Abstract
Chromosomal insertions are thought to be rare structural rearrangements. The current understanding of the underlying mechanisms of their origin is still limited. In this study, we sequenced 16 cases with apparent simple insertions previously identified by karyotyping and/or chromosomal microarray analysis. Using mate-pair genome sequencing (GS), we identified all 16 insertions and revised previously designated karyotypes in 75.0% (12/16) of the cases. Additional cryptic rearrangements were identified in 68.8% of the cases (11/16). The incidence of additional cryptic rearrangements in chromosomal insertions was significantly higher compared to balanced translocations and inversions reported in other studies by GS. We characterized and classified the cryptic insertion rearrangements into four groups, which were not mutually exclusive: (1) insertion segments were fragmented and their subsegments rearranged and clustered at the insertion site (10/16, 62.5%); (2) one or more cryptic subsegments were not inserted into the insertion site (5/16, 31.3%); (3) segments of the acceptor chromosome were scattered and rejoined with the insertion segments (2/16, 12.5%); and (4) copy number gains were identified in the flanking regions of the insertion site (2/16, 12.5%). In addition to the observation of these chromothripsis- or chromoanasynthesis-like events, breakpoint sequence analysis revealed microhomology to be the predominant feature. However, no significant correlation was found between the number of cryptic rearrangements and the size of the insertion. Overall, our study provide molecular characterization of karyotypically apparent simple insertions, demonstrate previously underappreciated complexities, and evidence that chromosomal insertions are likely formed by nonhomologous end joining and/or microhomology-mediated replication-based DNA repair.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chromosomal insertions are structural rearrangements that involve a chromosome segment translocated interstitially into another chromosome (inter-chromosomal insertion) or to a different region of the same chromosome or the other homolog (intra-chromosomal insertion) (Kang et al. 2010). In simple chromosomal insertions, three breaks are required: two on the donor chromosome and one at the insertion site on the acceptor chromosome. The simple one-way inter-chromosomal insertion is the most common form of this rare rearrangement; the prevalence by conventional cytogenetics was reported to be 1 in 10,000 live births (Van Hemel and Eussen 2000). However, their incidence is thought to be significantly lower than the other structural rearrangements such as balanced translocations [1/500 (Mackie Ogilvie and Scriven 2002)].
Typically, chromosomal insertions occur as balanced events; however, de novo unbalanced insertions resulting in copy-number gain of the insertion segment have been also reported in some studies (Mendez-Rosado et al. 2017). Carriers of balanced insertions are generally asymptomatic; however, their fertility may be affected. Their offspring are at risk of inheriting derivative chromosomes involving copy-number gains or losses (or copy-number variants, CNVs) (Kehrer et al. 2015; Mendez-Rosado et al. 2017). During meiosis, sister chromosomes involved in the insertion may form quadrivalents depending on the size of the insertion segment; reported cases of potential recombination involved relatively large insertion segments (with haploid autosomal length > 1.5%) (Van Hemel and Eussen 2000). Chromosome segregation following the formation of a quadrivalent enables potential recombination within the insertion segments to generate complex rearrangements resulting in copy number gains/losses. Careful genetic workup of such insertions may be essential for proper management such as by preimplantation genetic testing (Melotte et al. 2004).
Like other structural rearrangements, formation of chromosomal insertions typically occurs during gametogenesis or meiosis (Pellestor et al. 2011). Simple insertions were thought to be predominantly formed by three double-strand breaks (DSBs) and were repaired by nonhomologous end joining (NHEJ) mechanism(s) (Bauters et al. 2008). Breakpoint junction sequence patterns of NHEJ include blunt ends, short-micro/small insertions and microhomologies. A subset of NHEJ is mediated by sequence microhomologies on both sides of the breakpoint, thus they are termed microhomology-mediated end joining (Ottaviani et al. 2014). In contrast, complex rearrangements involving duplications/triplications have been proposed to be formed by chromothripsis-like chromoanasynthesis mechanisms through microhomology-mediated replication such as fork stalling and template switching or microhomology-mediated break-induced replication (FoSTeS/ MMBIR) with iterative template switching (Liu et al. 2011; Stephens et al. 2011). A recent study investigating inter-chromosomal complex insertions also suggested that these replicative repair mechanisms may be involved (Gu et al. 2016).
By a combination of chromosomal microarray analysis (CMA) and fluorescence in situ hybridization (FISH), the incidence of chromosomal insertions was estimated to be significantly higher than previously reported by karyotyping, from 1 in 10,000 to 1 in 500 (Kang et al. 2010) or 1 in 563 (Neill et al. 2011). However, the combined approach cannot detect balanced rearrangements, limiting a comprehensive assessment of all rearrangements associated with the insertions. Genome sequencing (GS) on the other hand, enables the detection of both balanced and unbalanced rearrangements (Choy et al. 2019). A recent study utilizing GS in 14,891 individuals across diverse global populations demonstrates insertions to be abundant in the human genome; however, the majority of them are mobile element insertions (such as LINE, ~ 6 kb in size) (Collins et al. 2020). In a large study of 273 cases with known balanced translocations and inversions detected by conventional cytogenetics, additional cryptic rearrangements were detected by GS in 21% of the cases (Redin et al. 2017). In addition, these complexities may lead to de novo cryptic copy-number gains/losses related to the inherited rearrangements in their offspring (de Pagter et al. 2015), emphasizing the importance for comprehensive delineation of the rearrangements. We have previously applied mate-pair GS on 1090 couples with recurrent miscarriages which also demonstrated that over 15% of cases with balanced translocations harbored additional rearrangements cryptic to karyotyping (Dong et al. 2019a).
Limited studies have applied GS to study and delineate chromosomal insertions (Kato et al. 2017). Herein, we applied mate-pair GS for 16 patients with simple insertions identified by conventional cytogenetics to further investigate their structural rearrangements, breakpoint features and possible mechanism(s) of formation.
Materials and methods
Ethics, consent and permissions
The study was approved by the Institutional Review Board for Human Subject Research at Baylor College of Medicine (IRB# H-25466) and the Joint Chinese University of Hong Kong—New Territories East Cluster Clinical Research Ethics Committee (CREC Ref. Nos. 2016.713 and 2017.108). Overall, 16 patients with simple insertions previously identified by karyotyping and/or CMA and FISH were recruited for this study. Mate-pair GS data from Samples 01, 02, 03, and 05 were reported in our previous study (Dong et al. 2019b). The major indications for karyotyping were infertility or history of abnormal pregnancies (Table 1). DNA samples from the other 12 cases, including 1F, 3M, 4M, 5M, and 6M were previously reported by a CMA study (Kang et al. 2010). The remaining seven de-identified DNA samples were newly included in this study. Most of them presented with developmental delay, dysmorphic features, or congenital abnormalities (Table 1).
