated in a humid chamber at 42 °C overnight. On the following day, high-stringency washes were performed including 30 min in 2× SSC/0.1 % sarkosyl followed by 2 × 40 min with 0.1× SSC/0.1 % sarkosyl at 55 °C. The sections were rinsed in 70 and 100 % ethanol, air dried, and exposed to a Kodak BioMax MR X-ray film (Sigma, Eching, Germany) for 40 days.
Gene expression was measured as the relative optical density (OD) of the autoradiographic signal using Adobe Photoshop software (Adobe Systems, San Jose, CA, USA). Three to four sections containing the region of interest were analyzed and averaged to yield a single value for each mouse. For measurements of the signal from whole brain, all sections were analyzed and averaged. Since no area in the brain was completely unlabeled and the expression of the enzymes varied between time points in all the brain areas, the measured autoradiographic signals were corrected for non-specific background signal, calculated from the background of the entire film. Size, shape and location of the different brain regions were confirmed using sections counterstained with 0.1 % hematoxylin (Applichem, Darmstadt, Germany) and 0.5 % eosin G (Roth, Karlsruhe, Germany). The signal from the striatum was measured in the most caudal region. The signal from the dentate gyrus (DG) was measured in its most rostral region. The signal from the medial habenula (MHb) was measured in all the sections containing this nucleus. The mouse brain atlas of Franklin and Paxinos (2008) served as reference. The data are expressed as means ± SD from at least three animals per time point and genotype. Sense probes of NTPDase1, -2 and -3 generated no specific signal. The sense probes of eN generated a signal in the hippocampus which, however, was much weaker than the signal generated by the antisense probes.
RT-PCR
For reverse transcriptase-PCR (RT-PCR), C3H mice were killed at CT18. The brains were dissected and cut into 1-mm-thick slices placed on an ice-cold plate. Specific brain regions (see “Results”) were punched out and immediately frozen in dry ice. In addition, a tissue sample was obtained from the liver. Total RNA was then isolated using the RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Frozen tissue samples were homogenized in RLT lysate buffer containing beta-mercaptoethanol (Sigma, Eching, Germany) with a rotor stator and then centrifuged at full speed. Supernatants were treated with 70 % ethanol to facilitate binding of total RNA to the RNeasy elution column. Contaminants were washed out using the RW1 and RPE buffers provided. RNA was eluted in RNase-free water and measured with the NanoDrop spectrophotometer (PEQLAB, Erlangen, Germany). Reverse transcription of RNA was carried out using the iScript cDNA synthesis kit (Bio-Rad, Munich, Germany) according to the manufacturer’s instructions. The cDNA quality and concentration were controlled with the NanoDrop spectrophotometer. PCR primers specific for NTPDase3 were designed using the Primer-BLAST tool from NCBI and tested for specificity with additional BLAST searches. The following primer pair was used in a PCR reaction for the generation of a 453-bp product; forward: 5′-GAG ACA GCA GCT CGT GAA GT-3′, reverse: 5′-CAA GGT GAA GGC GGT ACT GT-3′. The PCR conditions used were: 95 °C (30 s), 60.2 °C (60 s) and 72 °C (90 s) × 35 cycles. Products were analyzed on 2 % agarose gels. Absence of contamination was confirmed by running a negative control containing water instead of cDNA.
Data analysis of locomotor activity rhythms
All animals experienced a 12 h light (L):12 h dark (D) cycle for at least 1 week before experiments. Spontaneous locomotor activity was continuously recorded with an infrared recording system (Mouse-E-Motion, Hamburg, Germany) in 10-min intervals for at least 10 consecutive days under LD conditions (Pfeffer et al. 2012). Actograms and activity profiles with acrophases were generated using Clocklab software (Actimetrics, Wilmette, IL, USA) as previously described (Pfeffer et al. 2012).
Statistical analysis
All data were statistically analyzed using the BIAS program package (Ackermann 1991). Time-dependent variances were compared using the Kruskal–Wallis test followed by Conover–Iman (Bonferroni–Holm correction) multiple comparison test to assess differences between the time points examined. For analysis of locomotor activity rhythms, an unpaired t test was employed after confirming that variances were not significantly different. Differences were considered significant at p < 0.05.
Results
Localization of ectonucleotidase mRNA expression in the prosencephalon
By means of radioactive in situ hybridization, the mRNA expression of ecto-5′-nucleotidase and NTPDase1, -2 and -3 was allocated to different regions in the C3H and C57Bl mouse prosencephalon (Figs. 1, 2, 3; Table 1).
