Abstract
Knowledge of the types of membrane protein provides useful clues in deducing the functions of uncharacterized membrane proteins. An automatic method for efficiently identifying uncharacterized proteins is thus highly desirable. In this work, we have developed a novel method for predicting membrane protein types by exploiting the discrimination capability of the difference in amino acid composition at the N and C terminus through split amino acid composition (SAAC). We also show that the ensemble classification can better exploit this discriminating capability of SAAC. In this study, membrane protein types are classified using three feature extraction and several classification strategies. An ensemble classifier Mem-EnsSAAC is then developed using the best feature extraction strategy. Pseudo amino acid (PseAA) composition, discrete wavelet analysis (DWT), SAAC, and a hybrid model are employed for feature extraction. The nearest neighbor, probabilistic neural network, support vector machine, random forest, and Adaboost are used as individual classifiers. The predicted results of the individual learners are combined using genetic algorithm to form an ensemble classifier, Mem-EnsSAAC yielding an accuracy of 92.4 and 92.2% for the Jackknife and independent dataset test, respectively. Performance measures such as MCC, sensitivity, specificity, F-measure, and Q-statistics show that SAAC-based prediction yields significantly higher performance compared to PseAA- and DWT-based systems, and is also the best reported so far. The proposed Mem-EnsSAAC is able to predict the membrane protein types with high accuracy and consequently, can be very helpful in drug discovery. It can be accessed at http://111.68.99.218/membrane.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cell membrane proteins are important components of a cell. They carry out many of the functions that are imperative to the cell’s survival. Membrane proteins are classified into transmembrane proteins, which span across the cell membrane and anchored membrane proteins that are attached only with one side of the cell membrane. Membrane proteins are further classified into six types (Chou and Cai 2005a): Type-I transmembrane, Type-II transmembrane, multipass transmembrane protein, lipid chain-anchored membrane, GPI-anchored membrane, and peripheral membrane protein. Prediction of membrane protein types is supportive in drug discovery, disease diagnosis and so on. Generally, in case of protein classification, the first phase is to convert each protein sequence into a feature-based representation. In the second phase, the feature-based representation is provided to a classification model that yields the predicted protein types. The crystallization of membrane proteins is a difficult task and most of them will not dissolve in normal solvents. Therefore, very few membrane protein structures have been determined up till now. Mostly, NMR is employed for determining the three-dimensional structures of membrane proteins (Call et al. 2010; Pielak and Chou 2010), but unfortunately, it is both, time-consuming and costly. The number of templates for membrane proteins is also very limited. Therefore, it is highly desirable to develop a computational method, which can predict the features of membrane proteins based on their primary sequences alone. In this regards, ensemble classification is rapidly emerging due to its superiority over the individual classifiers to enhance the prediction performance of a learning system (Zhang and Zhang 2008).
Recently, several interesting feature extraction strategies based on amino acid sequences and ensemble classifications have been reported (Shen and Chou 2007; Nanni and Lumini 2008a). A number of efforts have been carried out to predict membrane protein types according to their sequence information. Chou and Elrod (1999) have used the covariant discriminant algorithm (CDA) to identify membrane protein types based on their amino acid (AA) composition. Using AAC some sequence information is lost. To avoid losing many important information hidden in protein sequences, the pseudo amino acid composition (PseAAC) has been proposed (Chou 2001; Chou and Cai 2005a) to replace the simple amino acid composition (AAC) for representing the sample of a protein review (Chou and Shen 2009a). Chou (2001) has also proposed the use of the CDA in conjunction with the PseAAC-based feature extraction. Cai et al. (2004) have used AAC and SVM for prediction of membrane protein types. Wang et al. (2004) have utilized weighted SVM and PseAAC, while Liu et al. (2005) have employed the Fourier spectrum and SVM. Chou and Cai (2005a) have proposed amphipathic PseAAC and CDA, the discrete wavelets transform (DWT) and cascaded neural network (Rezaei et al. 2008). Similarly, DWT and SVM Qiu et al. (2010) have also been employed for the prediction of membrane protein types. Other typical examples are (Chou and Cai 2005a; Chou and Shen 2007a; Nanni et al. 2010). Quite a few important features of proteins are hidden in their complicated sequences. Consequently, sequence analysis is an interesting approach for predicting protein attributes such as structural class (Chou 1995) and subcellular locations (Chou and Shen 2007b, c; 2010b). The present study is an attempt for proposing a new method for predicting membrane protein types based on the sequence information to provide a useful tool for relevant areas.