Chromosomal microarray analysis
CNV analysis was performed prior to this study. Two CMA platforms [8X60K Fetal DNA Chip v2.0 (Agilent Technologies) (Chau et al. 2019, 2020; Huang et al. 2014) and 4X180K array (Kang et al. 2010)] were used. Both were performed according to the manufacturers’ protocols and CNVs were analyzed via the CytoGenomics 5.0 software (Chau et al. 2019; Huang et al. 2014). As an example, detection of a copy number gain of 16.7 Mb in size from chromosome 1 was identified by CMA and FISH subsequently verified the insertion to chromosome 5 as depicted in Supplementary Figure S1.
DNA preparation and mate-pair genome sequencing
The genomic DNA was quantified with Qubit dsDNA HS Assay kit (Invitrogen, Carlsbad, CA). One microgram of DNA (OD260/OD280 > 1.8; OD260/OD230 > 2) from each case was sheared to fragment sizes ranging from 3 to 8 kb by a HydroShear device (Digilab, Inc., Hopkinton, MA) using the reported parameters (Dong et al. 2014).
The fragmented DNA was then subjected for mate-pair library construction (Dong et al. 2019b). In brief, the fragmented DNA (3–8 kb) was purified with Agencourt AmpureXP beads (Beckman Coulter, Brea, CA, USA), followed by end-repair, A-tailing and Ad1 adaptor ligation. 320 ng of adaptor-ligated DNA was amplified with Pfu Turbo Cx polymerase (Agilent Technologies, Santa Clara, CA). After purification, amplified products were pooled together from 12 cases. The amplified products were treated with Uracil Specific Excision Reagent (NEB, Ipswich, MA) and T4 DNA ligase (Enzymatics, Beverly, MA) to form double-stranded circularized (dsCir) DNA with a single nick on one of the strands. A nick translation polymerization step was performed with 1 pmol of dsCir DNA, Bst DNA Polymerase (Full Length, NEB), Klenow fragment (Enzymatics) and controlled dNTPs, followed by 3′branch ligation (Wang et al. 2019) (attached the 3′-end of Ad2 to the products). Subsequently, primer extension polymerization step was performed by incubation with the reaction mixture at 92 °C for 5 min, and cycled at 56 °C for 60 s, and 60 °C for 40 s. The products were purified and ligated to the 5′ end of Ad2 and amplified with Pfu Turbo Cx. DNA nanoballs were prepared from single-stranded circularized DNA and sequenced on an MGISEQ-2000 platform (MGI Tech Co., Ltd., Shenzhen, China) for a minimum of 70 million read pairs (PE 100 bp) per sample, equivalent to ~ 4.6 X sequencing read depth.
Detection of copy number variants and structural rearrangements
After data QC, the read pairs were aligned to the human reference genome (GRCh37/hg19) using the Burrows–Wheeler aligner (BWA) (Li and Durbin 2009). Uniquely aligned reads were used for further analysis.
CNV and structural rearrangement (or structural variant, SV) detection was performed according to previously reported methods (Dong et al. 2019a, b; Wang et al. 2020). For CNV detection, reads were classified into both adjustable sliding windows (50 kb with 5 kb increments) and non-overlapping windows (5 kb). Subsequently, the copy ratios of all windows were normalized by GC% and population-based data from our in-house dataset (Chau et al. 2020; Wang et al. 2020). CNVs were reported by the increment-rate-of-coverage module (Dong et al. 2016) at a resolution of 50 kb (homozygous/hemizygous deletion at 10 kb). For SV detection, chimeric read pairs (aligned to different chromosomes or to the same chromosome with a genomic distance > 10 kb) were selected for event clustering. Each potential event was then filtered against a dataset of systematic errors with optimized parameters. Candidate CNVs and SVs were filtered against our in-house dataset.
Variant verification
Rearrangement junction-specific PCR and Sanger sequencing were performed for the verification of structural rearrangements identified by mate-pair GS (Dong et al. 2014). Primers were designed using online software Primer3, Primer-Blast (NCBI) and in silico PCR (UCSC). PCR was performed in cases and controls, and the products were sequenced on an ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA). The sequences were aligned to the reference genome by BLAT (UCSC) for SV verification and breakpoint junction sequence feature analysis (Supplementary Figure S2).
Analysis of breakpoint junction features
The Sanger sequencing results of each rearrangement breakpoint junction was investigated for sequence microhomologies, small insertions, blunt end or homologies. Stretches of shared nucleotide sequence identity present at junctions of rearranged genomic segments are considered as microhomology (≤ 20 bp) or homology (> 20 bp) (Ankala et al. 2012). Meanwhile, repetitive elements flanking the breakpoint junctions were annotated using the RepeatMasker track on the UCSC genome browser webpage.
In addition, to investigate whether sequence similarity was a mediator of chromosomal insertions, sequence similarity of the flanking regions (300 bp upstream and 300 bp downstream) between the proximal and distal reference sequences was analyzed. The Needleman–Wunsch algorithm in the Biostrings package (Gu et al. 2015) was used to align the proximal and distal reference sequences and their similarity was calculated in 20 bp sliding windows as the percentage of aligned bases over the total count of non-gap sequences, in the genomic orientation of the breakpoint flanking sequences. A heatmap was generated for each breakpoint junction (Hijazi et al. 2020).
Annotation of non-B DNA motifs
75 bp of reference genomic sequences directly upstream and downstream of each rearrangement was investigated for the presence of non-B DNA motifs. Non-B DNA motifs (Bacolla et al. 2011; Cer et al. 2012) including (1) alpha-phased repeats; (2) direct repeats and slipped motifs; (3) g-quadruplex forming repeats; (4) inverted repeats and cruciform motifs; (5) mirror repeats and triplex motifs; (6) Z-DNA motifs and (7) short-tandem repeats were annotated for the sequences flanking each junction. In addition, we generated 500 pairs of random genome-wide breakpoint junctions for comparison of the frequency of non-B motifs with the average genome (Vissers et al. 2009). After excluding breakpoint junctions with flanking sequences (150 bp in total with method described above) located in unidentified (N) regions of the human reference genome (GRCh37/hg19), 394 rearrangements were obtained to represent the genome baseline for comparison.
Furthermore, the start sites of non-B DNA motifs in 200 kb flanks of the rearrangement breakpoint were counted. Heatmaps were generated by the counts of non-B DNA motifs in 10 kb windows.
Results
Mate-pair GS results were obtained for all cases. After alignment, a minimum of 60 million uniquely aligned read pairs were obtained which were comparable across each sample (with an insert size of ~ 5 kb). This resulted in approximately 100-fold physical coverage per case [read pairs (60 M) × insert size (5000 bp)/genome size (3G) = 100-fold]. In our previous study with low-coverage sequencing data from the 1000 Genomes Project, we demonstrated a minimum of physical coverage for detection of structural rearrangements to be 8.25 X (Dong et al. 2018). Therefore, the number of read pairs generated in this study was sufficient for our analysis. Mate-pair GS detected all 16 insertions identified previously by conventional G-banded chromosome analysis and/or CMA. The referral indications and the cytogenetic nomenclature of each case are shown in Table 1. Five cases have balanced chromosomal insertions and 11 cases have unbalanced chromosomal insertions (segmental duplication for the insertion locus). The insertion sizes detected by mate-pair GS ranged from 4.7 to 45.9 Mb.
