Abstract
The aim of this study was to characterize the Tunisian Fasciola spp. flukes by morphometric and molecular analyses. Flukes were collected from livers of sheep slaughtered in Sejnane slaughterhouses (Bizerte gouvernorate, Northwest Tunisia) between January and March 2021.
Five morphometric parameters were determined for all the liver flukes, as follows: (i) total body length (BL), (ii) distance between ventral sucker and the tail (VS-T), (iii) distance between oral sucker and ventral sucker (OS-VS), (iv) abdomen diameter (AD), (v) tail diameter (TD) and the body length to width ratio (BL/BW). Molecular identification of the fluke specimens was carried out by polymerase chain reaction, restriction fragment polymorphism (PCR-RFLP) of a 680 bp sequence of the internal transcribes spacer 1 (ITS1) gene and by amplification, sequencing, and phylogenetic analysis of a 500 bp sequence of the ITS2 gene. Morphometric measurements showed that the mean of the total body length of the adult flukes was 21.1 ± 2.7 mm with minimum and maximum lengths of 13 and 31 mm, respectively. The PCR-RFLP analysis revealed a single profile consisting of three bands of approximately 370, 100, and 60 bp. Fasciola sequences described in the present study (GenBank numbers: OQ457027 and OQ457028) showed 99.58–100% identity to Fasciola hepatica. In conclusion, the results of this study show that molecular and phylogenetic analyses confirm the presence of a single species of F. hepatica in the Sejnane region Northwest of Tunisia. However, further studies are needed to identify the occurrence of Fasciola species in other Tunisian regions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fasciolosis is a worldwide zoonosis caused by Fasciola trematode parasites, mainly Fasciola hepatica (Linnaeus, 1758) and Fasciola gigantica (Cobbold, 1856) (Itagaki et al. 2022). Liver flukes primarily infect domestic ruminants, although wild herbivores and other mammals, including humans, can be also infected (Mas-Coma et al., 2009; Sabourin et al., 2018; Evack et al., 2020; Omar et al., 2021). Of the two species, F. hepatica has a cosmopolitan distribution over the five continents with the exception of Antarctica (Abebe et al., 2010; Admassu et al., 2015), while F. gigantica is widespread in the tropical and subtropical regions of Africa and Asia (Robinson and John 2009). Moreover, F. hepatica and F. gigantica could coexist in areas where the climatic and ecological conditions allow the survival intermediate hosts of both species (Mas-Coma et al., 2009; Malatji et al., 2019). Consequently, cases of hybridization between F. hepatica and F. gigantica have been reported in areas where the geographical distribution of the two Fasciola species overlaps, such as in some African countries like Egypt and South Africa (Amer et al., 2011; Haridwal et al., 2021) and in Asia like Japan, Korea, China, and Vietnam (Itagaki et al., 2005a; Ichikawa and Itagaki, 2012; Anh et al., 2018; Itagaki et al., 2022). Fasciola hybrids result from hybridization between Fasciola species composed of both mixed and introgressive genotypes resulting from interspecific mating between them (Nguyen et al., 2018; Eliza et al., 2020).
It is important to distinguish between the two infections caused by F. hepatica and F. gigantica, respectively, as they have distinct pathological and epidemiological features and different control approaches. Distinction at the species level can be performed by morphological measurements of adult flukes based on body length and width, including the ratio between them (Periago et al., 2006, 2008; Diyana et al., 2020; Shykat et al., 2022), but this method does not allow identification in areas where the two species overlap due to the multiple variations in their morphological characteristics (Mas-Coma and Bargues 1997). Fasciola identification to the species level can be performed also by molecular tools. For this purpose, different molecular targets have been used, mainly nucleotide sequences of the first nuclear ribosomal internal transcribed spacer (ITS1), second nuclear ribosomal internal transcribed spacer (ITS2), and 28S ribosomal RNA genes (28S rRNA) (Marcilla et al., 2002; Itagaki et al., 2005a; Anh et al., 2018; Evack et al., 2020; Omar et al., 2021). Moreover, mitochondrial DNA (mtDNA) markers like cytochrome c oxidase 1 (cox1) and the NADH dehydrogenase subunit 1 (NAD1) genes have been developed for the phylogenetic studies and genetic variability of Fasciola species (Farjallah et al., 2009; Gupta and Bhardwaj, 2013; Bargues et al., 2017; Laatamna et al., 2021; Shykat et al., 2022). Furthermore, the coprological analyses were not able to differentiate between the eggs of the two Fasciola species; however, they only allow a diagnosis at the genus level (Mas-Coma et al., 2005).
