Abstract
Meiotic recombination is vital for maintaining the sequence diversity in human genome. Meiosis and recombination are considered the essential phases of cell division. In meiosis, the genome is divided into equal parts for sexual reproduction whereas in recombination, the diverse genomes are combined to form new combination of genetic variations. Recombination process does not occur randomly across the genomes, it targets specific areas called recombination “hotspots” and “coldspots”. Owing to huge exploration of polygenetic sequences in data banks, it is impossible to recognize the sequences through conventional methods. Looking at the significance of recombination spots, it is indispensable to develop an accurate, fast, robust, and high-throughput automated computational model. In this model, the numerical descriptors are extracted using two sequence representation schemes namely: dinucleotide composition and trinucleotide composition. The performances of seven classification algorithms were investigated. Finally, the predicted outcomes of individual classifiers are fused to form ensemble classification, which is formed through majority voting and genetic algorithm (GA). The performance of GA-based ensemble model is quite promising compared to individual classifiers and majority voting-based ensemble model. iRSpot-GAEnsC has achieved 84.46 % accuracy. The empirical results revealed that the performance of iRSpot-GAEnsC is not only higher than the examined algorithms but also better than existing methods in the literature developed so far. It is anticipated that the proposed model might be helpful for research community, academia and for drug discovery.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Meiotic recombination plays a preliminary role in the maintenance of sequence diversity in human genomes (Chen et al. 2013; Qiu et al. 2014a). The process of recombination is carried out in two steps. In first step, the genome is divided into two equal parts called daughter cells, which participate in sexual reproduction; this process is referred to as meiosis. In the second step, these diverse gametes are joined to form new combination of genetic variations, it is known as recombination. Recombination is very crucial to genetic variations and is considered a main driven force in these variations. In human chromosomes, it targets very narrow spots, which are called hotspots and coldspots. The region of chromosomes where the frequency of recombination is high is called hotspot and the region where the frequency is low recombination is referred to as coldspot. The identification of recombination spots is very essential to understand the reproduction and growth of the cells. A recent study demonstrated that meiotic recombination events occur in 1–2.5 kilo base regions rather than its random occurrence across a genome. A schematic drawing of the meiotic recombination pathway in a DNA system is illustrated in Fig. 1.
An illustration to show the process of meiotic recombination in a DNA system, adopted from Akbar et al. (2014)
The process of recombination is initiated by double-strand break (broken DNA ends) (Chou 2001a; Keeney 2008; Liu et al. 2012). The hotspots, coldspots and the pattern that is formed by these sites provide fundamental in-depth information on processes of human crossover and gene conversion. Due to large exploration of genome sequences, it is highly desired to develop a precise, consistent, robust and automated system for timely identification of recombination spots. A considerable progress has been made in this area; still need for further improvements in terms of accuracy exists. A series of efforts have been reported in the literature (Chen et al. 2013; Qiu et al. 2014a). Initially, recombination of spots has been predicted using nucleotide composition. However, the main issue in nucleotide composition was only considering little sequence into account where some important hereditary information was lost (Liu et al. 2012). However, the number of possible patterns for DNA sequence is extremely large. Thus, it is very difficult to incorporate the sequence-order information into a statistical predicator with such a large length. To compensate this problem, the concept of pseudo-amino acid composition (PseAAC) was introduced by Chou (Chou 2001a). Furth er, this concept of PseAAC was adopted by almost all fields of computational proteomics such as predicting protein subcellular localization (Lin et al. 2008, 2009a; Khan et al. 2011; Dehzangi et al. 2015; Mandal et al. 2015), protein structural class (Sahu and Panda 2010), DNA-binding proteins (Fang et al. 2008); identifying bacterial virulent proteins (Nanni et al. 2012), predicting metalloproteinase family (Beigi et al. 2011), protein folding rate (Guo et al. 2011), GABA(A) receptor proteins (Mohabatkar et al. 2011), protein super secondary structure (Zou et al. 2011), cyclin proteins (Mohabatkar 2010); classifying amino acids (Georgiou et al. 2009); predicting enzyme family class (Zhou et al. 2007), identifying risk type of human papillomaviruses (Esmaeili et al. 2010); predicting allergenic proteins (Mohabatkar et al. 2013); identifying G protein-coupled receptors and their types (Khan 2012) and discriminating outer membrane proteins (Hayat and Khan 2012a), among many others.
As demonstrated in series of recent publication and comprehensive review demonstrated in (Xu et al. 2013a, b, 2014a, b; He et al. 2015; Jia et al. 2015; Liu et al. 2015f) and in compliance with Chou’s 5-step rule (Chou 2011), to establish a really useful sequence-based statistical predictor for a biological system, we should follow the following five guidelines: (a) construct or select a valid benchmark dataset to train and test the predictor; (b) formulate the biological sequence samples with an effective mathematical expression that can truly reflect their intrinsic correlation with the target to be predicted; (c) introduce or develop a powerful algorithm (or engine) to operate the prediction; (d) properly perform cross-validation tests to objectively evaluate the anticipated accuracy of the predictor; (e) establish a user-friendly web server for the predictor that is accessible to the public.
In this study, we propose genetic algorithm (GA)-based ensemble model iRSpot-GAEnsC for identification of DNA recombination hotspots and coldspots. Numerical descriptors are extracted using two powerful sequence representation techniques namely: dinucleotide composition and trinucleotide composition. Various classification algorithms are investigated individually and finally their predicted outcomes are combined to form ensemble model. The ensemble model is formed using majority voting and GA. Leave-one-out test was applied to assess the performance of proposed model.