Comparison of the insertion breakpoints by mate-pair GS vs karyotyping
Chromosome 2 had significantly higher incidences (Z-score ≥ 2, P ≤ 0.02275) of both overall involvement in the insertions and higher incidence of being the donor chromosome in our cohort (Supplementary Figure S3). There were three cases with insertion segments from the short arm of chromosome 2, overlapping the 2p22.21p21 region (Tables 1 and 2): one case with an unbalanced insertion (25M) and two cases with balanced insertions (Sample02 and 50783). In addition, four cases had chromosome 6 as the acceptor chromosome with the highest incidence (4/16).
Cryptic complexities in chromosomal insertions
Mate-pair GS identified additional cryptic complexities in 11/16 (68.8%) cases (Tables 1 and 2), which were submicroscopic rearrangements of the insertions previously not identified by karyotyping. In the 11 cases with cryptic complexities, the insertion segment was fragmented into two or more subsegments and were rearranged at the insertion site or involved copy number variants of subsegments in the donor/acceptor chromosome. This indicated that most insertions are more complex. Among the 16 cytogenetically characterized simple insertions, 4 classes of cryptic rearrangements were observed:
-
1)
The insertion segment from the donor chromosome was fragmented into two or more subsegments and were rearranged at the insertion sites in 10 of the 16 cases (62.5%, Table 1 and Figs. 1, 2, 3 and 4 and S4). The incidence of this finding was higher than in a previously published cohort of complex insertions (Gu et al. 2016) (44.4%, 4/9, Chi-square test: P = 0.382677).
-
2)
One or more cryptic subsegments of the insertion were not inserted into the acceptor chromosome. Among the 11 cases with unbalanced insertions, there were three [3/11, 27.3%; 3M (Fig. 2a), 10F (Fig. 2b) and 31M (Fig. 1)] with such observations. This was also observed in 2/5 cases with balanced insertions (Sample05 and Sample03; Fig. 3a, b). For example, in Sample05 with a balanced insertion (Fig. 2a), a subsegment (seq[GRCh37] del(3)(q24) chr3:g.146055006_148300124del) from the donor chromosome was deleted, while another subsegment (seq[GRCh37] 3q21.1 chr3:g.122008143_122161592) remained on the derivative chromosome 3, and both of them were not inserted into chromosome 6.
-
3)
Segments of the acceptor chromosome were scattered and rejoined with the insertion segments in two cases (2/16, 12.5%; Sample02 and 5M, Fig. 4a, b). In Sample02, the region surrounding the insertion site on acceptor chromosome (chromosome 6) was broken into six subsegments and rearranged with the insertion subsegments. In 5M, the acceptor chromosome (chromosome 6) was also fragmented, resulting in a 14.6 kb segment (seq[GRCh37] 6q16.1 chr6:g.98433429_98447982) located between the insertion subsegments.
-
4)
Copy number gains were identified in the flanking regions of the insertion site. There were two cases with copy number gains identified in the flanking regions of the breakpoint junctions on the acceptor chromosome (2/16, 12.5%; samples 10F, Fig. 2b, and 25M, Supplementary Figure S5). For instance, an 11.7-kb segment originating from the insertion site was duplicated and flanked the entire insertion from 3p (indicated as a segment in purple shown in Fig. 2b).
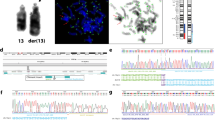
Composition of the derivative chromosome identified by mate-pair GS in case 31M. Chromosomal insertion schematic diagram illustrates the rearrangement identified by mate-pair GS. In the top panel, a snapshot from USCS Genome Browser shows start of the SHROOM4 gene and the two black arrows indicate the breakpoint junctions disrupting the gene. In the following panel, the color scheme depicts a normal short arm of the chromosome X in gray, while all subsegments are represented by purple, red, blue and yellow. The black vertical arrows show the genomic breakpoint junctions. In the middle panel, the diagram shows the fragmentation of the inserted segment and potential repairing process in a single event. In each subpanel, the upper chromosome is the derivative chromosome, while the lower chromosome is the normal chromosome indicating the location of the inserted segment(s). During the process, each potential step connecting different segments is shown by a dotted line. In the lowest panel, the resultant derivative chromosome Xq at 89.86 Mb location represented the rearranged insertion fragment. A black horizontal arrow within each subsegment indicated the sequence orientation. Note that a small segment seq[GRCh37] chrX:g.54853904_54900190 (gray bar shown between blue and yellow bars) was not included in the insertion
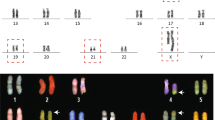
Cryptic segment from the insertion was not present in the insertion locus: 3M and 10F unbalanced insertions. ai G-banded chromosomes showing der(3)ins(3;1)(q23;p21.1p21.3) (left, as indicated by the blue arrow) and normal chromosome 3 (right). aii Fluorescent in situ hybridization (FISH) with probes RP11-465K10 (red) and control probe RP11-488L (green) showing an unbalanced insertion on chromosome 3 as indicated by the red arrow. aiii Part of the chromosome 1p depicted in red and part of the chromosome 3q in green. Different colors illustrate that only the subsegments in blue and yellow of the inserted fragment from chromosome 1p were rearranged and inserted to chromosome 3q at 143 Mb location, resulting in der(3) with 7.9 Mb and 2.8 Mb duplications. The two subsegments (in blue and yellow) were originally separated by a subsegment [seq[GRCh37]1p21.2(101733446_101983289)] in red. bi Case 10F with chromosome insertion from chromosome 3p25.3p21.31 to chromosome 3q27; the blue arrow shows the insertion segment. bii FISH with probe (RP11-1024P17) showed the inserted segment from chromosome 3p25.3 (red) to 3q27, while the green signal represents the centromeric probe of chromosome 3. biii Chromosome 3 represented in gray. Schematic color diagram showing the insertion from 3p25.3p21.31 to 3q27.3. Three subsegments (red, blue and yellow bars) were rearranged and inserted into 3q27.3. The resultant chromosome der(3) depicts a subsegment seq[GRCh37] chr3:g.4679248_46851722 (gray bar) that was not involved in the insertion loci, while the rearranged subsegments (in blue, yellow and red) were inserted into 3q27.3. In addition, an 11.7 kb segment in the flanking region of the breakpoint junction from the acceptor chromosome was duplicated (shown in purple) and present in both upstream and downstream of the insertions. In each figure, the breakpoint junctions are indicated by black vertical arrows, and the genomic orientation is indicated by a black horizontal arrow within each box
Cryptic segment from the insertion was not present in the insertion locus: Sample05 and Sample03 balanced insertions. Diagram of rearrangements detected in Sample05 (a) and Sample03 (b). a Parts of the original chromosomes 3 and 6 are depicted in the upper panel, while the derivative chromosomes after insertion are shown in the lower panel. Subsegments were painted different colors to indicate the original and final location of each subsegment. This complex insertion in the lower panel resulted a der(6) with the inserted yellow and brown subsegment in one direction, while the blue segment is in an inverted direction. The der(3) retained a 153 kb subsegment (in purple), while the 2.3 Mb segment in gray was deleted. b In the upper panel, the original locus of each DNA segment is shown with genomic coordinate (in Mb scale) indicated by a black vertical arrow. In the lower panel, the composition of each derivative chromosome is shown. The yellow segment from the donor chromosome remained, but with inverted orientation. Each rearranged segment is shown with the genomic orientation indicated by a horizontal arrow