Fasciolosis is one of the most serious parasitic infections of small ruminants in Tunisia, particularly in sheep reared in the northwest and southwest regions of the country. The mean prevalence of F. hepatica infection in the Sejnane region (Northwest Tunisia) was 70%, 65%, and 60% in tracer lambs, ewes, and lambs, respectively (Akkari et al., 2011), while the prevalence values were about 35% and 44% in sheep in Gafsa and Tozeur regions (southwest Tunisia), respectively (Ayadi et al., 1997; Hammami et al., 2005). Human fasciolosis appears to be under notified and/or diagnosed in Tunisia since only 36 cases were reported between 1940 and 2005 (Hammami et al., 2005). However, there is a lack of molecular studies and genetic characterization of Tunisian liver flukes (Farjallah et al., 2009; Amor et al., 2011). Moreover, morphometric characterization was never performed on Tunisian Fasciola populations. Therefore, the present study aimed to perform the first morphological and molecular study on Tunisian Fasciola spp. population collected from sheep livers in Sejnane slaughterhouse, Northwest Tunisia, using polymerase chain reaction restriction fragment polymorphism (PCR-RFLP) analysis of the ITS1 and sequence analysis of ITS2.
Materials and methods
Study area and sample collection
Between January and March 2021, monitoring activities were performed at the local sheep slaughterhouse in Sejnane (Northwest Tunisia). The Sejnane district belongs to the governorate of Bizerte, and it is situated in the Northwest of Tunisia at a mean altitude of 92 m (Fig 1). This locality is characterized by a Mediterranean climate with a mean annual rainfall of 507.5 mm and a mean annual temperature ranging from 10.7 to 30 °C during winter and summer, respectively (National Institute of Meteorology, Tunisia 2021). Moreover, this slaughterhouse is the only one in the Sejnane district, which includes the three main endemic regions of fasciolosis in northern Tunisia (Sejnane, Joumine, and Bazina).
A total of 382 adult liver flukes were collected from 66 sheep slaughtered in the study period. The mean number of flukes collected from each liver was 5.78 ± 2.01 with a maximum and minimum of 2 and 24 flukes per liver, respectively. After collection, the liver flukes were thoroughly washed with sterile distilled water and preserved in 70% ethanol at room temperature until further analysis.
Fasciola morphometric measurement
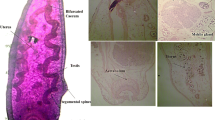
Of the total 382 liver flukes, 335 (87.7%) were subjected to morphometric analysis, as some specimens were damaged during collection. To characterize the Tunisian Fasciola liver flukes, several morphometric parameters, expressed in millimeters (mm), were estimated using a stereomicroscope equipped with a calibrated ocular micrometer. Therefore, the morphometric parameters included in the study were as follows: (i) total body length (BL), (ii) distance between ventral sucker and the tail (VS-T), (iii) distance between oral sucker and ventral sucker (OS-VS), (iv) abdomen diameter (AD), (v) tail diameter (TD) and the body length to width ratio (BL/BW) (Valero et al., 2001; Akhlaghi et al., 2017; Diyana et al., 2020) (Fig. 2, Table 1). The results are given as mean values, standard deviation (SD), and range of values (min and max) of all measured parameters.
To avoid any bias due to specimens’ preservation, all morphometric measurements were carried out in the day of flukes’ collection.
DNA extraction
A total of 66 liver flukes (i.e., one fluke for each liver) were selected for molecular analyses. DNA was extracted from approximately 50 mg of each fluke specimen, using the Wizard® Genomic DNA Purification Kit (Wizard Genomics DNA Promega, Madison, WI, USA) according to the manufacturer’s instructions, and DNA was stored at −20 °C until analysis. The PCR was performed for each sample using a pair of primers targeting the hypervariable regions V1–V3 coding for the 18S rRNA gene to detect the presence of eukaryotic DNA and to assess its quality as described by Wang et al. (2014) (Table 2).
ITS1-PCR and RFLP analysis
PCR was performed for the Fasciola spp. ITS1 gene, approximately 680 bp in length, coding for the 18S-5.8S rRNA, according to the protocol of Itagaki et al. (2005b). Briefly, a total volume of 25 μl containing 1x PCR buffer, 4.5 mM MgCl2, 1.0 mM of each dNTP, 0.75 U of Taq polymerase (Bioron GmbH, Germany), 10 μM of each primer (ITS1-F: TTGCGCTGATTACGTCCCTG and ITS1-R: TTGGCTGCGCTCTTCATCGAC) (Bio Basic, Canada Inc.), and 2 μL DNA sample was prepared A negative control consisting of nuclease-free water was added for each PCR run. The PCR program consisted of an initial denaturation at 94 °C for 10 min, followed by 25 cycles (94; 58 and 72 °C for 90 s each) and a final extension at 72 °C for 10 min (Table 2). PCR products were examined in a 2% w/v agarose gel, stained with ethidium bromide at a concentration of 0.5 μg/ml, and visualized under UV light.