In this study, we present Mem-EnsSAAC, a novel method for predicting membrane protein types. It is based on split amino acid composition (SAAC) based feature extraction and majority voting based ensemble classification. Our aim is to demonstrate that membrane protein types can be effectively predicted by exploiting the discrimination power of difference in amino acids at the N and C terminus. Three feature extraction strategies, namely, PseAA, DWT, SAAC and a hybrid version of these methods are analyzed for membrane-protein type prediction. K-nearest neighbor (KNN), probabilistic neural network (PNN), support vector machine (SVM), random forest (RF), and Adaboost are used as base learners for classification. The results of these feature extraction strategies are compared using the same classifiers. Our main goal is to first find a reasonable feature extraction strategy for membrane protein prediction. Second, we take an advantage of the discriminative power of the best feature extraction strategy and the classification capabilities of PNN, KNN, SVM, RF, and Adaboost for developing an effective and high throughput ensemble system for membrane protein prediction. We have compared Mem-EnsSAAC against other high-tech membrane protein predictors using an extensively used standard dataset (Chou and Cai 2005b). The results confirm that Mem-EnsSAAC outperforms the existing predictors. Especially, Mem-EnsSAAC yields high prediction performance for the membrane protein types for which the existing predictors yield average performance.
The next section describes materials and methods, followed by performance measures and results and discussion. Finally, the last section concludes the paper.
Materials and methods
Dataset
The two datasets; Dataset1 and Dataset2, used in this paper are the same as used in (Chou and Cai 2005a), and (Chou and Shen 2007a). Dataset1 has been developed from the SWISS-PROT data bank. Dataset1 represents six types of membrane protein. The dataset1 is passed from various processes. First, only those sequences are included in the dataset whose descriptions are clear. Secondly, only one protein sequence is included from those that are having the same name, but are from different species. Finally, sequences whose type is described by two or more types are not included because of lack of uniqueness. After the above screening procedures, the obtained dataset contains only 2,628 protein sequences in the training dataset and 3,160 sequences in the testing dataset.
In Dataset1, the training set contains 2,628 proteins, of which type-I transmembrane proteins are 372, type-II transmembrane proteins are 151, multipass transmembrane proteins are 1,903, lipid chain-anchored membrane proteins are 104, GPI-anchored membrane proteins are 68, and peripheral membrane proteins are 30. The analysis of the sequence identity, for each membrane protein types is conducted. Assume one sequence is N1 residues long and the other is N2 residues long (N1 > N2), and the maximum number of residues matched by sliding one sequence along the other is M. The sequence identity percentage between the two sequences is computed as (M/N1) × 100%. The average sequence identity percentages for type-I transmembrane proteins, type-II transmembrane proteins, multipass transmembrane proteins, lipid chain-anchored membrane proteins, GPI-anchored membrane proteins, and peripheral membrane proteins are 7.97, 7.94, 8.31, 7.94, 7.92, and 11.36%, respectively. These numbers have indicated that the majority of pairs in each of these types have very low sequence identity.
On the other hand, the independent dataset comprises 3,160 proteins, of which 462 are of type-I transmembrane proteins, 144 of type-II transmembrane proteins, 2,402 of multipass transmembrane proteins, 67 of lipid chain-anchored membrane proteins, 83 of GPI-anchored membrane proteins, and 2 of peripheral membrane proteins. None of the protein sequences in the testing dataset occurs in the training dataset. The resultant average sequence identity percentages are 8.34, 9.53, 8.55, 10.22, 11.75, and 5.00%, respectively, indicating that the sequence identity for majority of pairs in each of the six types in the independent dataset is also very low. Dataset2 has been downloaded from http://www.csbio.sjtu.edu.cn/bioinf/. Eight types of membrane proteins are defined in this dataset (Chou and Shen 2007a). First, those sequences annotated with “fragment” were excluded. Second, those sequences annotated with ambiguity were removed. The original dataset contains 3,249 membrane protein sequences. The training dataset consists of 610 single-pass type-I, 312 single-pass type-II, 24 single-pass type-III, 44 single-pass type-IV, 1,316 multipass, 151 lipid chain-anchored, 182 GPI anchored, and 610 peripheral membrane protein sequences. Redundancy is removed using 30% CD-HIT and only those sequences are included in the dataset that have less than 30% sequence identity. Dataset2 also contains some sequences whose length is less than 50. Therefore, we have removed those sequences from the dataset whose length is less than 50 amino acids. Furthermore, 25% similarity cutoff has also been applied to remove homology between the sequences and have included only those sequences in the dataset that have less than 25% identity. Finally, the obtained dataset consists of 2,978 membrane protein sequences in which 576 are single-pass type-I, 269 are single-pass type-II, 17 are single-pass type-III, 34 are single-pass type-IV, 1,285 are multipass, 97 are lipid chain-anchored, 154 are GPI anchored, and 546 are peripheral membrane protein sequences.
Feature extraction strategies
Discrete wavelet analysis
To analyze the various components of a signal, wavelet analysis is a useful tool because it is able to localize variation both in space and scale domains.