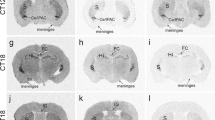
Distribution of the mRNA of the enzymes eN, NTPDase1 and NTPDase3 in coronal sections of the C3H mouse prosencephalon as revealed by radioactive in situ hybridization a, b, d, e, g, h Representative autoradiographs showing signals obtained with antisense probes for eN (a, b), NTPDase1 (d, e) and NTPDase3 (g, h). c, f, i Control for specificity with sense probes for eN (c), NTPDase1 (f) and NTPDase3 (i). a An intense signal for eN mRNA was apparent in striatum (S), hippocampus (HI) including the fasciola cinerea (FC) and meninges. b The lateral and capsular areas of the central amygdala and the interstitial nucleus of the posterior limb of the anterior commissure (Ce/IPAC) were more intensely labeled than the striatum, hippocampus and meninges. The indusium griseum (IG) was also strongly labeled. d, e Clear NTPDase1 mRNA expression specific to blood vessels (►), striatum (S) and the hypophysial pars tuberalis (PT). The ependymal cell layer of the third ventricle displayed a weaker signal (»). g Strong NTPDase3 mRNA expression in striatum (S), hippocampus (HI), medial habenula (^), cingulate cortex (Cg), piriform cortex (Pir) and zona incerta (*). A weaker signal was obtained in the lateral habenula (ɨ). h In addition to the strong expression in striatum, hippocampus, medial habenula, cingulate and piriform cortex, a strong signal was located in the ventromedial hypothalamus (●), arcuate nucleus (Arc) and medial posterior amygdala (◊). The sense probes (c, f, i) generated no signal with the exception of a faint labeling of eN mRNA in the hippocampus (c). Scale bar 1 mm
Distribution of the mRNA of the enzymes eN, NTPDase1 and NTPDase3 in coronal sections of the C57Bl mouse prosencephalon as revealed by radioactive in situ hybridization a, b, d, e, g, h Representative autoradiographs showing signals obtained with antisense probes for eN (a, b), NTPDase1 (d, e) and NTPDase3 (g, h). c, f, i Control for specificity with sense probes for eN (c), NTPDase1 (f) and NTPDase3 (i). a Strong eN mRNA expression was located in the striatum (S), choroid plexus (CP) and meninges while an even more intense signal was observed in the lateral and capsular areas of the central amygdala and the interstitial nucleus of the posterior limb of the anterior commissure (Ce/IPAC). b Intense signal in striatum, hippocampus and meninges. d, e Strong NTPDase1 mRNA expression in blood vessels (►), striatum (S) and the hypophysial pars tuberalis (PT). g NTPDase3 mRNA was strongly labeled in striatum (S), hippocampus (HI), medial habenula (^), cingulate cortex (Cg), piriform cortex (Pir) zona incerta (*) and in the premammillary nucleus (PM). A weaker signal was generated in the lateral habenula (ɨ). h Strong signal in striatum (S), hippocampus (HI), medial habenula (^), cingulate (Cg) and piriform cortex (Pir), ventromedial hypothalamus (●), arcuate nucleus (Arc) and medial posterior amygdala (◊). No labeling was observed in the sense probes (c, f, i) with the exception of a faint signal for eN mRNA in the hippocampus (c). Scale bar 1 mm
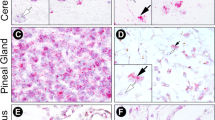
Distribution of NTPDase2 mRNA in coronal sections of C3H and C57Bl mouse prosencephalon as revealed by radioactive in situ hybridization a, b, d, e, g, h Representative autoradiographs showing signals obtained with antisense probes for NTPDase2 in the C3H mouse (a, b), C57Bl mouse (d, e), NTPDase2 knockout (KO) mouse (h), and in the corresponding wild-type (WT) (g). c, f, i Control for specificity with sense probes for NTPDase2 in the C3H (c), C57Bl (f) and KO mouse (i). a, b, d, e, g Specific hybridization signals were located in the dentate gyrus (DG), ependymal layer of the third ventricle (») and the subventricular zone of the lateral ventricles (*). The subfornical organ (SFO) displayed a particularly strong signal. In the medial habenula (^) the signal was weak and diffuse. h The signal was completely absent in the knockout mouse. c, f, i No signal was obtained with the sense probes. Scale bar 1 mm
Ecto-5′-nucleotidase, the dominant AMPase in the mouse brain, was highly expressed in the hippocampus (Figs. 1a, b, 2b). Strong expression was observed in the C3H mice in the hippocampal fasciola cinerea (Fig. 1a) and in the adjacent indusium griseum (Fig. 1b). In C57Bl mice, the fasciola cinerea was morphologically thinner and consequently the signal was much weaker. Notably, the indusium griseum was not stained at all in the C57Bl mice (Fig. 2b). Also, the striatum and meninges revealed strong signals (Figs. 1a, b, 2a, b). The mRNA expression was even stronger in the interstitial nucleus of the posterior limb of the anterior commissure (IPAC) and in the lateral and capsular areas of the central amygdala (CeL and CeC), but weaker in the medial central amygdala (CeM) (Figs. 1b, 2a). A weak, but distinct eN mRNA expression was observed in the medial posterior amygdala (MeP) and in the rostral part of the ventromedial hypothalamus (VMH). Interestingly, ecto-5′-nucleotidase (eN) expression was present in the choroid plexus (CP) of C57Bl mice, but was absent from the CP of C3H mice (Figs. 2a, 1a). Conversely, eN was expressed in the subfornical organ (SFO) of C3H mice but was lacking in the SFO of the C57Bl mice.
NTPDase1 mRNA was highly expressed in striatum, blood vessels (including the posterior and anterior cerebral arteries), hypophysial pars tuberalis (PT), the ependymal cell layer of the third ventricle (3V) (Figs. 1d, e, 2d, e), and the anteroventral thalamic nuclei (AV). A weaker expression of NTPDase1 was apparent in the SFO, paraventricular nucleus (PVN) and in the supraoptic nucleus (SON). Similar to the eN expression, moderate NTPDase1 expression was detected in the CP of C57Bl mice, but not in the CP of C3H mice (Figs. 2d, 1e). In contrast, NTPDase1 mRNA was expressed in the ventral posterior parvicellular nucleus of the thalamus (VPPC) in C3H mice but not in C57Bl mice. No clear NTPDase1 signal was obtained in the hippocampus (Figs. 1d, e, 2d).