Fasciola species were identified using an RFLP assay with the RsaI restriction enzyme. The reaction mixture consisted of 10 μL PCR product, 2 μl buffer, 1 μL RsaI enzyme, and distilled water for a final volume of 22 μL. Then, the mixture was incubated at 37 °C for 3 h (Nangru et al. 2022). The RFLP products were electrophorized in a 2.5% w/v agarose gel, stained with ethidium bromide at a concentration of 0.5 μg/mL, and then visualized under UV light. ITS1 profiles allowing species identification were obtained from the study by Ichikawa and Itagaki (2010).
ITS2 amplification and phylogenetic analysis
The genetic diversity of Tunisian Fasciola specimens was investigated by amplifying approximately a 500 bp of the ITS2 gene coding for the 5.8S and 28S rRNA according to the protocol of Itagaki et al. (2005a). The reaction volume consisted of 12.5 μl master mix (Takara, France), 6.25 pM of each primer (ITS2-F: TGTGTCGATGAAGAGCGCAG and ITS2-R: TGGTTAGTTTCTTTTCCTCCGC) (Eurofins, Germany), 5 μl of DNA, and distilled water to a volume of 25 μL. The amplification conditions were as follows: an initial denaturation at 95 °C for 10 min, followed by 35 cycles (94 °C at 60 s, 53 °C at 90 s, and 72 °C at 60 s) and a final extension at 72 °C for 10 min (Table 2). PCR products were electrophorized in a 1% w/v agarose gel, stained with ethidium bromide at a concentration of 0.5 μg/ml, and visualized under UV light.
All amplicons were purified using the Zymo Research DNA Clean & Concentration clean up kit (Genomics DNA Clean & Concentrator®, USA) according to the manufacturer’s instructions and sequenced in the forward and reverse directions with the same primers used for amplification. The chromatograms were evaluated and analyzed with 4peaks software (version 1.8). Multiple sequence alignment was performed using MEGA XI software (version 11.0.11) (Kumar et al., 2018). The NCBI Basic Local Alignment Search Tool (BLAST) was used to evaluate the level of identity with other reported sequences deposited in the GenBank database (Altschul et al. 1990). Phylogenetic trees of Tunisian Fasciola spp. were constructed using the neighbor-joining method (Saitou and Nei 1987) implemented in the software MEGA XI following 1000 bootstrap replications. Moreover, F. gigantica (GenBank accession number DQ385828) was used as an outgroup root.
Results
Fluke’s morphometric measurement
The morphometric characteristics of the isolated flukes from sheep are summarized in Table 1. The results showed that the mean of the total body length of the adult flukes was 21.1 ± 2.7 mm and the mean ratio length/width of the fluke body was 2.32 ± 0.96 mm. Moreover, the morphometric assessment revealed that the mean of abdomen diameter and the tail diameter of the Fasciola specimens were 10.14 ± 3.04 mm and 6 ± 2.22 mm, respectively. The morphological parameters of the adult Fasciola specimens isolated from sheep in Tunisia did not differ from those of F. hepatica isolated from sheep in Bolivia and Iran (Table 1).
ITS1-PCR and RFLP analysis
In ITS1-PCR amplification, all samples produced an amplicon of approximately 680 bp, indicating that they belong to the Fasciola genus. The negative control did not produce bands in any of the PCR reactions. The RFLP results generated a single profile consisting of three bands of approximately 370, 100, and 60 bp indicating that the amplicons belong to F. hepatica (Fig. 3). Neither F. gigantica nor an intermediate RFLP pattern was detected in all digested amplicons. Therefore, all specimens collected from sheep livers in the current study belonged to F. hepatica.
ITS2 amplification and phylogenetic analysis
The sequencing of 66 ITS2 PCR amplicon from Tunisian Fasciola spp. confirmed the presence of F. hepatica in sheep livers in the Sejnane region (Northwest Tunisia). Sequence alignment of the amplicons (approximately 500 bp in length) revealed two distinct variants (GenBank accession numbers OQ457027 and OQ457028). Both variants have two nucleotide differences at positions 15 (first variant) and 16 (second variant) (T/G and G/A, respectively), with 99.79% similarity between them. Of the 66 aligned sequences, 63.64% (42/66) belongs to the first variant and 36.36% (24/66) to the second variant. These results show that the first variant is more abundant than the second in the Tunisian F. hepatica specimens.
Phylogenetic trees of F. hepatica were constructed using the ITS2 rDNA gene sequence of our amplicons and those published in GenBank. Our F. hepatica amplicons showed 99.58–100% identity with the amplicons published in GenBank. The sequences of the two amplicons obtained in the present study fall into the same clade as amplicons from North Africa (Egypt and Libya), Asia (Japan, Vietnam, India, and Iran), Australia, and slightly distantly from amplicons from Switzerland and Iran. Our variants are clearly distinct from other Fasciola species such as F. gigantica (GenBank DQ385828) in sheep in Egypt (Fig. 4).