Fragmented acceptor chromosome: Sample02 and 5M. Diagram of rearrangements detected in Sample02 (a) and 5M (b). a Parts of the original chromosomes 2 and 6 are depicted in the upper panel, while the derivative chromosomes after insertion are shown in the lower panel. Subsegments were painted in different colors to indicate the original and final location of each subsegment. The genomic coordinates (in Mb scale) are indicated by black vertical arrows. The subsegments in blue, pink and yellow on chromosome 2p were inserted to chromosome 6q. The resulting der(6) in the lower panel revealed complex rearrangements. For example, the pink subsegment at seq[GRCh37] 2p16.1 chr2:g.5816279_58708053 was inserted in reverse orientation into 6q at the 114.5 Mb position, while the blue and yellow subsegments were rearranged and inserted at the 120 Mb position. The 200 kb segment in orange on chromosome 6q at 74.1 Mb location was moved distally to the 162.9 Mb location. (B) The inserted segment from chromosome 2 consists of yellow and blue subsegments and were inserted into chromosome 6 at 98.4 Mb position in the reverse orientation. The resultant der(6) was rearranged into three segments involving a small segment 14.6 kb seq[GRCh37] 6q16.1(98433429_98447982), located between the yellow and blue subsegments
Mate-pair GS identified 59 rearrangements breakpoints among these cases (Fig. 5a). The number of additional cryptic rearrangements found in our insertions cohort was significantly higher than other balanced chromosomal rearrangements (such as translocations and inversions) reported in the Developmental Genome Anatomy Project (DGAP, 65/248, Chi-square test: P = 0.00027) (Redin et al. 2017). Investigating chromosomal insertions by mate-pair GS in comparison with a combination of karyotyping/FISH and CMA identified significantly more cryptic complex rearrangements (8/40, Chi-square test P = 0.0005) (Gu et al. 2016). We then investigated whether the number of subsegment rearrangement was correlated with the size of the insertion. However, no significant correlation was found (Fig. 5b).
An overview of 16 cases with chromosomal insertions depicting the distribution of breakpoints and size of insertion segments. a The spectrum of chromosomal structural rearrangements detected by mate-pair GS in 16 cases with known chromosomal insertions. Each line represents a rearrangement junction and each color depicts an individual case. b The size of the insertion segment and the number of structural rearrangements identified in the acceptor chromosome are shown. The two red arrows show the two cases with the highest number of rearrangements; however, there is no correlation to the size of the insertion segment. P values of Pearson and Spearman correlation are shown accordingly
Gene disruption at rearrangement breakpoints
In 15/16 cases, at least one RefSeq gene was disrupted by the rearrangement breakpoints. In total, 31 genes were disrupted by rearrangement breakpoints in these cases, 9 out of them were OMIM disease-causing genes (Table 2). For instance, a male case 31M with unbalanced insertion (represented as a duplication) presented with moderate developmental delay and autism spectrum. A 4.6 Mb duplication and insertion dup(X)(p11.22p11.21) consisting of five subsegments was rearranged. Four of the five segments were inserted into the long arm of X chromosome resulting in a duplication of genomic content, one subsegment was not inserted (seq[GRCh37] Xq21.31 chrX:g.54853904_54900190, copy number neutral) (Fig. 1). Within the duplicated region, OMIM disease-causing genes SHROOM4, IQSEC2, SMC1A, HUWEI and FGD1 may associate with mental retardation. However, none of these have been reported with triplosensitivity by the ClinGen Dosage Sensitivity Curation. In our case, the gene SHROOM4 was disrupted. Whether gene disruption of SHROOM4 may contribute to the phenotype is currently unknown. Although a study reported disruption of this gene by balanced X;autosome translocations, which might explain the phenotypic presentations (mild to moderate mental retardation) of two unrelated female patients carriers (Hagens et al. 2006), suggesting that the disruption of SHROOM4 in our case might also contribute to the phenotype. In addition, among the five cases with balanced chromosomal insertions, a cryptic 2.2 Mb deletion seq[GRCh37] del(3)(q24) chr3:g.146055006_148300124del was detected in Sample05 (Fig. 3a) (Dong et al. 2019b). This 2.2 Mb deletion involves four OMIM annotated genes, including the ZIC1 gene, in which mutations are associated with structural brain anomalies with impaired intellectual development and craniosynostosis. Although this deletion was classified as pathogenic, no clinically recognized malformation was identified in this patient, possibly due to the incomplete penetrance of ZIC1 deletion (Ferraris et al. 2013).
Breakpoint junction features
GS in combination with junction-specific PCR and Sanger sequencing enabled the investigation of breakpoint features at the nucleotide level. Among the 59 rearrangements detected by mate-pair GS, 41 (69.5%) breakpoint junctions were fine mapped by junction-specific PCR and Sanger sequencing. The remaining 18 breakpoints could not be resolved to nucleotide level because of primer design failure or Sanger sequencing failure due to the limitation of current technologies.
Among the 41 pinpointed junctions, microhomology was the predominant type of breakpoint features in both groups, followed by blunt ends, and small insertions accounting for most of the breakpoint junctions (n = 40). In addition, SINEs and LINEs were found in 24% and 15% of the breakpoint junctions, respectively (Table 3). However, none of them had the same subcategory of repetitive elements or self-chain alignment in the proximal and distal reference sequences of the rearrangements (Supplementary Table S2). Sequence similarity between the proximal and distal reference strands was identified in only one junction (Supplementary Figure S6). Only one breakpoint junction had the same subcategory of repetitive element on both sides of the rearrangement (self-chain, Supplementary Table S2).
We further compared the breakpoint features between simple and complex insertions, but the results showed no significant differences (Table 4). In addition, we also compared the incidence of different breakpoint features with previously reported complex insertions (Gu et al. 2016) and balanced chromosomal rearrangements (translocations and inversions) (Redin et al. 2017). The results also showed no significant differences (Table 4). Furthermore, we subclassified the breakpoint junctions based on whether they are located on the donor or acceptor chromosomes, and no differences for each category between two groups were found (Table 5). However, the sample size was limited.
Non-B DNA motifs
To investigate whether the percentage of breakpoint junctions with non-B DNA motifs or the motif distribution was different from the genome, we obtained 394 genome-wide simulated “rearrangements” to represent the background for comparison (see “Materials and methods”).
As the number of breakpoint junctions in simple/complex insertions was limited, we involved the breakpoint junctions from the reported complex insertions (Gu et al. 2016) for the analysis. Interestingly, each group (either including only complex insertions or all insertions) showed a significant difference on the distribution of various types of the non-B motifs compared with the random generated data (Table 6).