NTPDase2 mRNA was detected in the triangular septal nuclei (TS), SFO, hippocampal dentate gyrus (DG), subventricular zone (SVZ) of the lateral ventricles, and ependymal cell layer of the 3V (Fig. 3a, b, d, e, g). A less intense signal was observed in the MHb (Fig. 3a), MeP, globus pallidus (GP), and in the cingulate cortex (Cg). A diffuse signal was obtained in the medial thalamus (both ventro- and dorsomedial thalamic nucleus). Interestingly, the signal from the SFO was markedly stronger in C57Bl mice than in the SFO of C3H mice (Fig. 3e, b). In the NTPDase2 knockout mouse, all mRNA signals were abrogated (Fig. 3h).
Notably, NTPDase3 was strongly and widely expressed in the mouse prosencephalon (Figs. 1g, h, 2g, h). Strong expression was observed in striatum, VMH, arcuate nucleus (Arc), anterodorsal thalamic nucleus (AD), MHb, MeP, piriform cortex (Pir), Cg, anterior paraventricular thalamic nucleus (PVA), zona incerta (ZI), subthalamic nucleus (STh), substantia nigra (SN), premammillary nucleus (PMD + PMV), and in the nucleus of Darkschewitsch (Dk). The mRNA signal was also strong in the hippocampus, including the FC. Interestingly, the staining in the hippocampal CA1 area was weaker than in CA2 and CA3; however, these differences were not evident in the most rostral region (Figs. 1g, h, 2g, h). A less intense signal was detected in the dorsomedial hypothalamus (DM), PVN, VPPC, ventral posterior thalamic nucleus (VP) and posterior thalamic nuclear group (Po), lateral habenula (LHb) (Figs. 1g, 2g), reticular thalamus (Rt) and in the SFO. As for eN and NTPDase1, NTPDase3 mRNA was strongly expressed in the CP of C57Bl mice, but not in the CP of C3H mice. Overall, the signal for NTPDase3 appeared to be stronger than that for the other enzymes.
RT-PCR
Since the ISH revealed an unexpectedly wide NTPDase3 distribution in the mouse brain, RT-PCR was performed to corroborate the ISH data. Also this method revealed widespread expression of NTPDase3 in the brain. NTPDase3 mRNA was absent from the hypohysial pars nervosa (PN) and the liver. The water control probe produced no signal (Fig. 4). Taken together, these data show that individual ectonucleotidases reveal differential and region-specific expression patterns in mouse brain and that the expression can vary between melatonin-proficient C3H and melatonin-deficient C57Bl mice.
Expression of NTPDase3 mRNA in various regions of the mouse brain as shown by RT-PCR. PD pars distalis, MHb medial habenula, Cor cortex, Thal thalamus, S striatum, IP interpeduncular nucleus, HI hippocampus, SCN suprachiasmatic nucleus, Med medulla oblongata. No signal was obtained in pars nervosa (PN), liver and water control (WC). Products were analyzed on two separate 2 % agarose gels
Time-dependent changes in ectonucleotidase mRNA expression in the prosencephalon
To analyze time-dependent changes in the expression of ectonucleotidase mRNA, ISH data obtained from mice sacrificed at four different time points of the day (CT00, CT06, CT12 and CT18) were analyzed (Figs. 5, 6). To avoid masking artifacts, animals were transferred to constant darkness 1 day before tissue sampling and circadian time (CT) 00 was defined as “lights on” in the previous light–dark schedule. In the mouse strain C3H, clear time-dependent differences in the expression (relative OD measurements) of all four enzymes were observed (Figs. 5a, b, 6a, b). The mRNA expression was low at the beginning of the subjective day (CT00), rose during the day and reached a peak in the middle of the subjective night (CT18). This expression pattern was equally obtained for whole brains (entire section surface) and individual brain regions with elevated expression levels. Interestingly, levels of NTPDase3 mRNA increased drastically from CT12 to CT18 (Fig. 6b). This elevation was more pronounced than the increases between the earlier time points. For eN and NTPDase2, the increase in mRNA expression was more prominent in whole brain than in individual regions depicting high mRNA expression (Figs. 5a, 6a). These data demonstrate that all ectonucleotidases reveal a prominent circadian pattern of expression.