Methods and materials
Dataset
To construct a promising computational model, there need some valid benchmark datasets to train the model effectively. For this purpose, we have used dataset S in this study, which has been taken from (Chen et al. 2013; Qiu et al. 2014a). This dataset contains 490 sequences for hotspot recombination and 591 sequences for coldspot recombination. The dataset S of both hotspots and coldspot sequences of recombination can be formulated by:
where S + is the subset for the hotspot recombination and S − is the subset of coldspot recombination, while the symbol “∪” shows union of both hotspot and coldspot recombination.
Feature extraction strategies
Feature extraction is considered one of the fundamental steps in machine learning process. In feature extraction phase, numerical attributes are extracted from biological sequences because statistical models require numerical descriptors for training. With the explosive growth of biological sequences generated in the post-genomic age, one of the most important but also most difficult problems in computational biology is how to formulate a biological sequence with a discrete model or a vector, yet still keep considerable sequence pattern information. This is because all the existing operation engines, such as SVM (Support Vector Machine) and NN Neural Network), can only handle vector but not sequence samples, as elaborated in (Chou 2015). However, a vector defined in a discrete model may completely lose all the sequence-order information. To avoid completely losing the sequence-order or pattern information for proteins, the pseudo-amino acid composition or PseAAC was proposed (Chou 2001a). Ever since the concept of pseudo-amino acid composition was proposed, it has penetrated into nearly all the areas of computational proteomics (Chen et al. 2015). Because of its successes to deal with protein/peptide sequences in computational proteomics, the concept of PseAAC has been recently extended to dealing with DNA/RNA sequences in computational genetics and genomics (Chen et al. 2012, 2014c, d, 2015; Feng et al. 2013; Liu et al. 2014, 2015a, b, c, d, e, f). Based on the concept of PseAAC, the “pseudo k-tuple nucleotide composition (PseKNC)” (Chen et al. 2014c, d; Liu et al. 2015c, e) was proposed in genome analysis. Owing to wide usage of PseAAC, recently the PseKNC was proposed and demonstrated the effectiveness in predicting nucleosome (Guo et al. 2014), identifying splicing sites (Chen et al. 2014b), identifying translation site (Chen et al. 2014a) and origin of replication (Li et al. 2015). Both PseAAC and PseKNC achieved very exciting results and have played very important roles in relevant fields. In this study, we are to use the concept of pseudo-components to predict the recombination spot in DNA. In practical applications, particularly in developing high-throughput tools for predicting various important attributes for biomacromolecules, many different descriptors to represent biological sequence samples have been developed and widely used, such as those by means of cellular automata image (Xiao et al. 2009), those by complexity measure factor (Xiao et al. 2011), and those by grey dynamic model (Lin et al. 2009b, 2012; Qiu et al. 2014c; Xiao et al. 2015), as well as a long list of the relevant references cited in a recent comprehensive review (Chou 2009). Two powerful DNA sequence representation approaches are used to extract high discriminative features.
Dinucleotide composition (DNC)
A DNA sequence is a polymer of four nucleotides namely adenine (A), cytosine (C), guanine (G) and thymine (T). Let us consider the following DNA sequence X with L residues long, i.e.,
where N1 is first position residue of DNA sequence, N2 the second position residue and NL the Lth position residue of the DNA sequence. Simple nucleotide composition has four values, which represents the occurrence frequency of these four nucleotides (Chou et al. 2012). It can be represented as:
where f(A)represents the occurrence frequency of nucleotide A, f(C) determines the occurrence frequency of nucleotide C, f(G) shows the occurrence frequency of nucleotide G and f(T) denotes the occurrence frequency of nucleotide T, whereas symbol T in the superscript represents the transpose operator. However, the main drawback of simple nucleotide composition is not preserving sequence-order information. To amalgamate the occurrence frequency along with sequence-order information, dinucleotide composition (DNC) was introduced. In DNC, the relative frequency of nucleotide pair is computed (Chen et al. 2014b). It can be demonstrated as:
where f(AA) represents the occurrence frequency of AA pair, f(AC) represents the occurrence frequency of AC pair and f(TT) denotes the occurrence frequency of TT pair. As a result, 4 × 4 = 16-D corresponding features are contained in feature space.
Trinucleotide Composition (TNC)
In dinucleotide composition, only two nucleotides are paired. In contrast, in trinucleotide composition (TNC), three nucleotides are combined. In TNC, the occurrence frequency of three nucleotides is calculated. It can be formulated as:
where f(AAA) shows the occurrence frequency of AAA in the DNA sequence, f(AAA) shows the occurrence frequency of AAC in the DNA sequence and so forth (Duda et al. 2012). It revealed that the corresponding feature space will contain 4 × 4 × 4 = 64 pairs of the nucleotides. Equation (6) is written into generalized form so the corresponding space X having 4k components, i.e.,
The above-mentioned procedure revealed that as the number of nucleotides in pair is increased the number of tuple increased (Chen et al. 2014a). The local or short range of sequence-order information is gradually included into information but the global order sequence information is not reflected by the formulation (Qiu et al. 2014a).
Classification algorithms
Classification is the subfield of data mining and machine learning in which the data are categorized into predefined classes. In this study, several supervised classification algorithms are utilized to select the best one for identification of hotspots and coldspots.
K-Nearest Neighbor (KNN)
KNN is widely used algorithm in the field of pattern recognition, machine learning and many other areas. KNN is simple but widely used algorithm for classification (Duda et al. 2012). KNN algorithm is also known as instance-based learning (Lazy learning) algorithm. It does not build classifier or model immediately but save all the training data samples and wait until new observation needs to be classified. Lazy learning nature of KNN makes it better than eager leaning, which constructs classifier before new observation needs to be classified. It is significant for dynamic data that change and update rapidly (Han and Kamber 2006). KNN algorithm has the following five steps;
Step 1: Provide feature space to KNN algorithm to train the system.