Discussion
Mate-pair GS enabled investigation of chromosomal insertions with/without copy number changes at nucleotide-level resolution. Among our 16 cases with simple chromosomal insertions, GS results not only revised the G-banded chromosome analysis results in 75.0% (12/16) of cases, but also enabled detection of additional complexities in 68.75% (11/16) of cases.
In this study, the proportion of cases with cryptic rearrangements in chromosomal insertions was higher than previously reported by a combination approach utilizing CMA and FISH/karyotyping (8/76) (Gu et al. 2016). Mate-pair GS can detect rearrangements regardless of copy number changes, emphasizing its utility for investigation of related balanced and unbalanced chromosome aberrations. In addition, the incidence of cryptic rearrangements was significantly higher in simple chromosomal insertions compared to previously reported balanced translocations and inversions (Dong et al. 2019a; Redin et al. 2017). We characterized and further classified four classes of cryptic rearrangements, which were not mutually exclusive. Among these four classes, insertion segments were fragmented and rearranged, but clustered at the insertion site in ten cases (10/16, 62.5%, class 1 cryptic rearrangements), half of which (5/10) were with cryptic subsegments not inserted to the insertion site (class 2). It indicated that a proportion of chromosomal insertions might not undergo a simple release of a segment from two breaks on the donor chromosome, but scattered into pieces and some of which were not inserted into the acceptor chromosome. This was also evidenced by the observation of multiple fragments from the acceptor chromosome at the insertion site in two cases (2/16, 12.5%, class 3) and flanking duplications in another two cases (2/16, 12.5%, class 4).
Our study provided evidence to support the potential underlying mechanisms of chromosomal insertions. Firstly, evidence of replication-based DNA repair mechanisms such as chromoanasynthesis (Liu et al. 2011; Stephens et al. 2011) included the identification of copy number gains in the flanking regions of the breakpoint junctions in the acceptor chromosome in cases 10F and 25M (Class 4, Supplementary Figure S5), echoing the observations from a previous study (Gu et al. 2016). Secondly, microhomology was the predominant type of breakpoint junction features (21/41, 51.2%) across all four classes of complex rearrangements. Apart from microhomology-mediated end joining, an alternative NHEJ (Wang and Xu 2017), breakpoint junction microhomology is also a feature of MMBIR and iterative template switching replication-based mechanisms (Liu et al. 2011; Stephens et al. 2011). Therefore, the involvement of replication-based mechanisms in class 1–3 could not be excluded. Since the observations of class 1 and 3 events were not mutually exclusive as they were differentiated by the observation of multiple segments from the acceptor chromosome, it is possible that they resulted from shattering of one or more chromosomes as in chromothripsis (Liu et al. 2011; Stephens et al. 2011). Lastly, we showed that the number of additional cryptic rearrangements was not associated with the size of the insertion (Fig. 5b). Furthermore, by reviewing flanking regions of the breakpoint junctions, a different non-B DNA motif distribution was observed in simple/complex insertion compared with the average genome, although the frequency was similar. It indicated that regions surrounding insertion breakpoints had similar capabilities to adopt the non-B DNA motifs as the genome (Vissers et al. 2009).
By comparing the patterns of breakpoint junctions from both simple and complex insertions with previously published cohorts of insertions (Gu et al. 2016) and translocations/inversions (Redin et al. 2017), no significant differences were found. This suggests that rare structural rearrangements including insertions, translocations and inversions are likely generated by the same mechanism(s).
Identification of disrupted genes and cryptic copy number changes can aid the clinical interpretation of the chromosomal insertions (Fig. 1). In this cohort, there were 26 OMIM genes disrupted at the breakpoint junctions, 9 of which were OMIM disease-causing genes (Table 2). For example, in the male case 31M with the unbalanced insertion presenting with moderate developmental delay and autism spectrum, disruption of SHROOM4 may contribute to the phenotype. However, further comprehensive investigation by including the analysis of single nucleotide variants may be warranted. Recent studies demonstrated chromosomal structural variants may contribute to human diseases by disrupting long-range loops, topologically associating domains, and promotor enhancer interactions (David et al. 2020; Lupiáñez et al. 2015). Future investigation of potential correlation of chromosomal rearrangements with such elements is desirable. Furthermore, chromothripsis-like events can be observed in asymptomatic subjects with abnormal pregnancy history such as recurrent miscarriage (Dong et al. 2019a) and these events can increase the risk of de novo cryptic copy number deletion/duplication related to the inherited rearrangements in the offspring (de Pagter et al. 2015). The ability of GS to comprehensively investigate complex rearrangements highlights its utility to investigate chromothripsis- or chromoanasynthesis-like events in chromosomal abnormalities, which would otherwise be missed by conventional cytogenetic approaches.
Lastly, although studying the genetic inheritance of the insertions particularly for cryptic rearrangements identified by mate pair sequencing is important for clinical interpretation, this study has potential limitations. Due to the original study design and consenting process, parental karyotyping results were available only in 5/16 cases. Particularly, mate-pair sequencing and other molecular assays could not be performed in the parental samples. As such, we cannot exclude the possibility of the patients' unbalanced insertions to be derived from a parent with a balanced insertion. The scope of our study was to investigate the sequence features of the related breakpoint junctions in chromosomal insertions identified by karyotyping and/or CMA. Therefore, we compared our breakpoint sequence features with previously published studies that included: (1) chromosomal rearrangements identified by karyotyping and (2) breakpoint sequence features that are available (Tables 4 and 6). Large studies provide datasets interrogating a large number of structural variants identified in patients with autism spectrum disorders and other common, complex diseases (Abel et al. 2020; Collins et al. 2017 ), which lay the foundation to understand the genomic architecture of structural rearrangements from megabase chromosomal segment to single nucleotide level. A future study is warranted to investigate the breakpoint sequence feature differences of our results with the ones provided in these databases. Systematic study on insertions including visible and cryptic rearrangements may provide insight into the complexity and underlying mechanism(s) triggering the pathogenicity of the insertions. Furthermore, although the large insert sizes by mate-pair GS ranged from 3 to 8 kb compared to traditional short read sequencing improved structural variation detection, unresolved rearrangements still remain. For instance, a subset of breakpoints (18/59) could not be resolved to the nucleotide-level by Sanger sequencing especially in repeat regions. In one case (24F), the complex rearrangements could not be completely assembled albeit the detection of six independent breakpoints by mate-pair GS. Future studies by long read sequencing will have advantages in fine mapping rearrangement breakpoints in previously inaccessible loci including repeat regions, atypical GC regions and enabling phasing of complex rearrangements. (Cretu Stancu et al. 2017; Logsdon et al. 2020).
Conclusion
Mate-pair genome sequencing enabled the precise breakpoint characterization of chromosome insertions and revision of the karyotypes of 75.0% of cases. It also revealed additional cryptic complex rearrangements of the insertion subsegment changes in 68.8% of the cases, significantly higher than those reported in reciprocal translocations and inversions. Overall, our study provided molecular characterization of karyotypically simple insertions, demonstrated previously underappreciated complexities, and evidenced that chromosomal insertions are likely formed by nonhomologous end joining and/or microhomology-mediated replication-based DNA repair.