Wavelet transform is defined as: “the signal f(t) is multiplied by a scaled and shifted version of the wavelet function Ψ(t) and then summed”. The transformed coefficients T(a, b) of the signal f(t) can be expressed as:
where a is a scale and b is a translation parameter. Both belong to the real numbers R(n), a > 0, t is the length of the sequence, and \( \psi \left( {\frac{t - b}{a}} \right) \) is the analyzing wavelet function. The transformed coefficients T(a, b) are found for both specific locations on the signal, t = b, and for specific wavelet periods (which are a function of a). It is used to plot T(a, b) against a and b in a surface plot known as a scalogram, which is particularly suited to the detection of singularities. DWT decomposes the amino acid sequences into coefficients at different dilations and then removes the noise component from the profiles, so it can provide us local information of the sequences. With these properties, DWT can more effectively reflect the sequence order effects. In addition, the DWT is an economical way to compute wavelet transform, because it is computed only on a dyadic grid of points, where the sub-sampling is at a different rate for different scales. In this work, the DWT uses a 0 = 2 and b 0 = 1, so that the results can lead to a binary dilation of 2−m and a dyadic translation of n2−m. Therefore,
here, m = 1, 2,… and n = 0, 1, 2,… The wavelet coefficients of the signal f(t) are obtained by following formula:
T(a, b) is divided into two parts, approximation coefficient A j(n), which is high scale and low frequency component and detail coefficient D j(n), which represent the low scale and high frequency components of the signal f(t). Approximation coefficient and detail coefficient for level j can be expressed as:
Additional detailed characteristics of the signal can be observed when the level j of the decomposition is increased. Since in the current work, we are dealing with protein sequences where variation in the AAC is studied with respect to position, the time variable t will be replaced by position variable. In this paper, first the protein sequence is converted into Hydrophobicity scale (Kyte and Doolittle 1982), because it appears to be a good consensus for the definition of the amino acids’ hydrophobic properties. The Kyte–Doolittle scale is usually used for identifying hydrophobic regions in proteins. Regions with a positive value are hydrophobic. This scale can be used for detecting both surface-exposed regions and transmembrane regions. It is then converted to a digital signal to generate several groups of wavelet coefficients. Digital wavelet signal is decomposed up to levels 4, to obtain the approximation component cA4 and detail components cD4, cD3, cD2, and cD1. Different characteristics are exploited to evaluate and to analyze each signal component, such as Shannon entropy, log entropy, energy entropy, variance, min, max and mean. So, total 35 features are extracted. These features are normalized using the Euclidean normalization which brings the values within a similar range.
Pseudo-amino acid composition
Amino acid composition of a protein is defined by 20 discrete values representing the normalize frequency of the 20 native amino acids in proteins. In AAC, proteins can thus be expressed in 20D vector (Chou and Zhang 1993, 1994; Chou 1995; Nakashima and Nishikawa 1986):
where p 1 , p 2 , p 3 , … , p 20 are the composition components of 20 amino acids P and T denote transposition. However, using the AAC for a protein representation will lose its sequence order and sequence-length. To compensate this problem, Chou (2001) and Nanni and Lumini (2008b) proposed to represent a protein sample by its pseudo-amino acid composition, which is defined in a (20 + λ)D space:
The first 20 components are the same as those in the basic AAC; where as p 20+1 p 20+λ are the correlation factors of an amino acid sequence in the protein chain determined on the bases of hydrophobicity and hydrophilicity (Chou 2001). In our study, λ = 21 means taking first 21 ranks of sequence-order correlations into consideration. Thus, a protein sample is represented by a (20 + λ)D = 62D vector.
Split amino acid composition
In SAAC-based method, the protein sequence is divided into different parts and composition of each part is calculated separately (Chou and Shen 2006a, b). In our SAAC model, we divide the membrane protein sequence into three parts; (i) 25 amino acids of N termini, (ii) 25 amino acids of C termini, and (iii) region between these two terminus. The resultant feature vector is a 60D instead of 20D as in case of AAC.
Ensemble classifier
Recently, ensemble classification has achieved reasonable attention due to their superiority over single classifier based systems. The advantage of the ensemble classification is that if individual classifiers are diverse, then they can make different errors, and when these classifiers are combined, the error can be reduced through averaging. The framework of ensemble classifier system has been developed by combining numerous basic learners together to reduce the variance caused by the peculiarities of a single training set and hence be able to learn a more expressive concept in classification than a single classifier (Shen and Chou 2007; Nanni and Lumini 2006, 2008a). In this paper, we have used five different learning mechanisms; SVM, PNN, KNN, Adaboost, and RF. SVM is a machine learning technique based on the statistical learning theory (Chou and Cai 2002; Cai et al. 2003; Khan et al. 2008a, b). KNN is a learning algorithm that is based on the concept of proximity in the feature space (Khan et al. 2008c). PNN is based on the Bayes theory. It estimates the likelihood of a sample being part of a learned category (Khan et al. 2010). The RF is a combination of tree predictors. Each tree depends on the values of a random vector sampled independently and with the same distribution for all trees in the forest (Breiman 2001). Adaboost on the other hand, tries to improve the prediction results by combining weak predictors’ together (Schapire et al. 1998). First, the individual classifiers are trained and their predictions are noted down. The ensemble classifier is then formed by fusing the predictions of the individual classifiers;
where the symbol ⊕ denotes the fusing operator and EnsC is the ensemble classifier as shown in Fig. 1.