Time- and strain-dependent differences in the expression patterns of eN and NTPDase1 mRNA in the mouse prosencephalon a eN expression in C3H mice, b NTPDase1 expression in C3H mice, c eN expression in C57Bl mice, d NTPDase1 expression in C57Bl mice. a, b In the C3H mouse strain, both enzymes revealed a clear time-dependent increase in expression starting at the beginning of the subjective day (CT00) and peaking in the middle of the subjective night (CT18) in all brain regions tested, including whole brain. c, d The time-dependent expression patterns of eN and NTPDase1 differed between the two mouse strains. In opposition to the gradual rise in expression throughout the day in the C3H mice, a gradual decrease during daytime occurred in both eN and NTPDase1 in C57Bl mice; however, eN expression peaked at the beginning of the day while NTPDase1 reached the maximum at CT18. Gene expression was measured as relative OD of the autoradiographic signal obtained from mice sacrificed at four different time points throughout the day (CT00, CT06, CT12 and CT18). Values are expressed as means ± SD (n ≥ 3 per time point and strain). *P < 0.05; **P < 0.01; ***P < 0.001; Kruskal–Wallis test
Time- and strain-dependent differences in the expression patterns of NTPDase2 and NTPDase3 mRNA in the mouse prosencephalon a NTPDase2 expression in C3H mice, b NTPDase3 expression in C3H mice, c NTPDase2 expression in C57Bl mice, d NTPDase3 expression in C57Bl mice. a, b In the C3H mouse strain, both enzymes exhibited in all brain regions tested a clear time-dependent expression pattern with an increase during the day to reach a peak in the middle of the subjective night (CT18). c In C57Bl mice, a similar pattern was observed for NTPDase2 mRNA levels; however, the time-dependent changes were not significantly different. d In C57Bl mice, time-dependent patterns of NTPDase3 mRNA expression were similar to those in C3H in all brain regions tested. Gene expression was measured as relative OD of the autoradiographic signal obtained from mice sacrificed at four different time points throughout the day (CT00, CT06, CT12 and CT18). Values are expressed as means ± SD (n ≥ 3 per time point and strain). *P < 0.05; **P < 0.01; ***P < 0.001; Kruskal–Wallis test
Strain-dependent temporal patterns of ectonucleotidase mRNA expression in the prosencephalon
In order to examine the possible influence of melatonin on the circadian pattern of ectonucleotidase mRNA expression, melatonin-proficient C3H mice were compared with melatonin-deficient C57Bl mice. The pattern of mRNA expression of eN and NTPDase1 clearly differed between the two mouse strains (Fig. 5a–d). The temporal expression pattern of eN in C57Bl mice was exactly opposite to that in C3H mice: eN expression was highest at CT00 (beginning of the subjective day) and declined throughout the day to a trough at CT18 (mid subjective night) (Fig. 5a, c). The NTPDase1 mRNA expression in the C57Bl mice gradually decreased during day, reached a trough at CT12 (beginning of the subjective night) and showed a sharp increase at CT18 (Fig. 5d). The temporal dynamics in NTPDase2 and NTPDase3 expression were similar in both the C3H and C57Bl mice; however, in C57Bl, the NTPDase2 expression did not significantly differ between the time points (Fig. 6a–d).
Strain-dependent differences in diurnal mouse locomotor activity
We further compared the locomotor activity rhythms of the two mouse strains in a standard photoperiod. The two mouse strains differed with respect to their daily activity profiles. The melatonin-deficient C57Bl mice were more active during day time (the inactive phase) than the melatonin-proficient C3H mice. This was evident both from the inspection of the actograms (Fig. 7a, b) and from the plots of the averages of their day time activity (Fig. 7c).
Comparison of the locomotor activity rhythms of C3H and C57Bl mice in a standard photoperiod a Double-plotted actogram of the spontaneous locomotor activity in LD of a representative C3H mouse. b Double plotted actogram of the spontaneous locomotor activity in LD of a representative C57Bl mouse. c The relative activity during the light phase of C3H and C57Bl mice. Values are expressed as means ± SEM (n = 6 per strain). ***P < 0.001; unpaired t test
Discussion
In this study, we have investigated the expression of ectonucleotidases, major regulators of extracellular purine levels in the brain, in melatonin-proficient C3H and melatonin-deficient C57Bl mice by means of radioactive in situ hybridization (ISH). Our results provide novel data on the spatial distribution and time-dependent changes in ectonucleotidase expression in the prosencephalon. Notably, NTPDase3 appeared to be more widely expressed than previously appreciated. In C3H mice, mRNA levels of all ectonucleotidases investigated were high during the active phase. Furthermore, striking differences in eN and NTPDase1 expression were observed between melatonin-proficient and melatonin-deficient mice, implying a role for melatonin in the control of the expression of these ectonucleotidases and consequently in the control of adenosine production in the brain.
Ectonucleotidase localization in the mouse prosencephalon
Consistent with previous enzyme histochemical and immunological data (Zimmermann et al. 1993; Cunha et al. 2000; Bjelobaba et al. 2007; Langer et al. 2008), our ISH studies revealed a strong ecto-5′-nucleotidase (eN) mRNA expression in the hippocampus including the fasciola cinerea (FC). Notably, the FC was much thicker in C3H than in C57Bl mice and thus the FC signal was much stronger in C3H. Furthermore, we detected strong expression in the indusium griseum (IG) of C3H but not of C57Bl. eN mRNA was also strongly expressed in the striatum. Since immunostaining and catalytic activity of eN was demonstrated in the globus pallidus (GP) (Langer et al. 2008; Augusto et al. 2013; Ena et al. 2013; Kulesskaya et al. 2013), it is reasonable to assume that the eN-expressing neurons in the striatum project to the GP. A remarkably strong expression (stronger than in the striatum or hippocampus) was observed in the IPAC as well as in the CeL and CeC, suggesting a role for adenosine as a neuromodulator in these areas. The clear signal we detected in the meninges was consistent with a previous report (Kulesskaya et al. 2013). In the choroid plexus (CP) previously described as eN-positive (Mills et al. 2008; Petrovic-Djergovic et al. 2012; Kulesskaya et al. 2013), we obtained an ISH signal only from C57Bl but not from C3H mice. Contrarily, eN mRNA was present in the subfornical organ (SFO) of C3H but not of C57Bl mice. Thus, eN expression reveals strain-related differences in specific brain regions which could be attributed to the presence or absence of melatonin, implying an involvement of melatonin in the control of ectonucleotidase expression and thereby in the purinergic signaling pathway. This hypothesis needs to be corroborated in mice with targeted deletion of the melatonin receptors.