Partial sequence ITS2 gene phylogenetic tree of Fasciola hepatica isolated from sheep liver (GenBank accession number: OQ457027 and OQ457028) and Fasciola hepatica amplicons deposited in GenBank. Amplicons described in this study are indicated with a black square, and numbers at the branches refer to bootstrap (1000 replications)
Discussion
The present study provides new insights into the liver fluke (Fasciola spp.) population infecting sheep in the Sejnane region (Northwest Tunisia). To our knowledge, this is the first morphometric study of adult liver flukes isolated from sheep in Tunisia, complemented by molecular characterization of the ITS1 gene (PCR-RFLP) and phylogenetic analysis based on the ITS2 gene.
The morphometric measurements showed that the mean body length of Tunisian Fasciola spp. was 21.1 ± 2.7 mm, with minimum and maximum lengths of 13 and 31 mm, respectively. The mean abdomen diameter of the flukes was 10.14 ± 3.04 mm, ranging from 2 to 18 mm. These results are consistent with those obtained in other studies conducted on F. hepatica from sheep in Bolivia and Iran. The mean length of F. hepatica was 16.10 ± 4.80 mm, and its mean width was 7.11 ± 2.27 mm in Bolivia (Valero et al., 2001), whereas F. hepatica measured approximately 21.1 ± 2 mm in length and 12 ± 1.5 mm in width in Iran (Yakhchali et al., 2015). Moreover, F. gigantica isolated from the same host species in Iran measured 34.1 ± 4 mm in length and 9 ± 0.1 mm in width, respectively (Yakhchali et al., 2015). Other studies throughout the world described the morphology of F. hepatica isolated from other different species such as cattle, buffaloes, and pigs (Valero et al., 2001; Diyana et al., 2020; Sumruayphol et al., 2020; Haridwal et al., 2021; Abdel-Fatah et al., 2022). On the other hand, they compared the morphology of F. hepatica and F. gigantica isolated from the same host species and found that the latter was longer and narrower than F. hepatica (Valero et al., 2001; El-Rahimy et al., 2012; Amer et al., 2016; Akhlaghi et al., 2017; Diyana et al., 2020; Sumruayphol et al., 2020; Shykat et al., 2022). Similarly, previous studies based on morphometric differences between the two Fasciola species have shown that the average distance between the posterior testis and the posterior border of the body was shorter in F. hepatica than in F. gigantica (Bergeon and Laurent 1970; Sahba et al., 1972). Similarly, Mas-Coma and Bargues (1997) have shown that the shoulders of F. hepatica are more developed, the cephalic cone is longer, and the caeca are less branched than in F. gigantica. Moreover, Dar et al. (2003) showed that the rediae of F. gigantica developing in Radix natalensis and Galba truncatula are morphologically different from those of F. hepatica. Morphological measurements are a simple tool for discriminating F. hepatica and F. gigantica in areas with low occurrence and without intermediate forms. In contrast, morphometric assessments are unable to discriminate between the two liver fluke species in areas (e.g., Egypt, Japan, Korea, Vietnam, China) where the presence of co-infection with both Fasciola species and their intermediate forms has already been confirmed (Itagaki et al., 2005a, 2005b; Ichikawa and Itagaki, 2012; Anh et al., 2018; Itagaki et al., 2022). Furthermore, the morphometric characteristics of adult Fasciola spp. and eggs have been shown to be critically influenced by the definitive host species (Valero et al., 2001), thus confirming the limitations of morphometric analyses in discriminating Fasciola species.