The process of how the ensemble classifier EnsC works by fusing the five base classifiers is as follows:
Suppose the predicted result of individual classifiers for the protein query P are
where C 1 , C 2 , …, C 5 are individual classifiers and S 1 , S 2 , …, S 6 are membrane protein types.
Finally, the output of the ensemble classifier combined through majority voting using GA is obtained as:
where C EnsC is the predicted result of the ensemble classifier, the Max represents choosing the maximum one and w 1 , w 2 , …, w 5 is the optimal weight of classifiers.
Performance measures
In the field of machine learning, several performance measures are used to evaluate the performance of learning algorithms. Performances of the classifiers are measured from the confusion matrix, which records both the correctly and incorrectly recognized examples for each class. We have different performance measures as described below:
Accuracy.
where TP, FN, TN, and FP are the number of true positive, false negative, true negative and false positive protein sequences, respectively.
Sensitivity/specificity.
Mathews correlation coefficient (MCC). MCC is a discrete version of Pearson’s correlation coefficient that takes values in the interval of [−1, 1]. A value of 1 means the classifier never makes any mistakes and a value of −1 means the classifier always makes mistake.
F-measure. F-measure is used for evaluating statistical tests. It depends on of precision p and recall r. P is the number of correct predictions divided by the number of all returned predictions, while r is the number of correct predictions divided by the number of predictions. The F-measure can be considered as a weighted average of the precision and recall. The best value of F-measure is 1 and worst is 0.
In case of unbalanced datasets, the F-measure is better suited when compared with the accuracy, because the accuracy becomes biased towards an overrepresented class. Thus, if all the predicted instances belong to this class the accuracy will still be higher. The F-measure can be easily generalized for multilabel classification (Tsoumakas and Katakis 2007). Let S is a dataset with M instances. Let U and V are the set of correct labels where i ∈ S, respectively. Then the recall and precision for label k are defined as:
Q-Statistics. To measure the diversity in ensemble classifiers the average value of Q-statistics (Nanni and Lumini 2006) is used. The Q-statistic of any two base classifiers C i and C j are defined as:
where, a and d represent the number of correct and incorrect prediction of both classifiers. However, b is the correct prediction of classifier first and incorrect prediction of classifier second and c is the correct prediction of classifier second and incorrect of first. The value of Q varies between −1 and 1. For statistically independent classifiers, the value of Q i,j is zero. For ensemble classifier, the average value of Q-statistics among all pairs of the L base classifiers is calculated as:
Results and discussion
To show that Mem-EnsSAAC is well suited for predicting membrane protein types, we have compared it against the other state of the art membrane protein predictors; employing CDA (Chou and Elrod 1999), PseAA (Chou 2001), AA and SVM (Cai et al. 2004), Fourier spectrum (Liu et al. 2005), Amphipathic PseAA and CDA (Chou and Cai 2005a), DWT and SVM (Rezaei et al. 2008; Qiu et al. 2010). The predictors have been chosen because most of them are quite recent and are available as online or as a stand-alone version. Statistical tests are conducted to measure the prediction performance of the predictors. Three test methods are used to evaluate the quality of the proposed prediction model: self-consistency, jackknife (leave-one out) and the independent dataset test. In self-consistency test, the model is trained and tested with the same dataset. However, the self-consistency test is sometime considered as first basic test because any algorithm whose self-consistency performance is poor may not be conceived as a good one. In case of jackknifing, each membrane protein in the dataset is in turn taken out and all the rule parameters are calculated based on the remaining proteins. During the process of jackknifing, both the training and testing datasets are actually open and a protein will move from one to the other in turn. While in independent dataset test, the model is trained on one dataset and tested on another dataset test. Among the three tests, the jackknife test is considered the most objective one (Chou and Shen 2007a, 2010a), and has been increasingly used by investigators (Zhang and Zhang 2008; Zhou et al. 2007). Therefore, in this study, we have also used the jackknife test.