In murine brain, NTPDase1 is considered as the major ectonucleotidase associated with microglia and the endothelial and smooth muscle cells of the vasculature (Braun et al. 2000; Langer et al. 2008). Accordingly, a strong ISH signal was observed in the blood vessels. This strong NTPDase1 expression further supports its thromboregulatory role via the regulation of ADP levels, a major platelet aggregation and thrombosis-inducing agent (Deaglio and Robson 2011). A clear mRNA signal was detected in the striatum consistent with the demonstration of NTPDase1 catalytic activity (Langer et al. 2008). New evidence for NTPDase1 expression was found in the thalamic AV, the ependymal cell layer of the 3V, the SFO, PVN, and SON and in the VPPC region. Interestingly, the latter was only labeled in C3H but not in C57Bl mice. In contrast, NTPDase1 was only expressed in the CP of C57Bl but not of C3H mice.
NTPDase2, a marker for progenitor cells in the adult mouse brain (Gampe et al. 2014), was observed in the subventricular zone of the lateral ventricles, the dentate gyrus and the medial habenula (Braun et al. 2003; Shukla et al. 2005; Gampe et al. 2012). Our ISH data were consistent with these results. However, we also demonstrated weak NTPDase2 expression in TS, MeP, GP, and Cg, while an extremely strong signal was observed from the SFO, a sensory circumventricular organ involved in the cardiovascular and metabolic regulation (Mimee et al. 2013). Interestingly, this signal was stronger in the C57Bl mice than in the C3H mice. Importantly, the NTPDase2 signal was completely abrogated in the knockout mouse. These data provide further evidence for the specificity of our ISH data already demonstrated by means of the sense probes.
To date, information on NTPDase3 expression in the brain is limited (Vorhoff et al. 2005; Belcher et al. 2006; Vongtau et al. 2011). NTPDase3-immunoreactive nerve fibers were observed throughout the brain, but specifically immunostained perikarya were scattered in two brain regions only, adjacent to the lateral hypothalamic area and in the perifornical nucleus (Belcher et al. 2006). In contrast, our ISH studies revealed a wide distribution of NTPDase3 mRNA in the mouse prosencephalon with strong signals in multiple brain regions, such as striatum, hippocampus, VMH, Arc, thalamic AD, MeP, Pir, Cg, PVA, ZI, STh, PMD and PMV, SN and in the Dk. Moderate NTPDase3 expression was observed in the PVN, hypothalamic DM, VPPC, VP and Po, Rt and SFO. Finally, as shown for eN and NTPDase1, NTPDase3 mRNA was expressed only in the CP of C57Bl mice, but not in the CP of C3H mice. To control for the unexpected ISH data, we additionally performed RT-PCR. This method revealed an equally widespread expression of NTPDase3 in all those regions of the prosencephalon which displayed positive ISH signals. The strong NTPDase3 expression in so many brain regions conforms to the wide distribution of NTPDase3-immunoreactive nerve fibers throughout the brain (Belcher et al. 2006) and emphasizes its significance in nucleotide signaling in the brain.
Time-dependent changes and strain-dependent differences in ectonucleotidase expression patterns
The expression of all four ectonucleotidases examined gradually increased during the subjective day and peaked during the dark (active) phase in C3H mice. These results are consistent with previous data describing elevated ectonucleotidase activity in rat blood serum (Detanico et al. 2010) and increased eN activity in rat cerebral cortex (Mackiewicz et al. 2003) during the dark phase. High ectonucleotidase expression at night may be essential for purine regulation during activity and in particular for the production and accumulation of adenosine. However, this hypothesis needs to be corroborated by analyses of protein levels and enzyme activity (see below).
The comparison between melatonin-proficient C3H and melatonin-deficient C57Bl mice revealed striking differences in the expression of eN. While in C3H mice, mRNA levels increased gradually during the day and peaked at night, in C57Bl mice, eN mRNA levels peaked in the morning and gradually decreased thereafter. Via the hydrolysis of AMP the enzyme eN plays a central role in the production of the neuromodulator adenosine, which supposedly has a key role in the regulation of sleep homeostasis. Extracellular adenosine levels increase throughout the brain during wakefulness and thereby increase sleepiness. During sleep, however, adenosine levels decrease (Virus et al. 1983; Radulovacki et al. 1984; Radulovacki 1985; Ticho and Radulovacki 1991; Porkka-Heiskanen et al. 2000). It has already been shown that the enzyme eN is very responsive to different treatments, such as thyroid hormones (Wink et al. 2003), docosahexaenoic acid (Thom et al. 2013), TGFβ (Chatterjee et al. 2014) and the monastrol-derived compound LaSOM 63 (Figueiró et al. 2014). The differences observed here between the melatonin-proficient and the melatonin-deficient mice may suggest that melatonin also influences eN expression and thereby adenosine production. Along with the strain-dependent differences in eN expression, we demonstrated differences in locomotor activity during daytime (i.e., the inactive phase): melatonin-deficient C57Bl mice showed significantly increased activity during the inactive phase as compared to melatonin-proficient C3H mice. It is tempting to speculate that, due to the temporal shift in eN mRNA expression and consequently in adenosine production, the sleep homeostasis of C57Bl mice is somewhat disturbed.