The PCR-RFLP analysis of Tunisian Fasciola spp. of the ITS1 gene using the restriction enzyme RsaI revealed a unique profile consisting of three bands of approximately 370, 100, and 60 bp, thus indicating that the fluke samples belonged to the F. hepatica species. Moreover, neither F. gigantica nor intermediate RFLP patterns were detected in all digested amplicons The same profile of F. hepatica that we obtained (360, 100, and 60 bp) in the present study was also found in other studies in different countries around the world (Ichikawa and Itagaki 2010; Itagaki et al. 2011; Diyana et al. 2020; Hasanpour et al. 2020). On the other hand, F. gigantica yielded RFLP patterns of approximately 370, 170, and 60 bp using the same technique and restriction enzyme (Ichikawa and Itagaki, 2010; Itagaki et al., 2011; Ichikawa and Itagaki, 2012; Anh et al., 2018; Diyana et al., 2020; Hasanpour et al., 2020). It is noteworthy that an intermediate form has been detected by this technique in Japan, Korea, and Vietnam giving a profile consisting of four bands of approximately 370, 170, 100, and 60 bp (Itagaki et al., 2005b; Ichikawa and Itagaki, 2012; Anh et al., 2018). Other studies used a PCR-RFLP assay based on the partial rDNA of ITS1 and different restriction enzymes from RsaI to differentiate and identify Fasciola species such as AvaII and DraII, while the latter has no restriction sites in F. gigantica amplicons (Marcilla et al., 2002; El-Rahimy et al., 2012; Yakhchali et al., 2015). Molecular approaches based on DNA analyses are useful tools for the identification and genetic characterization of parasites with similar morphology. Therefore, various molecular targets, primarily deoxyribonucleic acid (DNA) sequences of the ITS1, ITS2, and 28S rRNA genes, have been used to differentiate F. hepatica from F. gigantica (Adlard et al., 1993; Marcilla et al., 2002; Itagaki et al., 2005a; Anh et al., 2018; Evack et al., 2020; Omar et al., 2021). In addition, RFLP of amplified DNA is a suitable method for differentiating Fasciola ITS1, using various restriction enzymes such as RsaI, AvaII, and DraII, while the PCR-RFLP method relies on patterns produced by the effects of endonucleases on ITS genes to identify Fasciola species (Ichikawa and Itagaki, 2010; Dar et al., 2012; Yakhchali et al., 2015; Anh et al., 2018; Diyana et al., 2020; Hasanpour et al., 2020).
The presence of F. hepatica in the Sejnane region of northwestern Tunisia was confirmed by sequencing of the ITS2 amplicon. The analysis revealed two distinct variants (GenBank accession numbers OQ457027 and OQ457028) with a similarity of 99.79%. This difference between the two variants could not be related to the host species or the geographical origin of the samples, as they are the same (Alasaad et al., 2007). However, this variation could be explained by the movements of Fasciola-infected animals in different regions of Tunisia or even by the introduction of infected animals imported from abroad (Schwantes et al., 2020). These results would require the sequencing of samples isolated from other host species and/or other regions in order to determine whether there is Fasciola species diversity in Tunisia.
The two sequences obtained in the present study clustered to those from Africa, Asia, and Australia, sharing high homology (99.58–100%) with amplicons from other regions and hosts deposited in GenBank. Our amplicons showed 99.6–100% identity with others isolated from sheep in Libya, India, and Australia (GenBank accession numbers: MT025436, MH048706, MF678650, respectively), from goats in Egypt (GenBank accession number: MT423007), and from cattle in Japan, Egypt, Vietnam, and Iran (GenBank accession numbers: LC056929, AB510492, MN970007, KF866250, respectively).
The two variants differed slightly (99.58% identity) in cattle isolates from Switzerland and in sheep isolate from Iran (GenBank accession numbers: MK321602 and HQ199841, respectively). However, this study confirms that the amplicons were distinct from other Fasciola species such as F. gigantica in sheep from Egypt (GenBank accession number: DQ385828).
The molecular identity of Tunisian F. hepatica with other F. hepatica isolates from other countries bordering Tunisia could be explained by a genetic mixing between Fasciola ecotypes due to intensive international livestock trade and passive displacement of intermediate hosts.
Conclusion