Prediction performance using Jackknife test
In Table 1, prediction results using three feature extraction strategies: DWT, PseAA, and SAAC are shown. Column 2 shows the accuracy of individual and ensemble classifiers. The prediction performance of all the classifiers using DWT-based feature extraction is not at far with that of the PseAA, and SAAC. RF has obtained the maximum accuracy of 80.7% using DWT, which is very low as compared with that obtained through PseAA, and SAAC. It is observed that the prediction performance of SVM, PNN, KNN, RF, Adaboost, and Ensemble classifiers using DWT is not comparable that obtained through PseAA and SAAC. This might be because in the DWT-based approach, with an increase of the decomposition level j, additional detailed characteristics of the signal can be observed. However, after a certain level of decomposition, there is no given information, rather feature redundancy is observed due to the short length of the protein signal. On the other hand, decomposing a longer sequence with too low a decomposition scale will heavily omit detailed information (Wens et al. 2005). Therefore, one needs to choose an appropriate decomposition level. In the case of PseAA, the prediction performance of ensemble classifier is 14.6% higher than that of the DWT. This is because in case of PseAA, when the value of tiers increases, the performance of the classifier also improves. Thus, PseAA better represents the sequence-order information. However, in case of short length sequences, increasing the tier value will have no effects on performance. In case of SAAC-based model, the accuracy of Mem-EnsSAAC is 17.3 and 2.7% higher than that of the DWT and PseAA, while 0.1% less than hybrid, respectively. In Table 2 shows the performance of classifiers using hybrid features. Thus, hybrid feature-extraction strategy performs the best among the feature extraction strategies that we have used. The result demonstrates that SAAC is effective and useful for the prediction of membrane protein types because it uses the composition of three different parts of protein independently. The fact is that the independent analysis composition of different parts of a protein provides more information than that of the composition of the whole sequence. The main advantage of SAAC over other methods is that it assigns large weight to proteins, which have a signal at either the N or C terminus. Other performance measures such as sensitivity, specificity, MCC, F-measure, and Q-Statistics of the individual and ensemble classifiers are shown in columns 4–8 of Table 1. Sensitivity, specificity, MCC, F-measure, and Q-Statistics of the Mem-EnsSAAC are 91.0, 92.2, 0.75, 0.79, and 0.91%, respectively. In case of individual classifiers, SVM in combination with SAAC yields the best performance of 90.8% accuracy.
Performance on Independent dataset test
In Table 1, column 8 shows the accuracy of individual and ensemble classifiers using DWT, PseAA, and SAAC. Ensemble classifier using PseAA yields an improvement in accuracy of 7.5% as compared to that of when used in conjunction with DWT. The performance of Mem-EnsSAAC is still 10.6% and 3.1% higher than that of the DWT- and PseAA-based prediction. Sensitivity, specificity, MCC, F-measure, and Q-Statistics of the Mem-EnsSAAC using independent dataset test are 88.4, 92.4, 0.74, 0.79, and 0.95%, respectively. RF using SAAC has obtained 90.1% accuracy, which is the highest among the individual classifiers.
Prediction performance using hybrid feature-extraction strategies
The performance of individual and ensemble classifier using hybrid features is shown in Table 2. In this study, we have developed two versions of hybrid models, hybrid1 and hybrid2. The first one is the combination of SAAC and PseAA and the second is the combination of SAAC, PseAA, and DWT-based features. In case of hybrid1 model, SVM and PNN have achieved the highest accuracy, which is 91.6% as compared to other classifiers using jackknife test. But the MCC of the SVM is higher than that of PNN. It has been observed that the performance of SVM is better using hybrid1 features as compared to all other individual classifiers. In contrast, the performance of ensemble classifier is better than that of all individual classifiers, which is 92.4%. In case of independent dataset test, SVM has obtained the highest result among all individual classifiers. But the performance of ensemble classifiers is 0.1% higher than that of SVM. On the other hand, using hybrid2 model, the performances of the classifiers are affected due to DWT-based features. Among the individual classifiers, SVM yields the highest accuracy of 90.8% using jackknife test compared not only to the classifiers trained on the hybrid features but also to those trained on DWT, PseAA, and SAAC features. In case of independent dataset test, SVM again provides the highest accuracy of 89.7% among the individual classifiers. The ensemble classifier in both cases, i.e., using jackknife and independent test, has obtained the highest accuracy of 92.3 and 91.3%, respectively. The prediction performance of ensemble classifier for jackknife test using hybrid2 features is modestly better than that of using DWT, PseAA, and SAAC individually. However, using hybrid1 features, the performance of Mem-EnsSAAC is slightly better, round about 0.2%. It means that DWT-based feature, when added, has slightly degraded the overall performance of classifiers. However, the computational cost has increased due to the high dimensionality, 157D for hybrid2 versus 122D for hybrid1.
Neighborhood preserving embedding for feature selection
In order to reduce feature vector dimensionality, we have employed neighborhood preserving embedding (NPE) on hybrid models. The predicted results of individual and ensemble classifiers are shown in Table 2. NPE has performed well compared to principal component analysis (PCA), which aims at preserving the global Euclidean structure while the NPE aims at preserving the local neighborhood structure on the data manifold. Therefore, NPE is less sensitive to outliers than PCA. The performance of the classifiers is assessed using various dimensions of hybrid models, which is reduced by NPE. The best result is obtained on 100 dimensions. Here we have observed two things; one is that the performance of the classifiers decreases compared to using original hybrid models. But the other fact is that the dimensionality of the feature vector is reduced. Considerably in case of hybrid1 model and NPE, the highest accuracy is obtained by SVM using jackknife test and by PNN using independent dataset test. On the other hand, the performance of the ensemble classifier is better in both cases than that of all the individual classifiers. In case of hybrid2 model and NPE, SVM and PNN have obtained approximately equal accuracy using jackknife test, while in case of independent dataset test, PNN has yielded the highest success rates compared to other classifiers. Again using hybrid2 model, the ensemble classifier has obtained the highest results than that of all the individual classifiers. After the feature reduction, the dimensionality of the both hybrid models is 100D but still the performance of some classifiers using hybrid1 model is slightly higher than that of hybrid2 model. It is observed that the addition of DWT-based features has a slight negative impact on the discrimination capability of the PseAA and SAAC.