Interestingly, a drastic decrease from peak (CT18) to trough (CT00) in the mRNA levels of all four ectonucleotidases occurred in C3H mice. This decrease might result from a negative feedback loop in the transcription mechanism of these enzymes. Furthermore, the sharp increase in NTPDase3 expression upon onset of darkness (activity) might point to a significant role of this enzyme in the control of the arousal and feeding behavior, as suggested by Belcher et al. (2006).
The ectonucleotidase expression solely in the CP of C57Bl but not of C3H mice implies a specific role for purinergic signaling pathways that had developed in the melatonin-deficient C57Bl mice in this brain region.
In summary, this study provides a detailed description of the spatial distribution and temporal dynamics of the expression of four major ectonucleotidases in the mouse prosencephalon. Notably, expression of NTPDase3 was found to be more widely expressed than previously assumed, underlining its significance in nucleotide signaling in the brain. The comparison of C3H and C57Bl mouse strains uncovered conspicuous strain-dependent differences in eN and NTPDase1 expression and suggested an interplay between melatonin and purinergic signaling. The present study provides an important basis for further examination of the complexity of the purinergic system in the brain. It now needs to be tested whether and how the changes described here in mRNA expression levels are translated into changes in protein levels and enzyme activity. Further analyses should focus on strain- and time-dependent variations in the expression of other elements of the purinergic system, such as P2 receptors, adenosine receptors, adenosine deaminase and adenosine kinase.
Abbreviations
- AD:
-
Anterodorsal thalamic nucleus
- Arc:
-
Arcuate nucleus
- AV:
-
Anteroventral thalamic nucleus
- CA:
-
Cornu ammonis
- CeC and CeL:
-
Capsular and lateral central amygdala
- CeM:
-
Medial central amygdala
- Cg:
-
Cingulate cortex
- CP:
-
Choroid plexus
- CT:
-
Circadian time
- DG:
-
Dentate gyrus
- Dk:
-
Nucleus of Darkschewitsch
- DM:
-
Dorsomedial hypothalamic nucleus
- eN:
-
Ecto-5′-nucleotidase
- FC:
-
Fasciola cinerea
- GP:
-
Globus pallidus
- IG:
-
Indusium griseum
- IPAC:
-
Interstitial nucleus of the posterior limb of the anterior commissure
- ISH:
-
In situ hybridization
- KO:
-
Knock out
- LHb:
-
Lateral habenula
- LV:
-
Lateral ventricle
- MeP:
-
Posterior medial amygdala
- MHb:
-
Medial habenula
- NTPDase:
-
Nucleoside triphosphate diphosphohydrolase
- Pir:
-
Piriform cortex
- PMD:
-
Premammillary nucleus dorsal
- PMV:
-
Premammillary nucleus ventral
- Po:
-
Posterior thalamic nuclear group
- PT:
-
Hypophysial pars tuberalis
- PVA:
-
Anterior paraventricular thalamic nucleus
- PVN:
-
Hypothalamic paraventricular nucleus
- Rt:
-
Reticular thalamic nucleus
- SCN:
-
Suprachiasmatic nucleus
- SFO:
-
Subfornical organ
- sm:
-
Stria medullaris
- SN:
-
Substantia nigra
- SON:
-
Supraoptic nucleus
- STh:
-
Subthalamic nucleus
- SVZ:
-
Subventricular zone
- TS:
-
Triangular septal nucleus
- 3V:
-
Third ventricle
- VMH:
-
Ventromedial hypothalamic nucleus
- VP:
-
Ventral posterior thalamic nucleus
- VPPC:
-
Ventral posterior parvicellular thalamic nucleus
- WT:
-
Wild-type
- ZI:
-
Zona incerta
References
Abbracchio MP, Burnstock G, Verkhratsky A, Zimmermann H (2009) Purinergic signalling in the nervous system: an overview. Trends Neurosci 32:19–29
Ackermann H (1991) BIAS. A program package for biometrical analysis of samples. Comput Stat Data Anal 11:223–224 [current program version: 10.02 epsilon]
Augusto E, Matos M, Sevigny J, El-Tayeb A, Bynoe MS, Muller CE, Cunha RA, Chen J (2013) Ecto-5′-nucleotidase (CD73)-mediated formation of adenosine is critical for the striatal adenosine A2A receptor functions. J Neurosci 33:11390–11399
Belcher S, Zsarnovszky A, Crawford P, Hemani H, Spurling L, Kirley T (2006) Immunolocalization of ecto-nucleoside triphosphate diphosphohydrolase 3 in rat brain: implications for modulation of multiple homeostatic systems including feeding and sleep–wake behaviors. Neuroscience 137:1331–1346
Bjelobaba I, Stojiljkovic M, Pekovic S, Dacic S, Lavrnja I, Stojkov D, Rakic L, Nedeljkovic N (2007) Immunohistological determination of ecto-nucleoside triphosphate diphosphohydrolase1 (NTPDase1) and 5′-nucleotidase in rat hippocampus reveals overlapping distribution. Cell Mol Neurobiol 27:731–743
Braun N, Sévigny J, Robson SC, Enjyoji K, Guckelberger O, Hammer K, Di Virgilio F, Zimmermann H (2000) Assignment of ecto-nucleoside triphosphate diphosphohydrolase-1/cd39 expression to microglia and vasculature of the brain. Eur J Neurosci 12:4357–4366