The results from PCR-RFLP showed a unique profile consistent with that of F. hepatica, indicating that this technique is effective in differentiating the two species, F. hepatica and F. gigantica. Molecular analyses were in line with morphometric and phylogenetic analyses, indicating the presence of a single species of F. hepatica in the Sejnane region of northwestern Tunisia. Our results will be useful to support molecular profiling and comparison with other Fasciola specimens from different regions of Tunisia and other North Africa countries.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Abdel-Fatah OR, Arafa WM, Wahba AA, El-Dakhly KM (2022) Economic losses, morpho-molecular identification, and identity of Fasciola species recovered from Egypt. J Parasit Dis 46:1036–1046. https://doi.org/10.1007/s12639-022-01526-x
Abebe R, Abunna F, Mekuria S, Megersa B (2010) Fasciolosis: prevalence, financial losses due to liver condemnation and evaluation of a simple sedimen- tation diagnostic technique in cattle slaughtered at Hawassa Municipal abattoir, southern Ethiopia. Ethiop Vet J 14(1):39–51. https://doi.org/10.4314/evj.v14i1.63868
Adlard RD, Barker SC, Blair D, Cribb TH (1993) Comparison of the second internal transcribed spacer (Ribosomal DNA) from populations and species of fasciolidae (Digenea). Int J Parasitol 23:423–425. https://doi.org/10.1016/0020-7519(93)90022-Q
Admassu B, Shite A, Kinfe G (2015) A review on bovine fasciolosis, vol 7, pp 139–146. https://doi.org/10.5829/idosi.ejbs.2015.7.03.9558
Akhlaghi E, Mohammadi MA, Ziaali N et al (2017) Morphometric and molecular study of Fasciola isolates from ruminants in Iran. Turkiye parazitolojii Derg 41:192–197. https://doi.org/10.5152/tpd.2017.5214
Akkari H, Gharbi M, Darghouth MA (2011) Infestation of tracer lambs by Fasciola hepatica in Tunisia : determining periods for strategic anthelmintic treatments. OIE Rev Sci Tech 30:917–929
Alasaad S, Huang CQ, Li QY et al (2007) Characterization of Fasciola samples from different host species and geographical localities in Spain by sequences of internal transcribed spacers of rDNA. Parasitol Res 101:1245–1250. https://doi.org/10.1007/s00436-007-0628-2
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Amer S, Dar Y, Ichikawa M et al (2011) Parasitology International Identi fi cation of Fasciola species isolated from Egypt based on sequence analysis of genomic (ITS1 and ITS2) and mitochondrial (NDI and COI) gene markers. Parasitol Int 60:5–12. https://doi.org/10.1016/j.parint.2010.09.003
Amer S, ElKhatam A, Zidan S et al (2016) Identity of Fasciola spp. in sheep in Egypt. Parasites and Vectors 9(1):8. https://doi.org/10.1186/s13071-016-1898-2
Amor N, Farjallah S, Said K, Ben SB (2011) First report of Fasciola hepatica in Equus caballus host species from Tunisia based on the ribosomal internal transcribed spacer regions. Turkish J Vet Anim Sci 35:319–324. https://doi.org/10.3906/vet-1007-391
Anh DN, Anh LT, Tuan LQ et al (2018) Identification of Fasciola species isolates from Nghe An Province, Vietnam, based on ITS1 sequence of ribosomal DNA using a Simple PCR-RFLP method. J Parasitol Res 2018:1–6. https://doi.org/10.1155/2018/2958026
Ayadi A, Makni F, Ben Said M (1997) État actuel de la fasciolose en Tunisie. Bull la Société Française Parasitol 15:27–32
Bargues MD, Gayo V, Sanchis J et al (2017) DNA multigene characterization of Fasciola hepatica and Lymnaea neotropica and its fascioliasis transmission capacity in Uruguay, with historical correlation, human report review and infection risk analysis. PLoS Negl Trop Dis 11(2):e0005352
Bergeon P, Laurent M (1970) Différences entre la morphologie testiculaire de Fasciola hepatica et Fasciola gigantica. Rev d’élevage médecine vétérinaire des pays Trop 23:223. https://doi.org/10.19182/remvt.7700
Dar Y, Amer S, Mercier A et al (2012) Molecular identification of Fasciola spp. (Digenea: Fasciolidae) in Egypt. Parasite 19:177–182. https://doi.org/10.1051/parasite/2012192177
Dar Y, Vignoles P, Dreyfuss G, Rondelaud D (2003) Fasciola hepatica and Fasciola gigantica: comparative morphometric studies on the redial stage of both species. Parasitol Res 91:369–373. https://doi.org/10.1007/s00436-003-0966-7
Diyana JNA, Mahiza MIN, Latiffah H et al (2020) Occurrence, morphometric, and molecular investigation of cattle and buffalo liver adult fluke in Peninsular Malaysia main abattoirs. J Parasitol Res 2020:9–14. https://doi.org/10.1155/2020/5436846
El-Rahimy HH, Mahgoub AMA, El-Gebaly NSM et al (2012) Molecular, biochemical, and morphometric characterization of Fasciola species potentially causing zoonotic disease in Egypt. Parasitol Res 111:1103–1111. https://doi.org/10.1007/s00436-012-2938-2
Eliza N, Calvani D, Ichikawa-seki M et al (2020) Which species is in the faeces at a time of global livestock movements : single nucleotide polymorphism genotyping assays for the differentiation of Fasciola spp . q. Int J Parasitol. 50(2):91–101. https://doi.org/10.1016/j.ijpara.2019.12.002
Evack JG, Schmidt RS, Boltryk SD et al (2020) Molecular confirmation of a Fasciola gigantica × Fasciola hepatica hybrid in a Chadian bovine. J Parasitol 106(2):316–322. https://doi.org/10.1645/19-66