Step 2: Measure the distance using the Euclidean distance formula.
Step 3: Sort the Euclidean distance values as di ≤ di + 1, where i = 1, 2, 3…k.
Step 4: Apply voting or means according to the data nature.
Step 5: Number of nearest neighbor (value of K) depends upon the nature and volume of data provided to KNN. For huge data, the k value should be large and for small data, k value should be small.
Probabilistic neural network (PNN)
The probabilistic neural network (PNN) was first introduced by Specht in 1990 (Specht 1990). It is based on Bayes’ theorem. PNN provides an interactive way to interpret the structure of the network in terms of probability density function (Georgiou et al. 2004). PNN has a similar structure as feed-forward networks but it has four layers. The first layer in known as input layer, second layer is known as pattern layer, third layer is known as summation layer and the fourth layer is known as output layer (Khan et al. 2015). The first layer contains the input vector, which is connected to the input neurons and passed to the pattern layer. The dimension of the pattern layer and the number of samples presented to the network is equal in number. Pattern and input layers are connected to each other by exactly one neuron for each training data sample. The summation layer has the same dimension as the number of classes in the set of data samples. Finally, the decision layer predicts the novel sample into one of the predefined classes.
Random forest
Random forest (RF) is a well-known ensemble technique, which was proposed by Breiman (Breiman 2001; Lou et al. 2014). It is widely used for the pattern classification in the field of bioinformatics (Kumar et al. 2009). The prediction performance of RF is high (Kumar et al. 2009; Chou et al. 2012). The information provided by RF is on variable basis for classification (Ebina et al. 2011; Boulesteix et al. 2012; Touw et al. 2013). RF has a large number of decision trees and every tree produces a classification (Breiman 2001). The final result is obtained by combining the results of all the decision trees by means of voting (Jiang et al. 2007). In addition, RF selects the features randomly. Instead of using all the features for one single tree, it splits the features into different trees and then combines the result of each tree (Jiang et al. 2007).
Support vector machine (SVM)
Support vector machine is an effective method used for the classification of supervised pattern recognition process and was first introduced by Vapnik in 1995 (Vapnik 2000; Qiu et al. 2009; Gu et al. 2010). Later on, it was updated by Vapnik in 1998 (Hayat and Khan 2011). Originally it was developed for two class problems but later it was adopted for multiclass problems (Ahmad et al. 2015). In two class problems, SVM transfers data to the high-dimensional feature space and then determines the optimal hyper plane (Chen et al. 2014b). It is very good classifier for identifying linear as well as non-linear patterns (Akbar et al. 2014). SVM uses different types of kernel functions including but not limited to linear, polynomial, Gaussian [RBF] and sigmoid. In this study, the ‘OVO’ strategy was employed for making predictions using the popular radial base function (RBF) as a kernel function with parameters γ and ∁ (Qiu et al. 2014a). The regularization parameter ∁ and the kernel width parameter γ were determined via an optimization procedure using a grid search approach for identification of recombination hotspots and coldspots.
where in the above equation the parameter γ shows the width of the Gaussian function. The values for the above parameters of RBF are calculated using a grid search during the training phase of SVM model. In our work, LIBSVM package (Chang and Lin 2011) has been used to predict the hotspots and coldspots in the DNA sequence. This software is free for download and is available at http://www.csie.ntu.edu.tw/~cjlin/libsvm .
Generalize regression neural network
Generalize regression neural network (GRNN) is mostly used for function approximation. The structure and functionality of GRNN and PNN are similar. It has four layers, i.e., input layer, radial base layer, special linear layer and the output layer. The total number of neurons in the input and output of GRNN is equal to the dimension of the input and output vectors. GRNN is a well-suited network for small- and medium-sized datasets. The overall process of GRNN is carried out in three steps. In first step, a set of training data and target data is created. Next step of GRNN, the input data, target data and spread constant value are passed to newgrnn as arguments. Finally, the response of the network is noted by simulating it according to the data provided (Cherian and Sathiyan 2012).
Feed forward neural network
A feed forward neural network (FFBPNN) is an artificial neural network (ANN), which consists of N layers. The first layer of FFBPNN is connected to the input vector. The preceding layer has a connection with each subsequent layer. The resultant output is produced by final layer of the network. The training of FFBPNN is carried out using the following Eq. (11);
where in Eq. (10), xj(t) shows the input value of j to the neuron at time t, wjk(t) the weight that is assigned to input value by neuron k and b0 is the bias of k neuron at time t. In Eq. (11), Yk(t) is the output of neuron k and φ is the activation function (ALAllaf 2012). The FFBPNN has two special versions of network which are;
Fitting network
Fitting network (FitNet) is a type of FFBPNN. It is used to fit an input–output relationship (ALAllaf 2012). Levenberg–Marquardt algorithm is the default algorithm used for the training of the system. The algorithm divides the feature vector randomly into three sets: (i) the training data (ii) the validation set data and (iii) the test data. A fitting network with one hidden layer and enough number of neurons can fit any finite number of input and output relationship.
Pattern recognition network
The pattern recognition neural network (PatternNet) is also a type of FFBPNN. It is used for solving pattern recognition problems like DNA. It is trained in such a way that it takes feature vector and classifies it according to the target vector. The training of PatternNet is performed using Scaled Conjugate Gradient algorithm. At each training cycle, the sequences are presented to the network. The PatternNet divides the data into three groups; (i) the training set (ii) the validation set and (iii) the test set. The process of training the system through FitNet and PatternNet is same as discussed above but the main difference between FitNet and PatternNet is that both networks use different algorithms for training the system. FitNet uses Levenberg–Marquardt algorithm whereas PatternNet uses Scaled Conjugate Gradient algorithm for training.