Ensemble through genetic algorithm
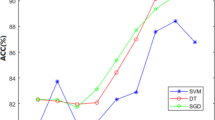
In order to improve the performance of the ensemble classifier, we have combined the prediction of the individual classifiers through optimization technique genetic algorithm (GA). The highest improvement has been reported using ensemble constructed through DWT-based features only. The improvement is observed in both cases; jackknife and independent dataset test and is 6.2 and 2.7%, respectively, as shown in Fig. 2. Its mean that there exist some diversity in the prediction of individual classifiers using DWT-based features. In each of feature extraction strategies, the performance of ensemble classifier through GA is better compared to ensemble classifier through simple majority voting. The highest accuracy 92.6% has been achieved by ensemble classifier using hybrid1 model, as shown in Fig. 3, while the success rate of ensemble classifier using SAAC is 92.4%. There is only 0.2% accuracy difference between using hybrid model1 and SAAC. But SAAC is 60D while hybrid1 model is 122D. We have thus observed that SAAC-based feature extraction strategy has performed well in discriminating membrane protein types as compared to rest of the individual feature extraction strategies.
Prediction performance for each membrane protein type and its biological significance
Prediction performances for each membrane protein type using ensemble classifier are shown in Table 3. In Table 3, columns 2–4 show DWT-based ensemble classifier prediction for each membrane protein types. Type-II, lipid-anchored, and GPI-anchored membrane proteins are predicted with an accuracy of 26.5, 30.8 and 13.2% respectively, while Type-I, multipass and peripheral membrane protein are predicted with an accuracy of 59.3, 90.1, and 40.0%, respectively. It has been observed that the overall accuracy of the DWT-based prediction is affected due to Type-II, lipid-anchored and GPI-anchored membrane prediction. In this study, wavelet signal is decomposed up to level 4, which may have introduced feature redundancy due to the short length of the protein signal. Because the average sequence length of Type-I, Type-II, multipass, lipid-anchored, GPI anchored, and peripheral is 772, 366, 521, 271, 456, and 467, respectively, it is obvious that length of Type-II and lipid-anchored is low as compared to the other membrane protein types. The other drawback is the unbalanced nature of dataset e.g., the number of training samples for multipass are higher in number as compared to other classes. Thus, the performance of classifiers is affected. In Table 3, columns 5–7 show the PseAA-based prediction using ensemble classifier. The ensemble classifier yields an accuracy of 91.9, 45.7, 97.4, 63.5, 25.0, and 20.0% for Type-I, Type-II, multipass, lipid anchored, GPI anchored, and peripheral, respectively. Almost, the same problem as with DWT occurs with PseAA because when the tier value increases, the performance of the short length sequence is affected. On the other hand, if the tier value decreases, information is lost in case of a lengthy sequence. Multipass class is dominant in being predicted accurately due to its large representation in training data.
In Table 3, columns 8–10 show the SAAC-based prediction performance of Mem-EnsSAAC for each membrane protein types. The accuracy for Type-I, Type-II, multipass, lipid anchored, GPI anchored, and peripheral membrane proteins are 93.8, 51.0, 98.0, 68.3, 36.7, and 36.6%, respectively. MCC and F-measure of each membrane protein types are provided in Table 3 and shown in Fig. 4. The performance of Hybrid-EnsC is shown in Table 3, columns 11–13. The obtained accuracy for Type-I, Type-II, multipass, lipid-anchored, GPI anchored, and peripheral membrane proteins is 91.6, 53.6, 97.8, 70.2, 33.8, and 33.3%, respectively.
The performance of the Mem-EnsSAAC is high for almost each type of membrane protein compared to that of using PseAA, DWT, and hybrid. This fact shows that the membrane protein types can be efficiently discriminated on the bases of differences in the amino acids at their N and C terminus. In all three feature-extraction strategies, the multipass class has received highest accuracy, MCC, and F-measures. This might be due to two reasons. First is that its sequence average length is higher than that of the Type-II, lipid-anchored, and GPI-anchored membrane protein. The second is that its number of sequences is 1903, which is high as compared to the other sequences. Membrane-bound proteins are of special interest to the drug discovery community as they constitute one-third of the genome and make up half of the pharmaceutically relevant drug targets. Defective membrane proteins are involved in diseases such as cancer, cardiovascular diseases and neurological diseases. Using the jackknife test F-measure, MCC and Q-Statistics are shown in Fig. 5.
Mem-EnsSAAC yields the highest MCC and F-measure of 0.76, 0.80, 0.73 and 0.79, respectively, using both jackknife and independent dataset test. On the other hand, accuracy, sensitivity, and specificity of all classifiers using PseAA composition, DWT, SAAC, and hybrid are shown in Fig. 6 with Mem-EnsSAAC offering highest sensitivity and specificity.