Farjallah S, Sanna D, Amor N et al (2009) Genetic characterization of Fasciola hepatica from Tunisia and Algeria based on mitochondrial and nuclear DNA sequences. Parasitol Res 105:1617–1621. https://doi.org/10.1007/s00436-009-1601-z
Gupta A, Bhardwaj A (2013) Mitochondrial DNA- a tool for phylogenetic and biodiversity search in equines. J Biodivers Endanger Species 01:1–8. https://doi.org/10.4172/2332-2543.s1-006
Hammami H, Hamed N, Ayadi A (2005) Epidemiological studies on Fasciola hepatica in Gafsa oases (South West of Tunisia). Parasite 14:261–264
Haridwal S, Malatji MP, Mukaratirwa S (2021) Food and Waterborne Parasitology Morphological and molecular characterization of Fasciola hepatica and Fasciola gigantica phenotypes from co-endemic localities in Mpumalanga and KwaZulu-Natal provinces of South Africa. Food Waterborne Parasitol 22:e00114. https://doi.org/10.1016/j.fawpar.2021.e00114
Hasanpour H, Falak R, Naddaf SR et al (2020) Molecular characterization of fasciola spp. From some parts of Iran. Iran J Public Health 49:157–166. https://doi.org/10.18502/ijph.v49i1.3062
Ichikawa M, Itagaki T (2012) Molecular analysis of aspermic Fasciola flukes from Korea on the basis of the nuclear ITS1 region and mitochondrial DNA markers and comparison with Japanese aspermic Fasciola flukes. J Vet Med Sci 74:899–904. https://doi.org/10.1292/jvms.11-0523
Ichikawa M, Itagaki T (2010) Discrimination of the ITS1 types of Fasciola spp. based on a PCR-RFLP method. Parasitol Res 106:757–761. https://doi.org/10.1007/s00436-010-1724-2
Itagaki T, Hayashi K, Ohari Y (2022) The causative agents of fascioliasis in animals and humans: parthenogenetic Fasciola in Asia and other regions. Infect Genet Evol 99:105248. https://doi.org/10.1016/j.meegid.2022.105248
Itagaki T, Ichinomiya M, Fukuda K et al (2011) Hybridization experiments indicate incomplete reproductive isolating mechanism between Fasciola hepatica and Fasciola gigantica. Parasitology 138:1278–1284. https://doi.org/10.1017/S0031182011000965
Itagaki T, Kikawa M, Sakaguchi K et al (2005a) Genetic characterization of parthenogenic Fasciola sp. in Japan on the basis of the sequences of ribosomal and mitochondrial DNA. Parasitology 131:679–685. https://doi.org/10.1017/S0031182005008292
Itagaki T, Kikawa M, Terasaki K et al (2005b) Molecular characterization of parthenogenic Fasciola sp. in Korea on the basis of DNA sequences of ribosomal ITS1 and mitochondrial NDI gene. J Vet Med Sci 67:1115–1118. https://doi.org/10.1292/jvms.67.1115
Kumar S, Stecher G, Li M et al (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Laatamna AE, Tashiro M, Zokbi Z et al (2021) Molecular characterization and phylogenetic analysis of Fasciola hepatica from high- plateau and steppe areas in Algeria. Parasitol Int 80:102234. https://doi.org/10.1016/j.parint.2020.102234
Malatji MP, Pfukenyi DM, Mukaratirwa S (2019) Fasciola species and their vertebrate and snail intermediate hosts in East and Southern Africa : a review. J Helminthol 94:1–11. https://doi.org/10.1017/S0022149X19000531
Marcilla A, Bargues MD, Mas-Coma S (2002) A PCR-RFLP assay for the distinction between Fasciola hepatica and Fasciola gigantica. Mol Cell Probes 16:327–333. https://doi.org/10.1006/mcpr.2002.0429
Mas-Coma S, Bargues MD (1997) Human liver flukes: a review. Res Rev Parasitol Parasitol 57:145–218
Mas-Coma S, Bargues MD, Valero MA (2005) Fascioliasis and other plant-borne trematode zoonoses. Int J Parasitol 35:1255–1278. https://doi.org/10.1016/j.ijpara.2005.07.010
Mas-Coma S, Valero MA, Bargues MD (2009) Chapter 2 Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Adv Parasitol 69:41–146. https://doi.org/10.1016/S0065-308X(09)69002-3
Nangru A, Maharana B, Vohra S, Kumar B (2022) Molecular identification of Theileria species in naturally infected sheep using nested PCR–RFLP. Parasitol Res 121(5):1487–1497. https://doi.org/10.1007/s00436-022-07489-5
National Institute of Meteorology, Tunisia 2021. https://www.meteo.tn/fr/institut-national-de-la-meteorologie. Consulted on March 2023
Nguyen TBN, Van DN, Nguyen TKL et al (2018) Distribution status of hybrid types in large liver flukes , Fasciola species (Digenea :Fasciolidae), from ruminants and humans in Vietnam. Korean J Parasitol 56:453–461. https://doi.org/10.3347/kjp.2018.56.5.453
Omar MA, Elmajdoub LO, Ali AO et al (2021) Original paper Genetic characterization and phylogenetic analysis of Fasciola species based on ITS2 gene sequence , with first molecular evidence of intermediate Fasciola from water buffaloes in Aswan. Egypt. Ann Parasitol 67(1):55–65. https://doi.org/10.17420/ap6701.312