Braun N, Sévigny J, Mishra SK, Robson SC, Barth SW, Gerstberger R, Hammer K, Zimmermann H (2003) Expression of the ecto-ATPase NTPDase2 in the germinal zones of the developing and adult rat brain. Eur J Neurosci 17:1355–1364
Burnstock G (2006) Purinergic signalling. Br J Pharmacol 147(Suppl 1):172–181
Burnstock G (2009) Purinergic signalling: past, present and future. Braz J Med Biol Res 42:3–8
Burnstock G, Verkhratsky A (2012) Purinergic signalling and the nervous system. Springer, Heidelberg
Chatterjee S, Thyagarajan K, Kesarwani P, Song JH, Soloshchenko M, Fu J, Bailey SR, Vasu C, Kraft AS, Paulos CM, Yu XZ, Mehrotra S (2014) Reducing CD73 expression by IL1β-Programmed Th17 cells improves immunotherapeutic control of tumors. Cancer Res 74:6048–6059
Cunha RA, Brendel P, Zimmermann H, Ribeiro JA (2000) Immunologically distinct isoforms of ecto-5′-nucleotidase in nerve terminals of different areas of the rat hippocampus. J Neurochem 74:334–338
Deaglio S, Robson SC (2011) Ectonucleotidases as regulators of purinergic signaling in thrombosis, inflammation, and immunity. Adv Pharmacol 61:301–332
Detanico BC, de Souza A, Medeiros LF, Rozisky JR, Caumo W, Hidalgo MPL, Battastini AMO, Torres ILS (2010) 24-hour temporal pattern of NTPDase and 5′-nucleotidase enzymes in rat blood serum. Chronobiol Int 27:1751–1761
Dunwiddie TV, Masino SA (2001) The role and regulation of adenosine in the central nervous system. Annu Rev Neurosci 24:31–55
Ena SL, De Backer J, Schiffmann SN, de Kerchove d’Exaerde A (2013) FACS array profiling identifies ecto-5′ nucleotidase as a striatopallidal neuron-specific gene involved in striatal-dependent learning. J Neurosci 33:8794–8809
Figueiró F, Mendes FB, Corbelini PF, Janarelli F, Jandrey EH, Russowsky D, Eifler-Lima VL, Battastini AM (2014) A monastrol-derived compound, LaSOM 63, inhibits ecto-5′nucleotidase/CD73 activity and induces apoptotic cell death of glioma cell lines. Anticancer Res 34:1837–1842
Franklin KBJ, Paxinos G (2008) The mouse brain in stereotaxic coordinates. Academic, New York
Gampe K, Hammer K, Kittel Á, Zimmermann H (2012) The medial habenula contains a specific nonstellate subtype of astrocyte expressing the ectonucleotidase NTPDase2. Glia 60:1860–1870
Gampe K, Stefani J, Hammer K, Brendel P, Pötzsch A, Enikolopov G, Enjyoji K, Acker-Palmer A, Robson SC, Zimmermann H (2014) NTPDase2 and purinergic signaling control progenitor cell proliferation in neurogenic niches of the adult mouse brain. Stem Cells 33:253–264
Goto M, Matsuo H, Iigo M, Furuse M, Korf HW, Yasuo S (2013) Melatonin-induced changes in the expression of thyroid hormone-converting enzymes in hypothalamus depend on the timing of melatonin injections and genetic background in mice. Gen Comp Endocrinol 186:33–40
Haskó G, Cronstein BN (2004) Adenosine: an endogenous regulator of innate immunity. Trends Immunol 25:33–39
Korf HW, von Gall C (2006) Mice, melatonin and the circadian system. Mol Cell Endocrinol 252:57–68
Korf HW, von Gall C, Stehle J (2003) The circadian system and melatonin: lessons from rats and mice. Chronobiol Int 20:697–710
Kukulski F, Lévesque SA, Sévigny J (2011) Impact of ectoenzymes on p2 and p1 receptor signaling. Adv Pharmacol 61:263–299
Kulesskaya N, Võikar V, Peltola M, Yegutkin GG, Salmi M, Jalkanen S, Rauvala H (2013) CD73 is a major regulator of adenosinergic signalling in mouse brain. PLoS ONE 8:e66896
Langer D, Hammer K, Koszalka P, Schrader J, Robson SC, Zimmermann H (2008) Distribution of ectonucleotidases in the rodent brain revisited. Cell Tissue Res 334:199–217
Mackiewicz M, Nikonova EV, Zimmerman JE, Galante RJ, Zhang L, Cater JR, Geiger JD, Pack AI (2003) Enzymes of adenosine metabolism in the brain: diurnal rhythm and the effect of sleep deprivation. J Neurochem 85:348–357
Mills JH, Thompson LF, Mueller C, Waickman AT, Jalkanen S, Niemela J, Airas L, Bynoe MS (2008) CD73 is required for efficient entry of lymphocytes into the central nervous system during experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A 105:9325–9330
Mimee A, Smith PM, Ferguson AV (2013) Circumventricular organs: targets for integration of circulating fluid and energy balance signals? Physiol Behav 121:96–102
Mutafova-Yambolieva VN, Durnin L (2014) The purinergic neurotransmitter revisited: a single substance or multiple players? Pharmacol Ther 144:162–191
Petrovic-Djergovic D, Hyman MC, Ray JJ, Bouis D, Visovatti SH, Hayasaki T, Pinsky DJ (2012) Tissue-resident ecto-5′ nucleotidase (CD73) regulates leukocyte trafficking in the ischemic brain. J Immunol 188:2387–2398