Ensemble classification
Ensemble classification has got a reasonable attention in the last decades. It has been successfully used to enhance the prediction power and widely applied for predicting protein subcellular location (Chou and Shen 2007a), predicting signal peptide (Chou and Shen 2007c), predicting subcellular location (Chou and Shen 2007b) and enzyme subfamily prediction (Chou 2005). The performance of ensemble classification approach is relatively better reported than the individual classifiers. The individual classifiers are diverse and can make different errors during the classification process but when these individual classifiers are combined, the errors can be reduced because the classification error of one algorithm is compensated by another algorithm (Hayat and Khan 2012a). The working of ensemble classification has been designed in such a way that it combines the results of different classification techniques and reduces the variance caused by anomaly in these single classification techniques. In this paper, seven different classification techniques have been used, which are GRNN, PNN, KNN, SVM, RF, PatternNet and FitNet. First, the individual classifiers are trained and tested. The individual predictions of each classification algorithm were then combined to form ensemble classifier. It can be represented as follows:
where EnsC shows the ensemble classifier and the symbol ⊕ shows the combination operator. The working of ensemble classifier EnsC by fusing the seven individual classifiers can be explained as: suppose that the predicted results of individual classifier for classification of DNA recombination hotspots and coldspots are:
where {C1, C2, C3, C4, C5, C6, C7}are the individual classifiers and {D1, D2} are the two classes of DNA recombination hotspots and coldspots (Hayat et al. 2012).
where
The output of the ensemble classifier using GA is obtained as:
where GAEnsC is the classification output of ensemble technique GA, Max shows the maximum result and the optimum weight of the individual classifier is w 1, w 2, w 3, … w 7. Majority voting-based ensemble classifier is a simple approach. In this approach, each classifier assigns equal weight, which represents all classifiers equally. However, the predictions of all classifiers are not in favor of all types of classes. Some classifiers are good for one class while others are good for other class. In such situation, the success rate of majority voting-based ensemble is not considerable. On the other hand, GA-based ensemble technique has the ability to automatically determine the appropriate weight for each classifier. It effectively finds the proper weights of all the eligible classes depending upon the prediction confidence. Initially, random weight assigns to each classifier. Further, the weight of the classifier is optimized on the basis of prediction confidence. Finally, those classifier outcomes whose confidence level is high are given more importance.
Frame work of proposed model
Looking at the importance of recombination spots, iRSpot-GAEnsC model is proposed for identification of hotspots and coldspots. Two powerful feature extraction methods called dinucleotide composition (DNC) and trinucleotide composition (TNC) were used to extract features from the dataset S. The extracted features were passed to seven different classification algorithms namely; GRNN, KNN, PNN, SVM, RF, PatternNet and FitNet. The best results of the individual classifiers were noted. The predicted results of the individual classifiers were combined to form ensemble model. Simple majority voting and optimization approach genetic algorithm were used to form ensemble model. The proposed model was trained on 64 features of TNC. Our model iRSpot-GAEnsC produced higher performance compared to the existing methods in the literature so far. The proposed model of ensemble classifier is shown in Fig. 2.
Performance measures
Several performance measures are applied in classification. These performance measures are used to measure the performance of the machine learning algorithms. Confusion matrix is used to record both the correct values and incorrect values for each class. There are different performance measures as given below.
-
I.
Accuracy
-
II.
Sensitivity
-
III.
Specificity
-
IV.
Mathews correlation coefficient (MCC)
where TP is True Positive, TN is False Negative, TN is True Negative and FP is False Positive.
-
V.
F-measure
The weighted average of precision and recall is known as F-measure. It is used for the evaluation of statistical methods. F-measure can be calculated as;
F-measure depends on two things; precision p and recall r, where
The resultant best value for F-measure is 1 and the worst value is 0.
-
VI.
G-mean
G-mean can be defined by two parameters called sensitivity (Sen) and specificity (Spe). G-mean is calculated by:
Sensitivity shows the performance of the positive class whereas specificity shows the performance of the negative class. G-mean incorporates the balanced performance of the learning algorithms between positive and negative class.
-
VII.
Q-statistics
Q-statistics is used to measure the diversity between two classification algorithms. The Q-statistics of any two classifiers C m and C n can be measured using the following formula:
where c is the correct prediction and w the wrong prediction of both classifiers. Likewise, a is the correct prediction of first classifier and b the incorrect prediction of second classifier and b the correct prediction of second classifier and incorrect prediction of the first classifier.
Although the four metrics (Eqs. 18, 19, 20, 21) were often used in literature to measure the prediction quality of a prediction method, they are no longer the best ones because of lacking intuitiveness and not easy to understand for most biologists, particularly the MCC (the Matthews correlation coefficient). To avoid this problem, we have adopted the following formulation proposed in the recent publication (Chou 2001b; Chou et al. 2011; Xu et al. 2013a; Qiu et al. 2014a, c).
The above-mentioned metrics given in Eqs. (27, 28, 29, 30) are valid only for single-label system. For multi-label systems whose existence has become more frequent in system biology (Chou et al. 2011) and system medicine (Xiao et al. 2013b), a completely different set of metrics as defined in (Chou 2013) is needed.
Results
Statistical methods are used to evaluate the predication performance of the classifiers. Mostly, three cross-validation tests that include independent dataset test, sub-sampling test and jackknife test are used for examining the performance of classifiers. However, among these tests, jackknife test is extensively applied because it always produces a unique result for a given dataset (Qiu et al. 2014a; Hayat and Tahir 2015). Therefore, the jackknife test has been increasingly and widely adopted by investigators to test the power of various predictors (Ding et al. 2009, 2012, 2014; Hayat and Khan 2012b; Zhang et al. 2012; Lin et al. 2013; Yuan et al. 2013; Lu et al. 2014). Hence, the jackknife cross-validation was utilized to examine the power of our method. Performance comparison of two feature spaces is discussed below.