Periago MV, Valero MA, El Sayed M et al (2008) First phenotypic description of Fasciola hepatica/Fasciola gigantica intermediate forms from the human endemic area of the Nile Delta. Egypt. Infect Genet Evol 8:51–58. https://doi.org/10.1016/j.meegid.2007.10.001
Periago MV, Valero MA, Panova M, Mas-Coma S (2006) Phenotypic comparison of allopatric populations of Fasciola hepatica and Fasciola gigantica from European and African bovines using a computer image analysis system (CIAS). Parasitol Res 99:368–378. https://doi.org/10.1007/s00436-006-0174-3
Robinson MW, John P (2009) Zoonotic helminth infections with particular emphasis on fasciolosis and other trematodiases. Philos Trans R Soc Lond., B, Biol Sci 364:2763–2776. https://doi.org/10.1098/rstb.2009.0089
Sabourin E, Alda P, Vázquez A et al (2018) Impact of human activities on fasciolosis transmission. Trends Parasitol 34(10):891–903. https://doi.org/10.1016/j.pt.2018.08.004
Sahba GH, Arfaa F, Farahmandian I, Jalali H (1972) Animal fascioliasis in Khuzestan, southwestern Iran. J Parasitol 58:712–716. https://doi.org/10.2307/3278298
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Schwantes JB, Quevedo P, D’Avila MF et al (2020) Fasciola hepatica in Brazil: genetic diversity provides insights into its origin and geographic dispersion. J Helminthol 94:1–7. https://doi.org/10.1017/S0022149X19000774
Shykat CA, Islam S, Ahmed F et al (2022) Current status of Fasciolosis of goat in Sylhet, Bangladesh: an integrated morphomolecular study. J Parasitol Res 2022:1–6. https://doi.org/10.1155/2022/6159388
Sumruayphol S, Siribat P, Dujardin JP et al (2020) Fasciola gigantica, F. Hepatica and Fasciola intermediate forms: geometric morphometrics and an artificial neural network to help morphological identification. PeerJ 8:e8597. https://doi.org/10.7717/peerj.8597
Valero MA, Darce NA, Panova M, Mas-Coma S (2001) Relationships between host species and morphometric patterns in Fasciola hepatica adults and eggs from the northern Bolivian Altiplano hyperendemic region. Vet Parasitol 102:85–100. https://doi.org/10.1016/S0304-4017(01)00499-X
Wang Y, Tian RM, Gao ZM et al (2014) Optimal eukaryotic 18S and universal 16S/18S ribosomal RNA primers and their application in a study of symbiosis. PLoS One 9:e90053. https://doi.org/10.1371/journal.pone.0090053
Yakhchali M, Malekzadeh-Viayeh R, Mani-Baran A, Mardani K (2015) Morphological and molecular discrimination of fasciola species isolated from domestic ruminants of urmia city, iran. Iran J Parasitol 10:46–55
Acknowledgements
The authors would like to thank the staff of the slaughterhouse of the Sejnane region (Tunisia) for their support for sampling. The authors would also like to thank the PREPARE4VBD project (European Union’s Horizon 2020 Research and Innovation Program under grant agreement no. 101000365) for supporting molecular analysis. Thanks also to the Mediterranean and Middle East Universities Network Agreement (MUNA) for collaboration in networking.
Funding
This work was supported by the Laboratoire d’Épidémiologie des Infections Enzootiques des Herbivores en Tunisie: Application à la Lutte (Ministère de l’Enseignement Supérieur et de la Recherche Scientifique, Tunisia) (LR16AGR01), the CGIAR Research Program on Livestock (CRP Livestock), and the Unit of Parasitology and Parasitic Diseases, Department of Veterinary Medicine and Animal Production, University of Naples Federico II.
Author information
Authors and Affiliations
Contributions
Ines Hammami: investigation, methodology, data analyses, writing—original draft. Lavina Ciuca: methodology, writing—review and editing. Maria Paola Maurelli: methodology, writing—review and editing. Rihab Romdhane: investigation. Limam Sassi: investigation. Mohamed Ridha Rjeibi: methodology. Nadia Farhat: investigation. Alain Kouam Simo: investigation, methodology. Laura Rinaldi: funding acquisition, writing—review and editing. Mourad Rekik: funding acquisition. Mohamed Gharbi: conceptualization, supervision, funding acquisition, writing—review and editing.
Corresponding author
Ethics declarations
Ethical approval
The present study was performed in a certified slaughterhouse under the supervision of an officially certified veterinarian by the Tunisian state.
Consent to participate
Informed consent was obtained from the slaughterhouse staff prior to sampling the Fasciola flukes.
Consent for publication
All authors read and consent to the publication of the manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Robin Flynn
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hammami, I., Ciuca, L., Maurelli, M.P. et al. First morphometric and molecular characterization of Fasciola spp. in Northwest Tunisia. Parasitol Res 122, 2467–2476 (2023). https://doi.org/10.1007/s00436-023-07933-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-023-07933-0