Data availability
The genome sequencing data of five samples used in this study have been made available on the CNGB Nucleotide Sequence Archive (CNSA: https://db.cngb.org/cnsa) under the accession number CNP0000078. For the other 12 cases, raw sequencing data have not been made available because most subjects did not consent to such sharing.
Code availability
NA.
References
Abel HJ, Larson DE, Regier AA, Chiang C, Das I, Kanchi KL, Layer RM, Neale BM, Salerno WJ, Reeves C, Buyske S, Genomics NCfCD, Matise TC, Muzny DM, Zody MC, Lander ES, Dutcher SK, Stitziel NO, Hall IM (2020) Mapping and characterization of structural variation in 17,795 human genomes. Nature 583:83–89. https://doi.org/10.1038/s41586-020-2371-0
Ankala A, Kohn JN, Hegde A, Meka A, Ephrem CL, Askree SH, Bhide S, Hegde MR (2012) Aberrant firing of replication origins potentially explains intragenic nonrecurrent rearrangements within genes, including the human DMD gene. Genome Res 22:25–34. https://doi.org/10.1101/gr.123463.111
Bacolla A, Wang G, Jain A, Chuzhanova NA, Cer RZ, Collins JR, Cooper DN, Bohr VA, Vasquez KM (2011) Non-B DNA-forming sequences and WRN deficiency independently increase the frequency of base substitution in human cells. J Biol Chem 286:10017–10026. https://doi.org/10.1074/jbc.M110.176636
Bauters M, Van Esch H, Friez MJ, Boespflug-Tanguy O, Zenker M, Vianna-Morgante AM, Rosenberg C, Ignatius J, Raynaud M, Hollanders K, Govaerts K, Vandenreijt K, Niel F, Blanc P, Stevenson RE, Fryns JP, Marynen P, Schwartz CE, Froyen G (2008) Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair. Genome Res 18:847–858. https://doi.org/10.1101/gr.075903.107
Cer RZ, Bruce KH, Donohue DE, Temiz NA, Mudunuri US, Yi M, Volfovsky N, Bacolla A, Luke BT, Collins JR, Stephens RM (2012) Searching for non-B DNA-forming motifs using nBMST (non-B DNA motif search tool). Curr Protoc Hum Genet 18(7):1–22. https://doi.org/10.1002/0471142905.hg1807s73
Chau MHK, Cao Y, Kwok YKY, Chan S, Chan YM, Wang H, Yang Z, Wong HK, Leung TY, Choy KW (2019) Characteristics and mode of inheritance of pathogenic copy number variants in prenatal diagnosis. Am J Obstet Gynecol 221:493.e1. https://doi.org/10.1016/j.ajog.2019.06.007
Chau MHK, Wang H, Lai Y, Zhang Y, Xu F, Tang Y, Wang Y, Chen Z, Leung TY, Chung JPW, Kwok YK, Chong SC, Choy KW, Zhu Y, Xiong L, Wei W, Dong Z (2020) Low-pass genome sequencing: a validated method in clinical cytogenetics. Hum Genet. https://doi.org/10.1007/s00439-020-02185-9
Choy KW, Wang H, Shi M, Chen J, Yang Z, Zhang R, Yan H, Wang Y, Chen S, Chau MHK, Cao Y, Chan OYM, Kwok YK, Zhu Y, Chen M, Leung TY, Dong Z (2019) Prenatal diagnosis of fetuses with increased nuchal translucency by genome sequencing analysis. Front Genet 10:761. https://doi.org/10.3389/fgene.2019.00761
Collins RL, Brand H, Redin CE, Hanscom C, Antolik C, Stone MR, Glessner JT, Mason T, Pregno G, Dorrani N, Mandrile G, Giachino D, Perrin D, Walsh C, Cipicchio M, Costello M, Stortchevoi A, An JY, Currall BB, Seabra CM, Ragavendran A, Margolin L, Martinez-Agosto JA, Lucente D, Levy B, Sanders SJ, Wapner RJ, Quintero-Rivera F, Kloosterman W, Talkowski ME (2017) Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol 18:36. https://doi.org/10.1186/s13059-017-1158-6
Collins RL, Brand H, Karczewski KJ, Zhao X, Alfoldi J, Francioli LC, Khera AV, Lowther C, Gauthier LD, Wang H, Watts NA, Solomonson M, O'Donnell-Luria A, Baumann A, Munshi R, Walker M, Whelan CW, Huang Y, Brookings T, Sharpe T, Stone MR, Valkanas E, Fu J, Tiao G, Laricchia KM, Ruano-Rubio V, Stevens C, Gupta N, Cusick C, Margolin L, Genome Aggregation Database Production T, Genome Aggregation Database C, Taylor KD, Lin HJ, Rich SS, Post WS, Chen YI, Rotter JI, Nusbaum C, Philippakis A, Lander E, Gabriel S, Neale BM, Kathiresan S, Daly MJ, Banks E, MacArthur DG, Talkowski ME (2020) A structural variation reference for medical and population genetics. Nature 581:444–451. https://doi.org/10.1038/s41586-020-2287-8
Cretu Stancu M, van Roosmalen MJ, Renkens I, Nieboer MM, Middelkamp S, de Ligt J, Pregno G, Giachino D, Mandrile G, Espejo Valle-Inclan J, Korzelius J, de Bruijn E, Cuppen E, Talkowski ME, Marschall T, de Ridder J, Kloosterman WP (2017) Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat Commun 8:1326. https://doi.org/10.1038/s41467-017-01343-4
David D, Freixo JP, Fino J, Carvalho I, Marques M, Cardoso M, Pina-Aguilar RE, Morton CC (2020) Comprehensive clinically oriented workflow for nucleotide level resolution and interpretation in prenatal diagnosis of de novo apparently balanced chromosomal translocations in their genomic landscape. Hum Genet 139:531–543. https://doi.org/10.1007/s00439-020-02121-x
de Pagter MS, van Roosmalen MJ, Baas AF, Renkens I, Duran KJ, van Binsbergen E, Tavakoli-Yaraki M, Hochstenbach R, van der Veken LT, Cuppen E, Kloosterman WP (2015) Chromothripsis in healthy individuals affects multiple protein-coding genes and can result in severe congenital abnormalities in offspring. Am J Hum Genet 96:651–656. https://doi.org/10.1016/j.ajhg.2015.02.005
Dong Z, Jiang L, Yang C, Hu H, Wang X, Chen H, Choy KW, Hu H, Dong Y, Hu B, Xu J, Long Y, Cao S, Chen H, Wang WJ, Jiang H, Xu F, Yao H, Xu X, Liang Z (2014) A robust approach for blind detection of balanced chromosomal rearrangements with whole-genome low-coverage sequencing. Hum Mutat 35:625–636. https://doi.org/10.1002/humu.22541
Dong Z, Zhang J, Hu P, Chen H, Xu J, Tian Q, Meng L, Ye Y, Wang J, Zhang M, Li Y, Wang H, Yu S, Chen F, Xie J, Jiang H, Wang W, Choy KW, Xu Z (2016) Low-pass whole-genome sequencing in clinical cytogenetics: a validated approach. Genet Med 18:940–948. https://doi.org/10.1038/gim.2015.199