Prediction performance of classifiers using DNC
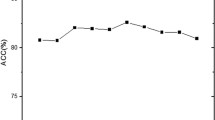
The success rates of individual and ensemble classifiers using DNC feature space are listed in Table 1. Among individual classifiers, RF achieved the highest accuracy among the classification algorithms. SVM and PatternNet obtained similar results. Likewise, KNN and PNN also yielded relatively similar accuracies. GRNN has achieved worse results compared to other classification algorithms. After that the individual classifier prediction are combined through majority voting and optimization technique GA. The outcome of majority voting-based ensemble was not reasonable. On the other hand, GA-based ensemble model obtained good results compared to individual and ensemble classifiers. Besides, accuracy, sensitivity, specificity and MCC, other performance measures such as F-measure, G-mean and Q-statistics are used to show more strength of proposed model. Q-statistics will show the diversity between individual classifiers. The accuracy of iRSpot-GAEnsC is shown Fig. 3.
Prediction performance of classifiers using TNC
The success rates of individual and ensemble classifiers using TNC feature space are reported in Table 2. Among individual classifiers, RF achieved the highest accuracy. SVM and PatternNet obtained somewhat comparable results. Likewise, KNN and PNN also yield relatively similar accuracies. GRNN has achieved the worse results compared to other classification algorithms. Further, the predicted outcomes of individual classifiers are combined through majority voting and optimization technique GA. The outcome of majority voting-based ensemble was not considerable. On the other hand, GA-based ensemble model obtained good results compared to individual and ensemble classifiers. The outcome of GA-based ensemble model is shown in Fig. 4.
Comparison of iRSpot-GAEnsC with existing methods
Comparison has been drawn between proposed model and already existing methods in the literature reported in Table 3. The pioneer work on this dataset has been carried out by Wei et al. (2013) (Chen et al. 2013) by introducing iRSpot-PseDNC predictor for identification of recombination hotspots and coldspots. Recently, Qiu et al. (2014) has developed iRSpot-TNCPseAAC model for the identification of recombination hotspots and coldspots (Qiu et al. 2014a). In contrast, our proposed model iRSpot-GAEnsC has achieved quite promising results compared to existing methods. The empirical results demonstrated that the performance of GA-based ensemble model is quite promising. This achievement has been ascribed with high variant features of TNC and optimization-based ensemble classification.
Discussion
In this study, a high-throughput computational model has been developed for identification of DNA recombination hotspots and coldspots. Two feature extraction methods including dinucleotide composition and trinucleotide composition were used to extract high discriminant features from DNA sequences. The performances of both feature spaces were evaluated using seven classification algorithms of different nature. These include GRNN, KNN, PNN, SVM, RF, PatternNet and FitNet. After examining the performance of individual classifiers, the predicted outcomes of individual classifiers are combined through simple majority voting and optimization approach genetic algorithm. Genetic algorithm-based ensemble model achieved quite promising results, which are higher than the performance of individual classifiers, and ensemble by majority voting. In addition, its performance is also higher than already existing methods reported in the literature so far. This remarkable achievement has been ascribed with high discriminated features of TNC and the ensemble strength of optimization method of GA. It is ascertained that our proposed model might be helpful in drug-related applications. As demonstrated in a series of recent publications (Xiao et al. 2013a; Ding et al. 2014; Qiu et al. 2014b; Xu et al. 2014b; Jia et al. 2015; Liu et al. 2015e, f) in developing new prediction methods, user-friendly and publicly accessible web servers enhance their impact (Chou 2015), we will make efforts in our future work to provide a web server for the prediction method of recombination hotspots and coldspots.
References
Ahmad S, Kabir M, Hayat M (2015) Identification of heat shock protein families and J-protein types by incorporating dipeptide composition into Chou’s general pseAAC. Comput Methods Programs Biomed. doi:10.1016/j.cmpb.2015.07.005
Akbar S, Ahmad A, Hayat M (2014) Identification of fingerprint using discrete wavelet transform in conjunction with support vector machine. IJCSI 11(Print):1694–0814
ALAllaf ONA (2012) Cascade-forward vs. function fitting neural network for improving image quality and learning time in image compression system. In: Proceedings of the world congress on engineering, pp 4–6
Beigi MM, Behjati M, Mohabatkar H (2011) Prediction of metalloproteinase family based on the concept of Chou’s pseudo amino acid composition using a machine learning approach. J Struct Funct Genomics 12:191–197
Boulesteix A, Bender A, Bermejo JL, Strobl C (2012) Random forest Gini importance favours SNPs with large minor allele frequency: impact, sources and recommendations. Brief Bioinform 13:292–304