The proposed method is also compared with state of art existing methods in Table 4. The proposed Mem-EnsSAAC provides the highest results in all the three tests, self-consistency, jackknife and independent dataset test obtaining an accuracy of 99.9, 92.4 and 92.2%, respectively.
The proposed Mem-EnsSAAC is 6.3% higher in case of jackknife and 1.6% in case of independent dataset test from highest performing membrane protein predictors. The achieved results are thus the highest, reported so far. A similar improvement is shown by the other performance measures. This effective performance improvement is due to the good discrimination capabilities of SAAC and the learning capability and robustness of the majority voting based ensemble approach.
Prediction performance using Dataset2
Dataset2 is also used to analyze the performance of the selected classification algorithms. The feature extraction strategy used for this dataset is SAAC and we have evaluated the performance of classifiers using jackknife test. In Table 5, the performance of individual and ensemble classifier for overall and each membrane protein type’s is shown and compared with that of MemType-2L (Chou and Shen 2007a) and (Mahdavi and Jahandideh 2011). Among the individual classifiers, SVM yields the highest accuracy of 84.2%. On the other hand, PNN and KNN yield an accuracy of 83.0 and 82.4%, respectively.
Thus in case of the individual classifiers, still the performance of SVM is better as compared to the rest of classifiers. The accuracy of the ensemble classifier is 86.2%, which accounts for its superiority as against the individual classifiers. The predicted results of Mem-EnsSAAC are higher than the predicted output of MemType-2L (Chou and Shen 2007a) and (Mahdavi and Jahandideh 2011), and are the best results reported so far.
Conclusions
In this study, the prediction of membrane protein types has been investigated. In this context, three feature extraction methods are analyzed in combination with several classification strategies. We have shown that the SAAC-based feature extraction yields better results than that of the PseAA and DWT. It has thus been observed that the membrane protein types can be efficiently discriminated based on the differences in the amino acid at their N and C terminus. Among the different individual classifiers, SVM in conjunction with the SAAC performs the best using jackknife test. While the RF yields better prediction performance using SAAC for independent dataset test. The proposed Mem-EnsSAAC predictor using SAAC provides the best-reported results using the same dataset so far. This shows that as against PseAA and DWT, the SAAC-based feature extraction has better discrimination capabilities in case of membrane protein types. Especially, the prediction accuracy of each membrane proteins types has been improved. The prediction performance of the different classifiers using the SAAC, is Mem-EnsSAAC > SVM > PNN > KNN > RF > Adaboost. The overall success rates obtained by the proposed Mem-EnsSAAC approach are 99.9, 92.4 and 92.2% using the self-consistency, jackknife and the independent dataset test, respectively. The prediction performance of Mem-EnsSAAC is promising and we hope that it will help the biologist to elucidate membrane protein types and their functions using protein sequence related information.
Since user-friendly and publicly accessible web-servers represent the future direction for developing practically more useful predictors (Chou and Shen 2009a), therefore, we have provided a free user friendly and publically accessible web-server for predicting membrane protein types Mem-EnsSAAC at http://111.68.99.218/membrane.
References
Breiman L (2001) Random forests. Mach Learn 45:5–32
Cai YD, Zhou GP, Chou KC (2003) Support vector machines for predicting membrane protein types by using functional domain composition. Biophys J 84:3257–3263
Cai YD, Ricardo PW, Jen CH, Chou KC (2004) Application of SVM to predict membrane protein types. J Theor Biol 226:373–376
Call ME, Wucherpfennig KW, Chou JJ (2010) The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nat Immunol 11:1023–1029
Chou KC (1995) A novel approach to predicting protein structural classes in a (20–1)-D amino acid composition space. Proteins Struct Funct Genet 21:319–344
Chou KC (2001) Prediction of protein subcellular attributes using pseudo-amino acid composition. Proteins Struct Funct Genet 43:246–255
Chou KC, Cai YD (2002) Using functional domain composition and support vector machines for prediction of protein subcellular location. J Biol Chem 277:45765–45769
Chou KC, Cai YD (2005a) Using GO-PseAA predictor to indentify membrane proteins and their types. Biochem Biophys Res Commun 327:845–847
Chou KC, Cai YD (2005b) Prediction of membrane protein types by incorporating amphipathic effects. J Chem Inf Model 45:407–413
Chou KC, Elrod DW (1999) Prediction of membrane protein types and subcellular locations. Proteins Struct Funct Genet 34:137–153
Chou KC, Shen HB (2006a) Predicting eukaryotic protein subcellular location by fusing optimized evidence-theoretic K-nearest neighbor classifiers. J Proteome Res 5:1888–1897