Dong Z, Wang H, Chen H, Jiang H, Yuan J, Yang Z, Wang WJ, Xu F, Guo X, Cao Y, Zhu Z, Geng C, Cheung WC, Kwok YK, Yang H, Leung TY, Morton CC, Cheung SW, Choy KW (2018) Identification of balanced chromosomal rearrangements previously unknown among participants in the 1000 genomes project: implications for interpretation of structural variation in genomes and the future of clinical cytogenetics. Genet Med 20:697–707. https://doi.org/10.1038/gim.2017.170
Dong Z, Yan J, Xu F, Yuan J, Jiang H, Wang H, Chen H, Zhang L, Ye L, Xu J, Shi Y, Yang Z, Cao Y, Chen L, Li Q, Zhao X, Li J, Chen A, Zhang W, Wong HG, Qin Y, Zhao H, Chen Y, Li P, Ma T, Wang WJ, Kwok YK, Jiang Y, Pursley AN, Chung JPW, Hong Y, Kristiansen K, Yang H, Pina-Aguilar RE, Leung TY, Cheung SW, Morton CC, Choy KW, Chen ZJ (2019a) Genome sequencing explores complexity of chromosomal abnormalities in recurrent miscarriage. Am J Hum Genet 105:1102–1111. https://doi.org/10.1016/j.ajhg.2019.10.003
Dong Z, Zhao X, Li Q, Yang Z, Xi Y, Alexeev A, Shen H, Wang O, Ruan J, Ren H, Wei H, Qi X, Li J, Zhu X, Zhang Y, Dai P, Kong X, Kirkconnell K, Alferov O, Giles S, Yamtich J, Kermani BG, Dong C, Liu P, Mi Z, Zhang W, Xu X, Drmanac R, Choy KW, Jiang Y (2019b) Development of coupling controlled polymerizations by adapter-ligation in mate-pair sequencing for detection of various genomic variants in one single assay. DNA Res. https://doi.org/10.1093/dnares/dsz011
Ferraris A, Bernardini L, Avramovska VS, Zanni G, Loddo S, Sukarova-Angelovska E, Parisi V, Capalbo A, Tumini S, Travaglini L, Mancini F, Duma F, Barresi S, Novelli A, Mercuri E, Tarani L, Italian CSG, Bertini E, Dallapiccola B, Valente EM (2013) Dandy-Walker malformation and Wisconsin syndrome: novel cases add further insight into the genotype-phenotype correlations of 3q23q25 deletions. Orphanet J Rare Dis 8:75. https://doi.org/10.1186/1750-1172-8-75
Gu S, Yuan B, Campbell IM, Beck CR, Carvalho CM, Nagamani SC, Erez A, Patel A, Bacino CA, Shaw CA, Stankiewicz P, Cheung SW, Bi W, Lupski JR (2015) Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3. Hum Mol Genet 24:4061–4077. https://doi.org/10.1093/hmg/ddv146
Gu S, Szafranski P, Akdemir ZC, Yuan B, Cooper ML, Magrina MA, Bacino CA, Lalani SR, Breman AM, Smith JL, Patel A, Song RH, Bi W, Cheung SW, Carvalho CM, Stankiewicz P, Lupski JR (2016) Mechanisms for complex chromosomal insertions. PLoS Genet 12:e1006446. https://doi.org/10.1371/journal.pgen.1006446
Hagens O, Dubos A, Abidi F, Barbi G, Van Zutven L, Hoeltzenbein M, Tommerup N, Moraine C, Fryns JP, Chelly J, van Bokhoven H, Gecz J, Dollfus H, Ropers HH, Schwartz CE, de Santos RCSD, Kalscheuer V, Hanauer A (2006) Disruptions of the novel KIAA1202 gene are associated with X-linked mental retardation. Hum Genet 118:578–590. https://doi.org/10.1007/s00439-005-0072-2
Hijazi H, Coelho FS, Gonzaga-Jauregui C, Bernardini L, Mar SS, Manning MA, Hanson-Kahn A, Naidu S, Srivastava S, Lee JA, Jones JR, Friez MJ, Alberico T, Torres B, Fang P, Cheung SW, Song X, Davis-Williams A, Jornlin C, Wight PA, Patyal P, Taube J, Poretti A, Inoue K, Zhang F, Pehlivan D, Carvalho CMB, Hobson GM, Lupski JR (2020) Xq22 deletions and correlation with distinct neurological disease traits in females: further evidence for a contiguous gene syndrome. Hum Mutat 41:150–168. https://doi.org/10.1002/humu.23902
Huang J, Poon LC, Akolekar R, Choy KW, Leung TY, Nicolaides KH (2014) Is high fetal nuchal translucency associated with submicroscopic chromosomal abnormalities on array CGH? Ultrasound Obstet Gynecol 43:620–624. https://doi.org/10.1002/uog.13384
Kang SH, Shaw C, Ou Z, Eng PA, Cooper ML, Pursley AN, Sahoo T, Bacino CA, Chinault AC, Stankiewicz P, Patel A, Lupski JR, Cheung SW (2010) Insertional translocation detected using FISH confirmation of array-comparative genomic hybridization (aCGH) results. Am J Med Genet A 152A:1111–1126. https://doi.org/10.1002/ajmg.a.33278
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O'Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Genome Aggregation Database C, Neale BM, Daly MJ, MacArthur DG (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581:434–443. https://doi.org/10.1038/s41586-020-2308-7
Kato T, Ouchi Y, Inagaki H, Makita Y, Mizuno S, Kajita M, Ikeda T, Takeuchi K, Kurahashi H (2017) Genomic characterization of chromosomal insertions: insights into the mechanisms underlying chromothripsis. Cytogenet Genome Res 153:1–9. https://doi.org/10.1159/000481586
Kehrer M, Liehr T, Benkert T, Singer S, Grasshoff U, Schaeferhoff K, Bonin M, Weichselbaum A, Tzschach A (2015) Interstitial duplication of chromosome region 1q25.1q25.3: report of a patient with mild cognitive deficits, tall stature and facial dysmorphisms. Am J Med Genet A 167A:653–656. https://doi.org/10.1002/ajmg.a.36943
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, Bacino CA, Campos-Acevedo LD, Delgado MR, Freedenberg D, Garnica A, Grebe TA, Hernandez-Almaguer D, Immken L, Lalani SR, McLean SD, Northrup H, Scaglia F, Strathearn L, Trapane P, Kang SH, Patel A, Cheung SW, Hastings PJ, Stankiewicz P, Lupski JR, Bi W (2011) Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146:889–903. https://doi.org/10.1016/j.cell.2011.07.042
Logsdon GA, Vollger MR, Eichler EE (2020) Long-read human genome sequencing and its applications. Nat Rev Genet. https://doi.org/10.1038/s41576-020-0236-x
Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, Santos-Simarro F, Gilbert-Dussardier B, Wittler L, Borschiwer M, Haas SA, Osterwalder M, Franke M, Timmermann B, Hecht J, Spielmann M, Visel A, Mundlos S (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161:1012–1025. https://doi.org/10.1016/j.cell.2015.04.004