Breiman L (2001) Random forests. Machine Learning 45:5–32
Chang CC, Lin CJ (2011) LIBSVM: a library for support vector machines. TIST 2:27
Chen W, Lin H, Feng PM, Ding C, Zuo YC, Chou KC (2012) iNuc-PhysChem: a sequence-based predictor for identifying nucleosomes via physicochemical properties. PLoS One 7:e47843
Chen W, Feng PM, Lin H, Chou KC (2013) iRSpot-PseDNC: identify recombination spots with pseudo dinucleotide composition. Nucleic Acids Res:gks1450
Chen W, Feng PM, Deng EZ, Lin H, Chou KC (2014a) iTIS-PseTNC: a sequence-based predictor for identifying translation initiation site in human genes using pseudo trinucleotide composition. Anal Biochem 462:76–83
Chen W, Feng PM, Lin H, Chou KC (2014b) iSS-PseDNC: identifying Splicing Sites Using Pseudo Dinucleotide Composition. BioMed Res Int 2014:12
Chen W, Lei TY, Jin DC, Lin H, Chou KC (2014c) PseKNC: a flexible web server for generating pseudo K-tuple nucleotide composition. Anal Biochem 456:53–60
Chen W, Zhang X, Brooker J, Lin H, Zhang L, Chou K (2014d) PseKNC-General: a cross-platform package for generating various modes of pseudo nucleotide compositions. Bioinformatics:btu602
Chen W, Lin H, Chou KC (2015) Pseudo nucleotide composition or PseKNC: an effective formulation for analyzing genomic sequences. Mol BioSyst
Cherian M, Sathiyan SP (2012) Neural Network based ACC for Optimized safety and comfort. Int J Comp Appl 42
Chou KC (2001a) Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins: structure. Function, and Bioinformatics 43:246–255
Chou KC (2001b) Using subsite coupling to predict signal peptides. Protein Eng 14:75–79
Chou KC (2005) Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 21:10–19
Chou KC (2009) Pseudo amino acid composition and its applications in bioinformatics, proteomics and system biology. Curr Proteomics 6:262–274
Chou K-C (2011) Some remarks on protein attribute prediction and pseudo amino acid composition. J Theor Biol 273:236–247
Chou KC (2013) Some remarks on predicting multi-label attributes in molecular biosystems. Mol BioSyst 9:1092–1100
Chou KC (2015) Impacts of bioinformatics to medicinal chemistry. Med Chem 11:218–234
Chou KC, Shen HB (2007a) Euk-mPLoc: a fusion classifier for large-scale eukaryotic protein subcellular location prediction by incorporating multiple sites. J Proteome Res 6:1728–1734
Chou KC, Shen HB (2007b) Recent progress in protein subcellular location prediction. Anal Biochem 370:1–16
Chou KC, Shen HB (2007c) Signal-CF: a subsite-coupled and window-fusing approach for predicting signal peptides. Biochem Biophys Res Commun 357:633–640
Chou KC, Wu ZC, Xiao X (2011) iLoc-Euk: a multi-label classifier for predicting the subcellular localization of singleplex and multiplex eukaryotic proteins. PLoS One 6:e18258
Chou KC, Wu ZC, Xiao X (2012) iLoc-Hum: using the accumulation-label scale to predict subcellular locations of human proteins with both single and multiple sites. Mol BioSyst 8:629–641
Dehzangi A, Heffernan R, Sharma A, Lyons J, Paliwal K, Sattar A (2015) Gram-positive and Gram-negative protein subcellular localization by incorporating evolutionary-based descriptors into Chou’s general PseAAC. J Theor Biol 364:284–294
Ding H, Luo L, Lin H (2009) Prediction of cell wall lytic enzymes using Chou’s amphiphilic pseudo amino acid composition. Protein Pept Lett 16:351–355
Ding C, Yuan LF, Guo SH, Lin H, Chen W (2012) Identification of mycobacterial membrane proteins and their types using over-represented tripeptide compositions. J Proteomics 77:321–328
Ding H, Deng EZ, Yuan LF, Liu L, Lin H, Chen W, Chou KC (2014) iCTX-Type: a sequence-based predictor for identifying the types of conotoxins in targeting ion channels. BioMed research international 2014
Duda RO, Hart PE, Stork DG (2012) Pattern classification. Wiley
Ebina T, Toh H, Kuroda Y (2011) DROP: an SVM domain linker predictor trained with optimal features selected by random forest. Bioinformatics 27:487–494
Esmaeili M, Mohabatkar H, Mohsenzadeh S (2010) Using the concept of Chou’s pseudo amino acid composition for risk type prediction of human papillomaviruses. J Theor Biol 263:203–209
Fang Y, Guo Y, Feng Y, Li M (2008) Predicting DNA-binding proteins: approached from Chou’s pseudo amino acid composition and other specific sequence features. Amino Acids 34:103–109
Feng PM, Chen W, Lin H, Chou KC (2013) iHSP-PseRAAAC: identifying the heat shock protein families using pseudo reduced amino acid alphabet composition. Anal Biochem 442:118–125
Georgiou V, Pavlidis N, Parsopoulos K, Alevizos PD, Vrahatis M (2004) Optimizing the performance of probabilistic neural networks in a bioinformatics task. In: Proceedings of the EUNITE 2004 Conference, pp 34–40
Georgiou D, Karakasidis TE, Nieto J, Torres A (2009) Use of fuzzy clustering technique and matrices to classify amino acids and its impact to Chou’s pseudo amino acid composition. J Theor Biol 257:17–26
Gu Q, Ding YS, Zhang TL (2010) Prediction of G-protein-coupled receptor classes in low homology using Chou’s pseudo amino acid composition with approximate entropy and hydrophobicity patterns. Protein Pept Lett 17:559–567
Guo J, Rao N, Liu G, Yang Y, Wang G (2011) Predicting protein folding rates using the concept of Chou’s pseudo amino acid composition. J Comput Chem 32:1612–1617