Chou KC, Shen HB (2006b) Hum-PLoc: a novel ensemble classifier for predicting human protein Subcellular localization. Biochem Biophys Res Commun 347:150–157
Chou KC, Shen HB (2007a) Memtype-2L: a web server for predicting membrane proteins and their types by incorporating evolution information through Pse-PSSM. Biochem Biophys Res Commun 360:339–345
Chou KC, Shen HB (2007b) Euk-mPLoc: a fusion classifier for large-scale eukaryotic protein subcellular location prediction by incorporating multiple sites. J Proteome Res 6:1728–1734
Chou KC, Shen HB (2007c) Review: recent progresses in protein subcellular location prediction. Anal Biochem 370:1–16
Chou KC, Shen HB (2009a) Review: recent advances in developing web-servers for predicting protein attributes. Nat Sci 2:63–92. http://www.scirp.org/journal/NS/
Chou KC, Shen HB (2010a) Cell-PLoc 2.0: an improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat Sci 2:1090–1103. http://www.scirp.org/journal/NS/
Chou KC, Shen HB (2010b) A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0. PLoS ONE 5:e9931
Chou JJ, Zhang CT (1993) A joint prediction of the folding types of 1490 human proteins from their genetic codons. J Theor Biol 161:251–262
Chou KC, Zhang CT (1994) Predicting protein folding types by distance functions that make allowances for amino acid interactions. J Biol Chem 269:22014–22020
Khan A, Tahir SF, Majid A, Choi Tae-Sun. (2008a) Machine learning based adaptive watermark decoding in view of an anticipated attack. Pattern Recognit 41:2594–2610
Khan A, Tahir SF, Choi TS (2008b) Intelligent extraction of a digital watermark from a distorted image. IEICE Trans Inf Syst. E91-D 7:2072–2075
Khan A, Khan FM, Choi TS (2008c) Proximity based GPCRs prediction in transform domain. Biochem Biophys Res Commun 371:411–415
Khan A, Majid A, Choi TS (2010) Predicting protein subcellular location: exploiting amino acid based sequence of feature spaces and fusion of diverse classifiers. Amino Acids 38:347–350
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Liu H, Wang M, Chou KC (2005) Low-frequency Fourier spectrum for predicting membrane protein types. Biochem Biophys Res Commun 336:737–739
Mahdavi A, Jahandideh S (2011) Application of density similarities to predict membrane protein types based on pseudo amino acid composition. J Theor Biol 276:132–137
Nakashima H, Nishikawa AO (1986) The folding type of a protein is relevant to the amino acid composition. J Biochem 99:152–162
Nanni L, Lumini A (2006) Ensemblator: an ensemble of classifiers for reliable classification of biological data. Pattern Recognit Lett 28:622–630
Nanni L, Lumini A (2008a) Genetic programming for creating Chou’s pseudo amino acid based features for submitochondria localization. Amino Acids 34:653–660
Nanni L, Lumini A (2008b) An ensemble of support vector machines for predicting the membrane proteins type directly from the amino acid sequences. Amino Acids 35(3):573–580
Nanni L, Brahnam S, Lumini A (2010) High performance set of PseAAC and sequence based descriptors for protein classification. J Theor Biol 266(1):1–10
Pielak RM, Chou JJ (2010) Flu channel drug resistance: a tale of two sites. Protein Cell 1:246–258
Qiu JD, Sun XU, Huang JH, Liang RP (2010) Prediction of the types of membrane proteins based on discrete wavelet transform and support vector machines. Protien J 29:114–119
Rezaei MA, Maleki PA, Karami Z, Asadabadi EB, Sherafat MA, Moghaddam KA, Fadaie M, Forouzanfar M (2008) Prediction of membrane protein types by means of wavelet analysis and cascaded neural network. J Theor Biol 255:817–820
Schapire RE, Freund Y, Bartlett P, Lee WS (1998) Boosting the margin a new explanation for the effectiveness of voting methods. Ann Stat 26:1651–1686
Shen HB, Chou KC (2007) Using ensemble classifier to identify membrane protein types. Amino Acids 32:483–488
Tsoumakas G, Katakis I (2007) Multi-label classification: an overview. Int J Data Wareh Min 3:1–13
Wang M, Yang J, Liu GP, Xu ZJ, Chou KC (2004) Weighted-support vector machines for predicting membrane protein types based on pseudo amino acid composition. Protein Eng Des Sel 17:509–516
Wens Z, Wang K, Li M, Nie F (2005) Analyzing functional similarity of protein sequences with discrete wavelet transform. Comput Biol Chem 29:220–228
Zhang CX, Zhang JS (2008) RotBoost: a technique for combining Rotation Forest and AdaBoost. Pattern Recognit Lett. doi:10.1016/j.patrec.2008.03.006
Zhou XB, Chen C, Li ZC, Zou XY (2007) Using Chou’s amphiphilic pseudo-amino acid composition and support vector machine for prediction of enzyme subfamily classes. J Theor Biol 248:546–551
Acknowledgments
This work is supported by the Higher Education Commission of Pakistan under the indigenous PhD scholarship program 17-5-3 (Eg3-045)/HEC/Sch/2006).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hayat, M., Khan, A. & Yeasin, M. Prediction of membrane proteins using split amino acid and ensemble classification. Amino Acids 42, 2447–2460 (2012). https://doi.org/10.1007/s00726-011-1053-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-011-1053-5