Mackie Ogilvie C, Scriven PN (2002) Meiotic outcomes in reciprocal translocation carriers ascertained in 3-day human embryos. Eur J Hum Genet 10:801–806. https://doi.org/10.1038/sj.ejhg.5200895
Melotte C, Debrock S, D'Hooghe T, Fryns JP, Vermeesch JR (2004) Preimplantation genetic diagnosis for an insertional translocation carrier. Hum Reprod 19:2777–2783. https://doi.org/10.1093/humrep/deh539
Mendez-Rosado LA, Lantigua A, Galarza J, Hamid Al-Rikabi AB, Ziegler M, Liehr T (2017) Unusual de novo partial trisomy 17p12p11.2 due to unbalanced insertion into 5p13.1 in a severely affected boy. J Pediatr Genet 6:165–168. https://doi.org/10.1055/s-0037-1599195
Neill NJ, Ballif BC, Lamb AN, Parikh S, Ravnan JB, Schultz RA, Torchia BS, Rosenfeld JA, Shaffer LG (2011) Recurrence, submicroscopic complexity, and potential clinical relevance of copy gains detected by array CGH that are shown to be unbalanced insertions by FISH. Genome Res 21:535–544. https://doi.org/10.1101/gr.114579.110
Ottaviani D, LeCain M, Sheer D (2014) The role of microhomology in genomic structural variation. Trends Genet 30:85–94. https://doi.org/10.1016/j.tig.2014.01.001
Pellestor F, Anahory T, Lefort G, Puechberty J, Liehr T, Hedon B, Sarda P (2011) Complex chromosomal rearrangements: origin and meiotic behavior. Hum Reprod Update 17:476–494. https://doi.org/10.1093/humupd/dmr010
Redin C, Brand H, Collins RL, Kammin T, Mitchell E, Hodge JC, Hanscom C, Pillalamarri V, Seabra CM, Abbott MA, Abdul-Rahman OA, Aberg E, Adley R, Alcaraz-Estrada SL, Alkuraya FS, An Y, Anderson MA, Antolik C, Anyane-Yeboa K, Atkin JF, Bartell T, Bernstein JA, Beyer E, Blumenthal I, Bongers EM, Brilstra EH, Brown CW, Bruggenwirth HT, Callewaert B, Chiang C, Corning K, Cox H, Cuppen E, Currall BB, Cushing T, David D, Deardorff MA, Dheedene A, D'Hooghe M, de Vries BB, Earl DL, Ferguson HL, Fisher H, FitzPatrick DR, Gerrol P, Giachino D, Glessner JT, Gliem T, Grady M, Graham BH, Griffis C, Gripp KW, Gropman AL, Hanson-Kahn A, Harris DJ, Hayden MA, Hill R, Hochstenbach R, Hoffman JD, Hopkin RJ, Hubshman MW, Innes AM, Irons M, Irving M, Jacobsen JC, Janssens S, Jewett T, Johnson JP, Jongmans MC, Kahler SG, Koolen DA, Korzelius J, Kroisel PM, Lacassie Y, Lawless W, Lemyre E, Leppig K, Levin AV, Li H, Li H, Liao EC, Lim C, Lose EJ, Lucente D, Macera MJ, Manavalan P, Mandrile G, Marcelis CL, Margolin L, Mason T, Masser-Frye D, McClellan MW, Mendoza CJ, Menten B, Middelkamp S, Mikami LR, Moe E, Mohammed S, Mononen T, Mortenson ME et al (2017) The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat Genet 49:36–45. https://doi.org/10.1038/ng.3720
Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal S, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Quail MA, Burton J, Swerdlow H, Carter NP, Morsberger LA, Iacobuzio-Donahue C, Follows GA, Green AR, Flanagan AM, Stratton MR, Futreal PA, Campbell PJ (2011) Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144:27–40. https://doi.org/10.1016/j.cell.2010.11.055
Van Hemel JO, Eussen HJ (2000) Interchromosomal insertions. Identification of five cases and a review. Hum Genet 107:415–432. https://doi.org/10.1007/s004390000398
Vissers LE, Bhatt SS, Janssen IM, Xia Z, Lalani SR, Pfundt R, Derwinska K, de Vries BB, Gilissen C, Hoischen A, Nesteruk M, Wisniowiecka-Kowalnik B, Smyk M, Brunner HG, Cheung SW, van Kessel AG, Veltman JA, Stankiewicz P (2009) Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum Mol Genet 18:3579–3593. https://doi.org/10.1093/hmg/ddp306
Wang H, Xu X (2017) Microhomology-mediated end joining: new players join the team. Cell Biosci 7:6. https://doi.org/10.1186/s13578-017-0136-8
Wang L, Xi Y, Zhang W, Wang W, Shen H, Wang X, Zhao X, Alexeev A, Peters BA, Albert A, Xu X, Ren H, Wang O, Kirkconnell K, Perazich H, Clark S, Hurowitz E, Chen A, Xu X, Drmanac R, Jiang Y (2019) 3' Branch ligation: a novel method to ligate non-complementary DNA to recessed or internal 3'OH ends in DNA or RNA. DNA Res 26:45–53. https://doi.org/10.1093/dnares/dsy037
Wang H, Dong Z, Zhang R, Chau MHK, Yang Z, Tsang KYC, Wong HK, Gui B, Meng Z, Xiao K, Zhu X, Wang Y, Chen S, Leung TY, Cheung SW, Kwok YK, Morton CC, Zhu Y, Choy KW (2020) Low-pass genome sequencing versus chromosomal microarray analysis: implementation in prenatal diagnosis. Genet Med 22:500–510. https://doi.org/10.1038/s41436-019-0634-7
Funding
This project is supported by the National Natural Science Foundation of China (31801042, 81741004 and 81741009), the Health and Medical Research Fund (04152666, 07180576), and Direct Grant (2019.051).
Author information
Authors and Affiliations
Contributions
SWC, ZD, MHKC, TYL and KWC designed the study. SWC and MHKC collected the samples. MHKC, YZ performed mate-pair sequencing. ZD, MHKC, PD, XK, PS and YK performed the analysis and data interpretation. MHKC, PD and YZ conducted the validation. ZD, MHKC, PS, SWC and KWC wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest or competing interest.
Ethics approval
The study was approved by the institutional review boards of each collaborative site.
Informed consent
Written informed consent for sample storage and genetic analyses of peripheral bloods and invasive diagnostic samples was obtained from each participant.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dong, Z., Chau, M.H.K., Zhang, Y. et al. Deciphering the complexity of simple chromosomal insertions by genome sequencing. Hum Genet 140, 361–380 (2021). https://doi.org/10.1007/s00439-020-02210-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-020-02210-x