Guo SH, Deng EZ, Xu LQ, Ding H, Lin H, Chen W, Chou KC (2014) iNuc-PseKNC: a sequence-based predictor for predicting nucleosome positioning in genomes with pseudo k-tuple nucleotide composition. Bioinformatics:btu083
Han J, Kamber M (2006) Data Mining, Southeast, Asia edn. Concepts and Techniques, Morgan kaufmann
Hayat M, Khan A (2011) Predicting membrane protein types by fusing composite protein sequence features into pseudo amino acid composition. J Theor Biol 271:10–17
Hayat M, Khan A (2012a) Discriminating outer membrane proteins with fuzzy K-nearest neighbor algorithms based on the general form of Chou’s PseAAC. Protein Pept Lett 19:411–421
Hayat M, Khan A (2012b) Mem-PHybrid: hybrid features-based prediction system for classifying membrane protein types. Anal Biochem 424:35–44
Hayat M, Tahir M (2015) PSOFuzzySVM-TMH: identification of transmembrane helix segments using ensemble feature space by incorporated fuzzy support vector machine. Mol BioSyst
Hayat M, Khan A, Yeasin M (2012) Prediction of membrane proteins using split amino acid and ensemble classification. Amino Acids 42:2447–2460
He X, Han K, Hu J, Yan H, Yang JY, Shen HB, Yu DJ (2015) TargetFreeze: Identifying Antifreeze Proteins via a Combination of Weights using Sequence Evolutionary Information and Pseudo Amino Acid Composition. J Membrane Biol:1–10
Jia J, Liu Z, Xiao X, Liu B, Chou KC (2015) iPPI-Esml: an ensemble classifier for identifying the interactions of proteins by incorporating their physicochemical properties and wavelet transforms into PseAAC. J Theor Biol 377:47–56
Jiang P, Wu H, Wang W, Ma W, Sun X, Lu Z (2007) MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res 35:W339–W344
Keeney S (2008) Spo11 and the formation of DNA double-strand breaks in meiosis. In: Recombination and meiosis. Springer, pp 81–123
Khan A (2012) Identifying GPCRs and their types with Chou’s pseudo amino acid composition: an approach from multi-scale energy representation and position specific scoring matrix. Protein Pept Lett 19:890–903
Khan A, Majid A, Hayat M (2011) CE-PLoc: an ensemble classifier for predicting protein subcellular locations by fusing different modes of pseudo amino acid composition. Comput Biol Chem 35:218–229
Khan ZU, Hayat M, Khan MA (2015) Discrimination of acidic and alkaline enzyme using Chou’s pseudo amino acid composition in conjunction with probabilistic neural network model. J Theor Biol 365:197–203
Kumar KK, Pugalenthi G, Suganthan P (2009) DNA-Prot: identification of DNA binding proteins from protein sequence information using random forest. J Biomol Struct Dyn 26:679–686
Li WC, Deng EZ, Ding H, Chen W, Lin H (2015) iORI-PseKNC: a predictor for identifying origin of replication with pseudo k-tuple nucleotide composition. Chemometr Intell Lab Syst 141:100–106
Lin H, Ding H, Guo FB, Zhang AY, Huang J (2008) Predicting subcellular localization of mycobacterial proteins by using Chou’s pseudo amino acid composition. Protein Pept Lett 15:739–744
Lin H, Wang H, Ding H, Chen YL, Li QZ (2009a) Prediction of subcellular localization of apoptosis protein using Chou’s pseudo amino acid composition. Acta Biotheor 57:321–330
Lin WZ, Xiao X, Chou KC (2009b) GPCR-GIA: a web-server for identifying G-protein coupled receptors and their families with grey incidence analysis. Protein Engineering Design and Selection:gzp057
Lin WZ, Fang JA, Xiao X, Chou KC (2012) Predicting secretory proteins of malaria parasite by incorporating sequence evolution information into pseudo amino acid composition via grey system model
Lin H, Chen W, Yuan LF, Li ZQ, Ding H (2013) Using over-represented tetrapeptides to predict protein submitochondria locations. Acta Biotheor 61:259–268
Liu G, Liu J, Cui X, Cai L (2012) Sequence-dependent prediction of recombination hotspots in Saccharomyces cerevisiae. J Theor Biol 293:49–54
Liu B, Xu J, Lan X, Xu R, Zhou J, Wang X, Chou KC (2014) iDNA-Prot| dis: Identifying DNA-binding proteins by incorporating amino acid distance-pairs and reduced alphabet profile into the general pseudo amino acid composition
Liu B, Fang L, Liu F, Wang X, Chen J, Chou KC (2015a) Identification of real microRNA precursors with a pseudo structure status composition approach. PLoS One 10:e0121501
Liu B, Liu F, Fang L, Wang X, Chou KC (2015b) repDNA: a Python package to generate various modes of feature vectors for DNA sequences by incorporating user-defined physicochemical properties and sequence-order effects. Bioinformatics 31:1307–1309
Liu B, Fang L, Liu F, Wang X, Chou KC (2015b) iMiRNA-PseDPC: microRNA precursor identification with a pseudo distance-pair composition approach. J Biomol Struct and Dynamics:1–13
Liu Z, Xiao X, Qiu WR, Chou KC (2015d) iDNA-Methyl: identifying DNA methylation sites via pseudo trinucleotide composition. Anal Biochem 474:69–77
Liu B, Liu F, Fang L, Wang X, Chou KC (2015d) repRNA: a web server for generating various feature vectors of RNA sequences. Mole Genet Genomics:1–9
Liu B, Liu F, Wang X, Chen J, Fang L, Chou KC (2015e) Pse-in-One: a web server for generating various modes of pseudo components of DNA, RNA, and protein sequences. Nucleic Acids Research:gkv458
Lou W, Wang X, Chen F, Chen Y, Jiang B, Zhang H (2014) Sequence based prediction of DNA-binding proteins based on hybrid feature selection using random forest and Gaussian Naïve Bayes. PLoS One 9:e86703