Pfeffer M, Rauch A, Korf HW, von Gall C (2012) The Endogenous Melatonin (MT) signal facilitates reentrainment of the circadian system to light-induced phase advances by acting upon MT2 receptors. Chronobiol Int 29:415–429
Porkka-Heiskanen T, Strecker R, McCarley R (2000) Brain site-specificity of extracellular adenosine concentration changes during sleep deprivation and spontaneous sleep: an in vivo microdialysis study. Neuroscience 99:507–517
Radulovacki M (1985) Role of adenosine in sleep in rats. Rev Clin Basic Pharm 5:327–339
Radulovacki M, Virus RM, Djuricic-Nedelson M, Green RD (1984) Adenosine analogs and sleep in rats. J Pharmacol Exp Ther 228:268–274
Robson SC, Sévigny J, Zimmermann H (2006) The E-NTPDase family of ectonucleotidases: Structure function relationships and pathophysiological significance. Purinergic Signal 2:409–430
Shukla V, Zimmermann H, Wang L, Kettenmann H, Raab S, Hammer K, Sévigny J, Robson SC, Braun N (2005) Functional expression of the ecto-ATPase NTPDase2 and of nucleotide receptors by neuronal progenitor cells in the adult murine hippocampus. J Neurosci Res 80:600–610
Thom VT, Wendel M, Deussen A (2013) Regulation of ecto-5′-nucleotidase by docosahexaenoic acid in human endothelial cells. Cell Physiol Biochem 32:355–366
Ticho SR, Radulovacki M (1991) Role of adenosine in sleep and temperature regulation in the preoptic area of rats. Pharmacol Biochem Behav 40:33–40
Vandenbeuch A, Anderson CB, Parnes J, Enjyoji K, Robson SC, Finger TE, Kinnamon SC (2013) Role of the ectonucleotidase NTPDase2 in taste bud function. Proc Natl Acad Sci U S A 110:14789–14794
Virus RM, Djuricic-Nedelson M, Radulovacki M, Green RD (1983) The effects of adenosine and 2′-deoxycoformycin on sleep and wakefulness in rats. Neuropharmacology 22:1401–1404
von Gall C, Duffield GE, Hastings MH, Kopp MD, Dehghani F, Korf HW, Stehle JH (1998) CREB in the mouse SCN: a molecular interface coding the phase-adjusting stimuli light, glutamate, PACAP, and melatonin for clockwork access. J Neurosci 18:10389–10397
von Gall C, Lewy A, Schomerus C, Vivien-Roels B, Pevét P, Korf HW, Stehle JH (2000) Transcription factor dynamics and neuroendocrine signalling in the mouse pineal gland: a comparative analysis of melatonin-deficient C57BL mice and melatonin-proficient C3H mice. Eur J Neurosci 12:964–972
Vongtau HO, Lavoie EG, Sévigny J, Molliver DC (2011) Distribution of ecto-nucleotidases in mouse sensory circuits suggests roles for nucleoside triphosphate diphosphohydrolase-3 in nociception and mechanoreception. Neuroscience 193:387–398
Vorhoff T, Zimmermann H, Pelletier J, Sévigny J, Braun N (2005) Cloning and characterization of the ecto-nucleotidase NTPDase3 from rat brain: Predicted secondary structure and relation to other members of the E-NTPDase family and actin. Purinergic Signal 1:259–270
Wicht H, Korf HW, Ackermann H, Ekhart D, Fischer C, Pfeffer M (2014) Chronotypes and rhythm stability in mice. Chronobiol Int 31:27–36
Wink MR, Tamajusuku AS, Braganhol E, Casali EA, Barreto-Chaves ML, Sarkis JJ, Battastini AM (2003) Thyroid hormone upregulates ecto-5′- nucleotidase/CD73 in C6 rat glioma cells. Mol Cell Endocrinol 205:107–114
Yasuo S, Yoshimura T, Ebihara S, Korf HW (2009) Melatonin transmits photoperiodic signals through the MT1 melatonin receptor. J Neurosci 29:2885–2889
Zimmermann H (2000) Extracellular metabolism of ATP and other nucleotides. Naunyn Schmiedebergs Arch Pharmacol 362:299–309
Zimmermann H, Vogel M, Laube U (1993) Hippocampal localization of 5′-nucleotidase as revealed by immunocytochemistry. Neuroscience 55:105–112
Zimmermann H, Zebisch M, Sträter N (2012) Cellular function and molecular structure of ecto-nucleotidases. Purinergic Signal 8:437–502
Zylka MJ (2011) Pain-relieving prospects for adenosine receptors and ectonucleotidases. Trends Mol Med 17:188–196
Acknowledgments
This work has been supported by the Dr. Senckenbergische Stiftung, Frankfurt am Main. We would like to thank Dr. Hanns Ackermann (Institute for Biostatistics und Mathematic Modelling, Goethe University, Frankfurt am Main, Germany) for his kind help with the statistical analyses.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Homola, M., Pfeffer, M., Fischer, C. et al. Expression of ectonucleotidases in the prosencephalon of melatonin-proficient C3H and melatonin-deficient C57Bl mice: spatial distribution and time-dependent changes. Cell Tissue Res 362, 163–176 (2015). https://doi.org/10.1007/s00441-015-2179-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-015-2179-7