Lu J, Huang G, Li HP, Feng KY, Chen L, Zheng MY, Cai YD (2014) Prediction of cancer drugs by chemical–chemical interactions. PLoS One 9
Mandal M, Mukhopadhyay A, Maulik U (2015) Prediction of protein subcellular localization by incorporating multiobjective PSO-based feature subset selection into the general form of Chou’s PseAAC. Med Biol Eng Comput 53:331–344
Mohabatkar H (2010) Prediction of cyclin proteins using Chou’s pseudo amino acid composition. Protein Pept Lett 17:1207–1214
Mohabatkar H, Mohammad Beigi M, Esmaeili A (2011) Prediction of GABA receptor proteins using the concept of Chou’s pseudo-amino acid composition and support vector machine. J Theor Biol 281:18–23
Mohabatkar H, Mohammad Beigi M, Abdolahi K, Mohsenzadeh S (2013) Prediction of allergenic proteins by means of the concept of Chou’s pseudo amino acid composition and a machine learning approach. Med Chem 9:133–137
Nanni L, Lumini A, Gupta D, Garg A (2012) Identifying bacterial virulent proteins by fusing a set of classifiers based on variants of Chou’s pseudo amino acid composition and on evolutionary information. IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB) 9:467–475
Qiu JD, Huang JH, Liang RP, Lu XQ (2009) Prediction of G-protein-coupled receptor classes based on the concept of Chou’s pseudo amino acid composition: an approach from discrete wavelet transform. Anal Biochem 390:68–73
Qiu WR, Xiao X, Chou KC (2014a) iRSpot-TNCPseAAC: identify recombination spots with trinucleotide composition and pseudo amino acid components. Int J Mol Sci 15:1746–1766
Qiu WR, Xiao X, Lin WZ, Chou KC (2014b) iMethyl-PseAAC: Identification of protein methylation sites via a pseudo amino acid composition approach. BioMed Res Int 2014
Qiu WR, Xiao X, Lin WZ, Chou KC (2014c) iUbiq-Lys: prediction of lysine ubiquitination sites in proteins by extracting sequence evolution information via a gray system model. J Biomol Struct Dynamics:1–12
Sahu SS, Panda G (2010) A novel feature representation method based on Chou’s pseudo amino acid composition for protein structural class prediction. Comput Biol Chem 34:320–327
Specht DF (1990) Probabilistic neural networks. Neural networks 3:109–118
Touw WG, Bayjanov JR, Overmars L, Backus L, Boekhorst J, Wels M, van Hijum SA (2013) Data mining in the Life Sciences with Random Forest: a walk in the park or lost in the jungle? Brief Bioinform 14:315
Vapnik V (2000) The nature of statistical learning theory. Springer
Xiao X, Wang P, Chou KC (2009) GPCR-CA: a cellular automaton image approach for predicting G-protein-coupled receptor functional classes. J Comput Chem 30:1414–1423
Xiao X, Wang P, Chou KC (2011) Quat-2L: a web-server for predicting protein quaternary structural attributes. Mol Diversity 15:149–155
Xiao X, Min JL, Wang P, Chou KC (2013a) iGPCR-Drug: a web server for predicting interaction between GPCRs and drugs in cellular networking. PLoS One 8:e72234
Xiao X, Wang P, Lin WZ, Jia JH, Chou KC (2013b) iAMP-2L: a two-level multi-label classifier for identifying antimicrobial peptides and their functional types. Anal Biochem 436:168–177
Xiao X, Hui MJ, Liu Z, Qiu WR (2015) iCataly-PseAAC: Identification of enzymes catalytic sites using sequence evolution information with grey model GM (2, 1). The J Memb Biol:1–9
Xu Y, Ding J, Wu LY, Chou KC (2013a) iSNO-PseAAC: predict cysteine S-nitrosylation sites in proteins by incorporating position specific amino acid propensity into pseudo amino acid composition. PLoS ONE 8:e55844
Xu Y, Shao XJ, Wu LY, Deng NY, Chou KC (2013b) iSNO-AAPair: incorporating amino acid pairwise coupling into PseAAC for predicting cysteine S-nitrosylation sites in proteins. PeerJ 1:e171
Xu R, Zhou J, Liu B, He Y, Zou Q, Wang X, Chou KC (2014a) Identification of DNA-binding proteins by incorporating evolutionary information into pseudo amino acid composition via the top-n-gram approach. Journal of Biomolecular Structure and Dynamics:1–11
Xu Y, Wen X, Wen LS, Wu LY, Deng NY, Chou KC (2014b) iNitro-Tyr: Prediction of nitrotyrosine sites in proteins with general pseudo amino acid composition
Yuan LF, Ding C, Guo SH, Ding H, Chen W, Lin H (2013) Prediction of the types of ion channel-targeted conotoxins based on radial basis function network. Toxicol In Vitro 27:852–856
Zhang YN, Yu DJ, Li SS, Fan YX, Huang Y, Shen HB (2012) Predicting protein-ATP binding sites from primary sequence through fusing bi-profile sampling of multi-view features. BMC Bioinform 13:118
Zhou XB, Chen C, Li ZC, Zou XY (2007) Using Chou’s amphiphilic pseudo-amino acid composition and support vector machine for prediction of enzyme subfamily classes. J Theor Biol 248:546–551
Zou D, He Z, He J, Xia Y (2011) Supersecondary structure prediction using Chou’s pseudo amino acid composition. J Comput Chem 32:271–278
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors. References
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kabir, M., Hayat, M. iRSpot-GAEnsC: identifing recombination spots via ensemble classifier and extending the concept of Chou’s PseAAC to formulate DNA samples. Mol Genet Genomics 291, 285–296 (2016). https://doi.org/10.1007/s00438-015-1108-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-015-1108-5